Etodolac Single Dose Metabolic Profile Elucidation: Pharmacokinetics and Adverse Events in Healthy Volunteers
Abstract
:1. Introduction
2. Results
2.1. Volunteers’ Clinical and Laboratory Characteristics
2.2. Determination of Etodolac Pharmacokinetics
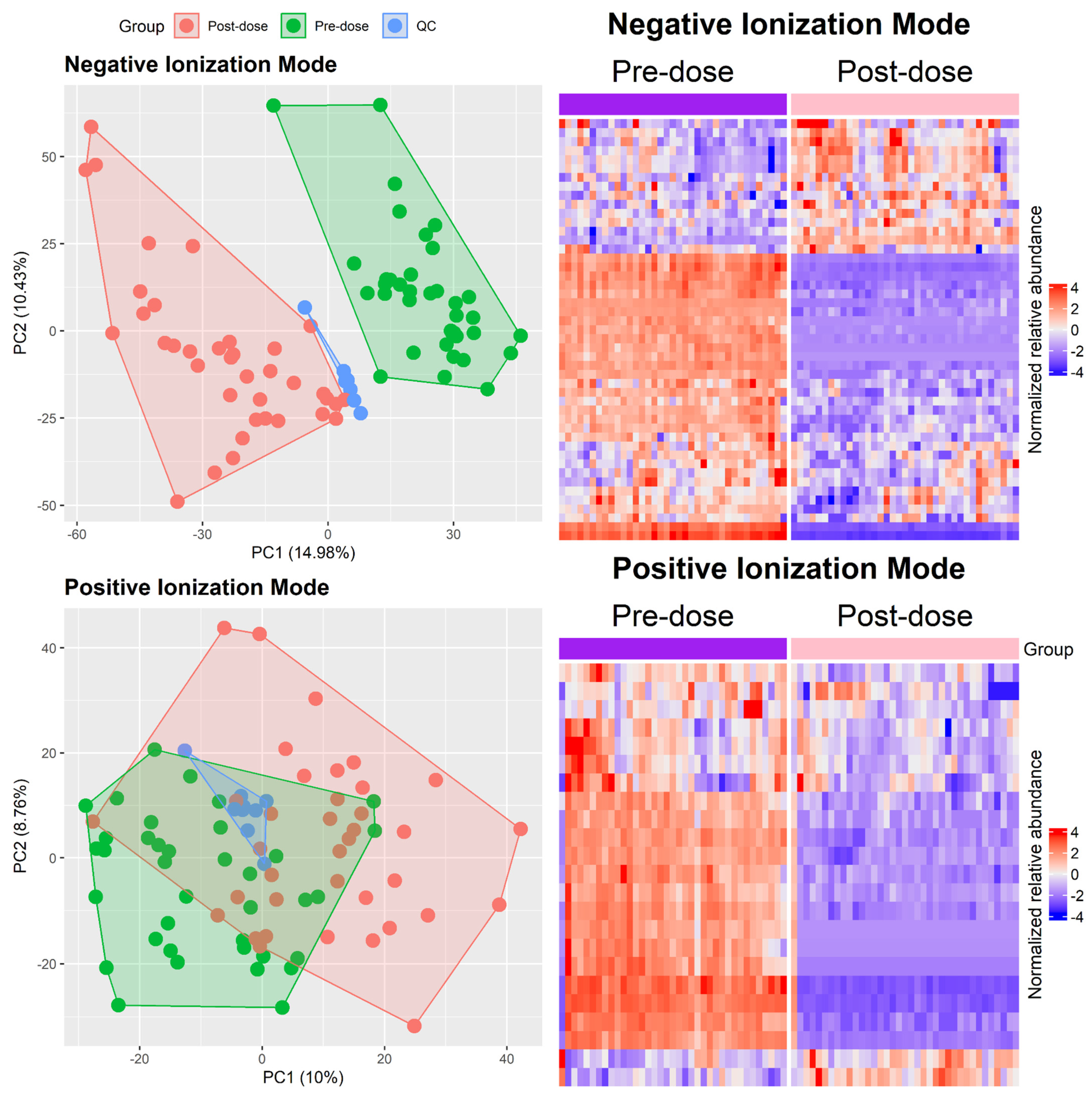
2.3. Metabolomic Analysis
2.4. Enrichment Analysis
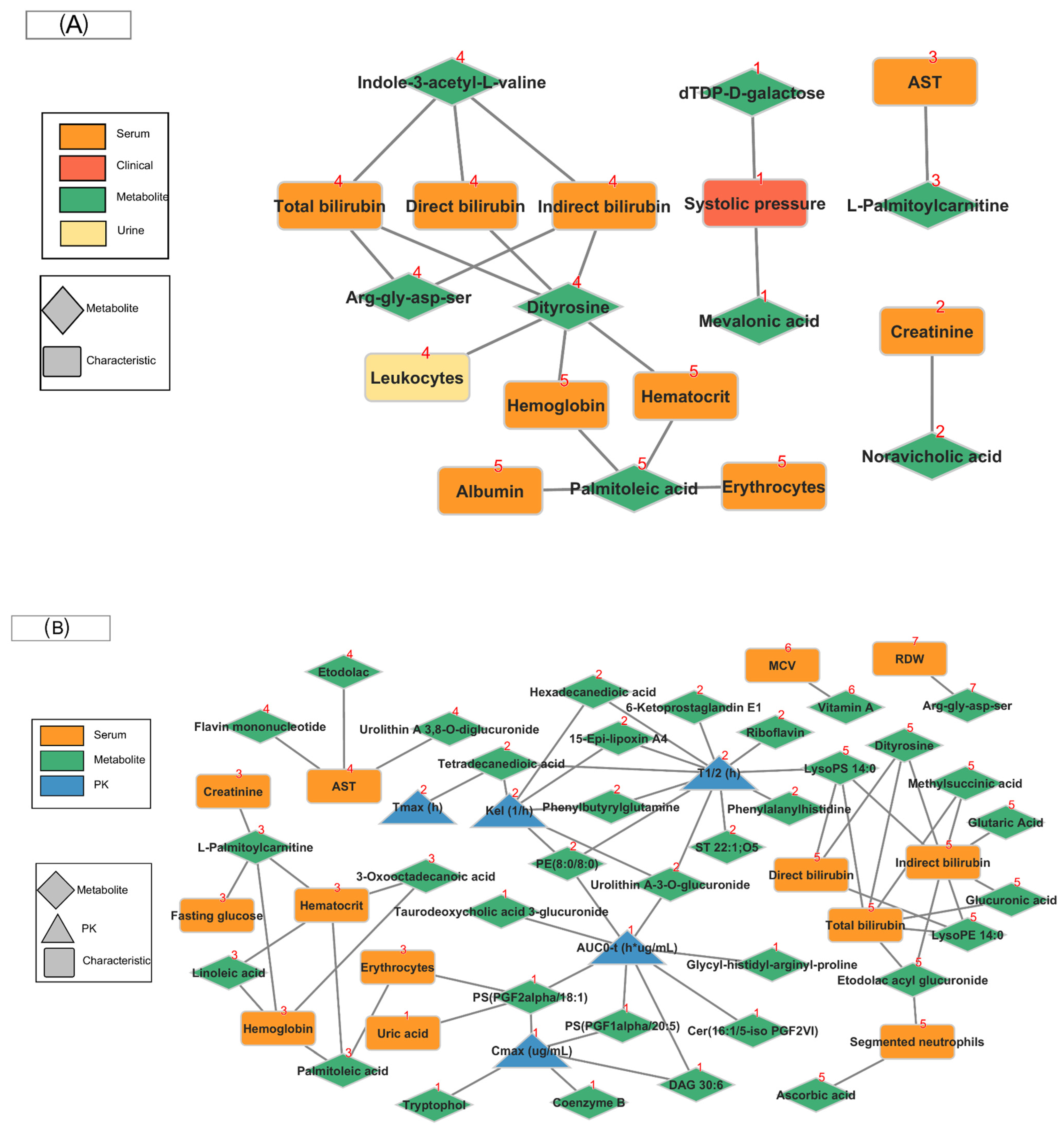
2.5. Community Network
2.6. Integrating Pathway and Network Results
3. Discussion
3.1. Mapping Predictive Associations
3.1.1. Association with Pharmacokinetic Parameters
3.1.2. Laboratory/Clinical Parameters
3.2. Exploring Explanatory Parameters
3.2.1. Pharmacokinetic Parameters
3.2.2. Laboratory/Clinical Characteristics
3.2.3. Adverse Event-Related Association
3.3. Limitations and Conclusions
4. Materials and Methods
4.1. Study Design and Healthy Human Subjects
4.2. Ethics
4.3. Determination of Pharmacokinetic Profile
4.4. Metabolomic Profile
4.4.1. Data Processing and Selection of Features
4.4.2. Identification of Metabolites
4.5. Enrichment Pathway Analysis
4.6. Community Network Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anekar:, A.A.; Hendrix, J.M.; Cascella, M. WHO Analgesic Ladder; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK554435/ (accessed on 30 August 2024).
- Gaddala, P.; Choudhary, S.; Sethi, S.; Sainaga Jyothi, V.G.; Katta, C.; Bahuguna, D.; Singh, P.K.; Pandey, M.; Madan, J. Etodolac utility in osteoarthritis: Drug delivery challenges, topical nanotherapeutic strategies and potential synergies. Ther. Deliv. 2024, 15, 977–995. [Google Scholar] [CrossRef]
- ANVISA. Consultas—Agência Nacional de Vigilância Sanitária. 2024. Available online: https://consultas.anvisa.gov.br/#/medicamentos/q/?nomeProduto=etodolaco (accessed on 2 December 2024).
- Davies, N.M.; Skjodt, N.M. Choosing the Right Nonsteroidal Anti-Inflammatory Drug for the Right Patient. Clin. Pharmacokinet. 2000, 38, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, W.W.; Khoder, M.; Abdelkader, H.; Singer, R.; Alany, R.G. Interactions of Cyclodextrins and their Hydroxyl Derivatives with Etodolac: Solubility and Dissolution Enhancement. Curr. Drug Deliv. 2024, 21, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Abramson, S.; Weissmann, G. The mechanisms of action of nonsteroidal antiinflammatory drugs. Clin. Exp. Rheumatol. 1989, 7 (Suppl. S3), S163–S170. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, K.; Kubo, M.; Imai, S.; Toyoda, F.; Maeda, T.; Okumura, N.; Matsuura, H.; Matsusue, Y. The COX-2 Selective Blocker Etodolac Inhibits TNFα-Induced Apoptosis in Isolated Rabbit Articular Chondrocytes. Int. J. Mol. Sci. 2013, 14, 19705–19715. [Google Scholar] [CrossRef]
- Vane, J.R.; Botting, R.M. Mechanism of action of nonsteroidal anti-inflammatory drugs. Am. J. Med. 1998, 104, 2S–8S; discussion 21S–22S. [Google Scholar] [CrossRef] [PubMed]
- Porat, D.; Dukhno, O.; Partook-Maccabi, M.; Vainer, E.; Cvijić, S.; Dahan, A. Selective COX-2 inhibitors after bariatric surgery: Celecoxib, etoricoxib and etodolac post-bariatric solubility/dissolution and pharmacokinetics. Int. J. Pharm. 2023, 645, 123347. [Google Scholar] [CrossRef]
- Currie, G.M. Pharmacology, Part 2: Introduction to Pharmacokinetics. J. Nucl. Med. Technol. 2018, 46, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Bowling, F.G.; Thomas, M. Analyzing the metabolome. Methods Mol. Biol. 2014, 1168, 31–45. [Google Scholar]
- Ribeiro, H.; Rodrigues, I.; Napoleão, L.; Lira, L.; Marques, D.; Veríssimo, M.; Andrade, J.P.; Dourado, M. Non-steroidal anti-inflammatory drugs (NSAIDs), pain and aging: Adjusting prescription to patient features. Biomed. Pharmacother. 2022, 150, 112958. [Google Scholar] [CrossRef]
- Song, I.D.; Kang, J.S.; Kim, H.J.; Kim, S.M.; Zhao, D.X.; Kim, S.H.; Chun, M.Y.; Lee, K.H. Quantification of Etodolac in Human Plasma for Pharmacokinetics and Bioequivalence Studies in 27 Korean Subjects. Drug Metab. Lett. 2017, 10, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Benet, L.Z. Pharmacokinetics of sustained-release etodolac. Rheumatol. Int. 1993, 13 (Suppl. S2), S3–S5. [Google Scholar] [CrossRef] [PubMed]
- Brocks, D.R.; Jamali, F. Etodolac clinical pharmacokinetics. Clin. Pharmacokinet. 1994, 26, 259–274. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B.L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Craig, M.; Yarrarapu, S.N.S.; Dimri, M. Biochemistry, Cholesterol; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK513326/ (accessed on 14 December 2024).
- Farag, M.A.; Gad, M.Z. Omega-9 fatty acids: Potential roles in inflammation and cancer management. J. Genet. Eng. Biotechnol. 2022, 20, 48. [Google Scholar] [CrossRef] [PubMed]
- Astudillo, A.M.; Meana, C.; Bermúdez, M.A.; Pérez-Encabo, A.; Balboa, M.A.; Balsinde, J. Release of Anti-Inflammatory Palmitoleic Acid and Its Positional Isomers by Mouse Peritoneal Macrophages. Biomedicines 2020, 8, 480. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.S.; Cheng, Y.M.; Yang, S.M. Impairments of the biological properties of serum albumin in patients on haemodialysis. Nephrology 2007, 12, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Malencik, D.A.; Anderson, S.R. Dityrosine as a product of oxidative stress and fluorescent probe. Amino Acids 2003, 25, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.E.; Rieckmann, M.; Sedding, D.; Hause, G.; Meister, A.; Mäder, K.; Lucas, H. Towards the Development of Long Circulating Phosphatidylserine (PS)- and Phosphatidylglycerol (PG)-Enriched Anti-Inflammatory Liposomes: Is PEGylation Effective? Pharmaceutics 2021, 13, 282. [Google Scholar] [CrossRef]
- Matsumoto, A.; Takahashi, Y.; Nishikawa, M.; Sano, K.; Morishita, M.; Charoenviriyakul, C.; Saji, H.; Takakura, Y. Role of Phosphatidylserine-Derived Negative Surface Charges in the Recognition and Uptake of Intravenously Injected B16BL6-Derived Exosomes by Macrophages. J. Pharm. Sci. 2017, 106, 168–175. [Google Scholar] [CrossRef]
- Hirayama, C. Tryptophan metabolism in liver disease. Clin. Chim. Acta 1971, 32, 191–197. [Google Scholar] [CrossRef]
- Teunis, C.; Nieuwdorp, M.; Hanssen, N. Interactions between Tryptophan Metabolism, the Gut Microbiome and the Immune System as Potential Drivers of Non-Alcoholic Fatty Liver Disease (NAFLD) and Metabolic Diseases. Metabolites 2022, 12, 514. [Google Scholar] [CrossRef]
- Das, U.N. Essential Fatty Acids and Their Metabolites in the Pathobiology of Inflammation and Its Resolution. Biomolecules 2021, 11, 1873. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Khan, A.; Jee, S.H.; Lee, H.S.; Hwang, M.S.; Koo, Y.E.; Park, Y.H. High resolution metabolomics to determines the risk associated with bisphenol A exposure in humans. Environ. Toxicol. Pharmacol. 2018, 58, 1–10. [Google Scholar] [CrossRef]
- Kikut, J.; Komorniak, N.; Ziętek, M.; Palma, J.; Szczuko, M. Inflammation with the participation of arachidonic (AA) and linoleic acid (LA) derivatives (HETEs and HODEs) is necessary in the course of a normal reproductive cycle and pregnancy. J. Reprod Immunol. 2020, 141, 103177. [Google Scholar] [CrossRef] [PubMed]
- Mota-Zamorano, S.; Robles, N.R.; Lopez-Gomez, J.; Cancho, B.; González, L.M.; Garcia-Pino, G.; Navarro-Pérez, M.L.; Gervasini, G. Plasma and urinary concentrations of arachidonic acid-derived eicosanoids are associated with diabetic kidney disease. EXCLI J. 2021, 20, 698–708. [Google Scholar] [PubMed]
- Shastak, Y.; Pelletier, W. From Metabolism to Vitality: Uncovering Riboflavin’s Importance in Poultry Nutrition. Animals 2023, 13, 3554. [Google Scholar] [CrossRef]
- Powers, H.J. Riboflavin (vitamin B-2) and health12. Am. J. Clin. Nutr. 2003, 77, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Wehrle, C.J.; Panconesi, R.; Satish, S.; Maspero, M.; Jiao, C.; Sun, K.; Karakaya, O.; Allkushi, E.; Modaresi Esfeh, J.; Whitsett Linganna, M.; et al. The Impact of Biliary Injury on the Recurrence of Biliary Cancer and Benign Disease after Liver Transplantation: Risk Factors and Mechanisms. Cancers 2024, 16, 2789. [Google Scholar] [CrossRef]
- Perl, E.; Waxman, J.S. Retinoic Acid Signaling and Heart Development. Subcell. Biochem. 2020, 95, 119–149. [Google Scholar] [PubMed]
- Cañete, A.; Cano, E.; Muñoz-Chápuli, R.; Carmona, R. Role of Vitamin A/Retinoic Acid in Regulation of Embryonic and Adult Hematopoiesis. Nutrients 2017, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Tougou, K.; Gotou, H.; Ohno, Y.; Nakamura, A. Stereoselective glucuronidation and hydroxylation of etodolac by UGT1A9 and CYP2C9 in man. Xenobiotica 2004, 34, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.A.R.; Cardoso, M.R.; Rezende, L.M.; Lin, J.Q.; Guimaraes, F.; Silva, G.R.P.; Murgu, M.; Priolli, D.G.; Eberlin, M.N.; Tata, A.; et al. Multiplatform Investigation of Plasma and Tissue Lipid Signatures of Breast Cancer Using Mass Spectrometry Tools. Int. J. Mol. Sci. 2020, 21, 3611. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Kind, T.; Cajka, T.; Hazen, S.L.; Tang, W.W.; Kaddurah-Daouk, R.; Irvin, M.R.; Arnett, D.K.; Barupal, D.K.; Fiehn, O. Systematic Error Removal Using Random Forest for Normalizing Large-Scale Untargeted Lipidomics Data. Anal. Chem. 2019, 91, 3590–3596. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 3 September 2024).
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef]
- Sanches, P.H.; Oliveira, D.C.D.; Reis, I.G.D.; Fernandes, A.M.; Silva, A.A.; Eberlin, M.N.; Carvalho, P.O.; Duarte, G.H.; Porcari, A.M. Fitting Structure-Data Files (.SDF) Libraries to Progenesis QI Identification Searches. J. Braz. Chem. Soc. 2023, 34, 1013–1019. [Google Scholar] [CrossRef]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [PubMed]
- Braisted, J.; Patt, A.; Tindall, C.; Sheils, T.; Neyra, J.; Spencer, K.; Eicher, T.; Mathé, E.A. RaMP-DB 2.0: A renovated knowledgebase for deriving biological and chemical insight from metabolites, proteins, and genes. Bioinformatics 2023, 39, btac726. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Agrawal, A.; Balcı, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next generation pathway database. Nucleic Acids Res. 2024, 52, D679–D689. [Google Scholar] [CrossRef] [PubMed]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef] [PubMed]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins Dos Santos, V.A.P.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Székely, G.J.; Rizzo, M.L.; Bakirov, N.K. Measuring and testing dependence by correlation of distances. Ann. Stat. 2007, 35, 2769–2794. [Google Scholar] [CrossRef]
- Csárdi, G.; Nepusz, T.; Traag, V.; Horvát, S.; Zanini, F.; Noom, D.; Müller, K.; Salmon, M.; Antonov, M.; Initiative, C.Z. igraph: Network Analysis and Visualization. 2024. Available online: https://cran.r-project.org/web/packages/igraph/index.html (accessed on 3 September 2024).


| Parameters | Molecule (ID HMDB/PubChem) | Biological/Chemical Class/Function/Pathway |
|---|---|---|
| Predictive T0 (Baseline metabolism pre-dose) | ||
| Laboratory/Clinical characteristics | ||
| Systolic pressure | dTDP-D-galactose (HMDB0006876) | Cholesterol biosynthesis = WP1795 Cholesterol synthesis disorders = WP5193. |
| Mevalonic acid (HMDB0000227) | Cholesterol metabolism (WP5304) | |
| Aspartate aminotransferase seric | L-Palmitoylcarnitine (HMDB0000222) | Carnitine metabolism (R-HSA-200425) An ester of carnitine that facilitates the transfer of fatty acids from the cytosol to the mitochondria during fatty acid oxidation. |
| Total bilurubin Direct bilirubin Indirect bilirrubin Leukocites urine | Dityrosine (HMDB0006045) | Dityrosine has also been found to be involved in oxidative/nitrative stress under a wide range of conditions and in a variety of biological systems. |
| Albumin seric Hemoglobin seric Erythrocytes seric Hematocrit seric | Palmitoleic acid (HMDB0003229) | Omega-9 fatty acid synthesis (WP4724) |
| Effect Tmax (Metabolism post-dose) | ||
| Pharmacokinetic parameters | ||
| AUC0-t (h·μg/mL) Cmax (μg/mL) | PS(PGF1alpha/20:5) (HMDB0282870) | Phospholipids with anti-inflammatory properties |
| PS(PGF2alpha/18:1) (HMDB0281557) | Phospholipids with anti-inflammatory properties | |
| Kel (1/h), T1/2 (h) | 15-Epi-lipoxin A4 (HMDB0012587) | Tryptophan metabolism (WP465) |
| Riboflavin (HMDB0000244) | Riboflavin Metabolism = map00740 | |
| 6-Ketoprostaglandin E1 (HMDB0004241) | Eicosanoid metabolism via cyclooxygenases (COX) | |
| Laboratory/Clinicals characteristics | ||
| Creatinine seric Fasting glucose seric Hemoglobin seric hematocrit seric Erythrocytes seric | L-Palmitoylcarnitine (HMDB0000222) | L-palmitoylcarnitine is an acylcarnitine. The acyl groups, organic acids and fatty acids are transported from the cytosol to the mitochondria for degradation to produce energy. |
| Linoleic acid (HMDB0000673) | Belongs to the class of organic compounds known as linoleic acids and derivatives. | |
| Palmitoleic acid (HMDB0003229) | Peptide hormone metabolism = R-HSA-2980736, sSynthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) = R-HSA-381771/WP2728; free fatty acid receptors = R-HSA-444209, Omega-9 fatty acid synthesis = WP4724 | |
| 3-Oxooctadecanoic acid (HMDB0010736) | It belongs to the class of organic compounds called long-chain fatty acids. | |
| Transaminase oxaloacetic acid (TGO) seric | Etodolac (HMDB0014887) | Drug |
| Flavin mononucleotide (HMDB0001520) | Metabolism of nitric oxide: NOS3 activation and regulation (WP1850), riboflavin metabolism = map00740, cytosolic iron-sulfur cluster assembly = WP2690, metabolism of cofactors = WP4990, riboflavin and CoQ disorders = WP5037, vitamin B12 disorders = WP4271 | |
| Total bilirubin seric Indirect bilirubin seric Leukocytes seric Segmented neutrophils seric | Glucuronic acid (HMDB0000127) | Phosphatidylinositol phosphate metabolism = map00562 |
| Etodolac acyl glucuronide (HMDB0060916) | Metabolite drug | |
| Ascorbic acid (HMDB0000044) | Carnitine synthesis = R-HSA-71262/WP4996, catecholamine biosynthesis = R-HSA-209905, vitamin C (ascorbate) metabolism = R-HSA-196836, effects of nitric oxide = WP1995, metabolism of amine-derived hormones = WP3548, vitamin B12 metabolism = WP1533, pluripotent stem cell differentiation pathway = WP2848 | |
| Mean corpuscular volume | Vitamin A (HMDB0000305) | Metabolism of fat-soluble vitamins = R-HSA-6806667, signaling by retinoic acid = R-HSA-5362517, retinoid metabolism and transport = R-HSA-975634, triglyceride metabolism = R-HSA-8979227, digestion and absorption = R-HSA-8963743, vitamins A and D—action mechanisms = WP4342 |
| Red cell distribution width | Arg-gly-asp-ser (HMDB0248573- PubChem 107775) | Tetrapeptide may play an important role in the cell attachment domain of fibronectin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Luquez, K.; Reis Silveira, A.M.; Sánchez-Vinces, S.; Rosini Silva, A.A.; Barreto, J.; Lemos de Brito, R.B.S.; Garcia, C.d.M.; Vieira, A.L.; Antonio, M.A.; de Oliveira Carvalho, P. Etodolac Single Dose Metabolic Profile Elucidation: Pharmacokinetics and Adverse Events in Healthy Volunteers. Pharmaceuticals 2025, 18, 82. https://doi.org/10.3390/ph18010082
Sánchez-Luquez K, Reis Silveira AM, Sánchez-Vinces S, Rosini Silva AA, Barreto J, Lemos de Brito RBS, Garcia CdM, Vieira AL, Antonio MA, de Oliveira Carvalho P. Etodolac Single Dose Metabolic Profile Elucidation: Pharmacokinetics and Adverse Events in Healthy Volunteers. Pharmaceuticals. 2025; 18(1):82. https://doi.org/10.3390/ph18010082
Chicago/Turabian StyleSánchez-Luquez, Karen, Anne Michelli Reis Silveira, Salvador Sánchez-Vinces, Alex Ap. Rosini Silva, Joyce Barreto, Rhubia Bethania Socorro Lemos de Brito, Caroline de Moura Garcia, Ana Lais Vieira, Marcia Ap. Antonio, and Patrícia de Oliveira Carvalho. 2025. "Etodolac Single Dose Metabolic Profile Elucidation: Pharmacokinetics and Adverse Events in Healthy Volunteers" Pharmaceuticals 18, no. 1: 82. https://doi.org/10.3390/ph18010082
APA StyleSánchez-Luquez, K., Reis Silveira, A. M., Sánchez-Vinces, S., Rosini Silva, A. A., Barreto, J., Lemos de Brito, R. B. S., Garcia, C. d. M., Vieira, A. L., Antonio, M. A., & de Oliveira Carvalho, P. (2025). Etodolac Single Dose Metabolic Profile Elucidation: Pharmacokinetics and Adverse Events in Healthy Volunteers. Pharmaceuticals, 18(1), 82. https://doi.org/10.3390/ph18010082







