Antistaphylococcal Triazole-Based Molecular Hybrids: Design, Synthesis and Activity
Abstract
:1. Introduction
2. Results and Discussion
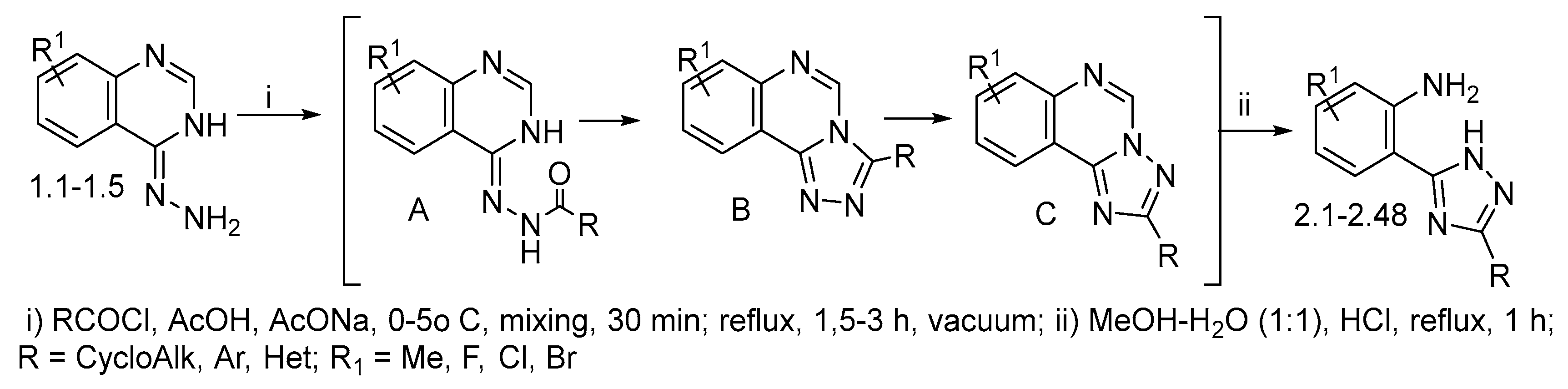
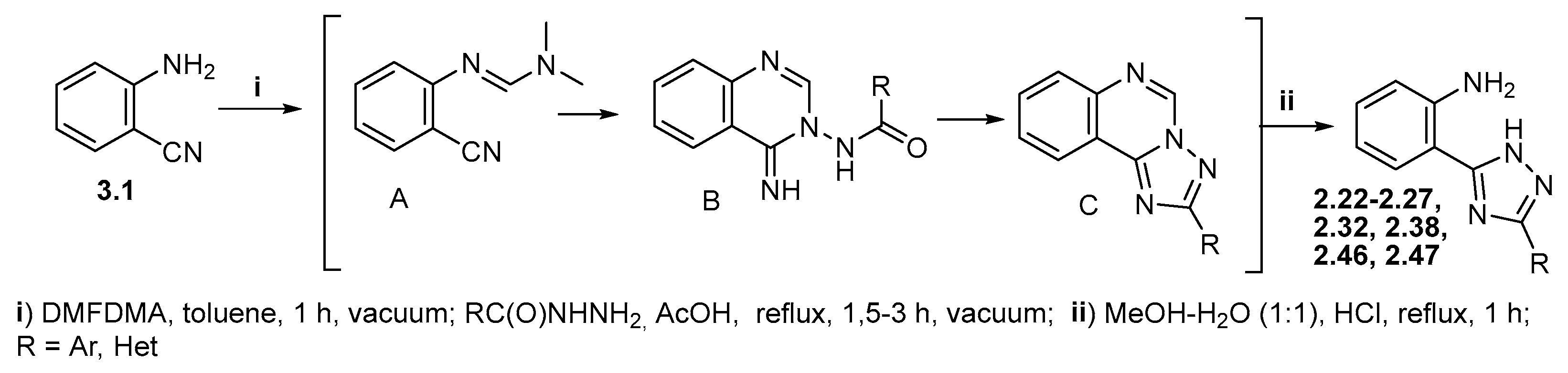
2.1. Chemical Studies
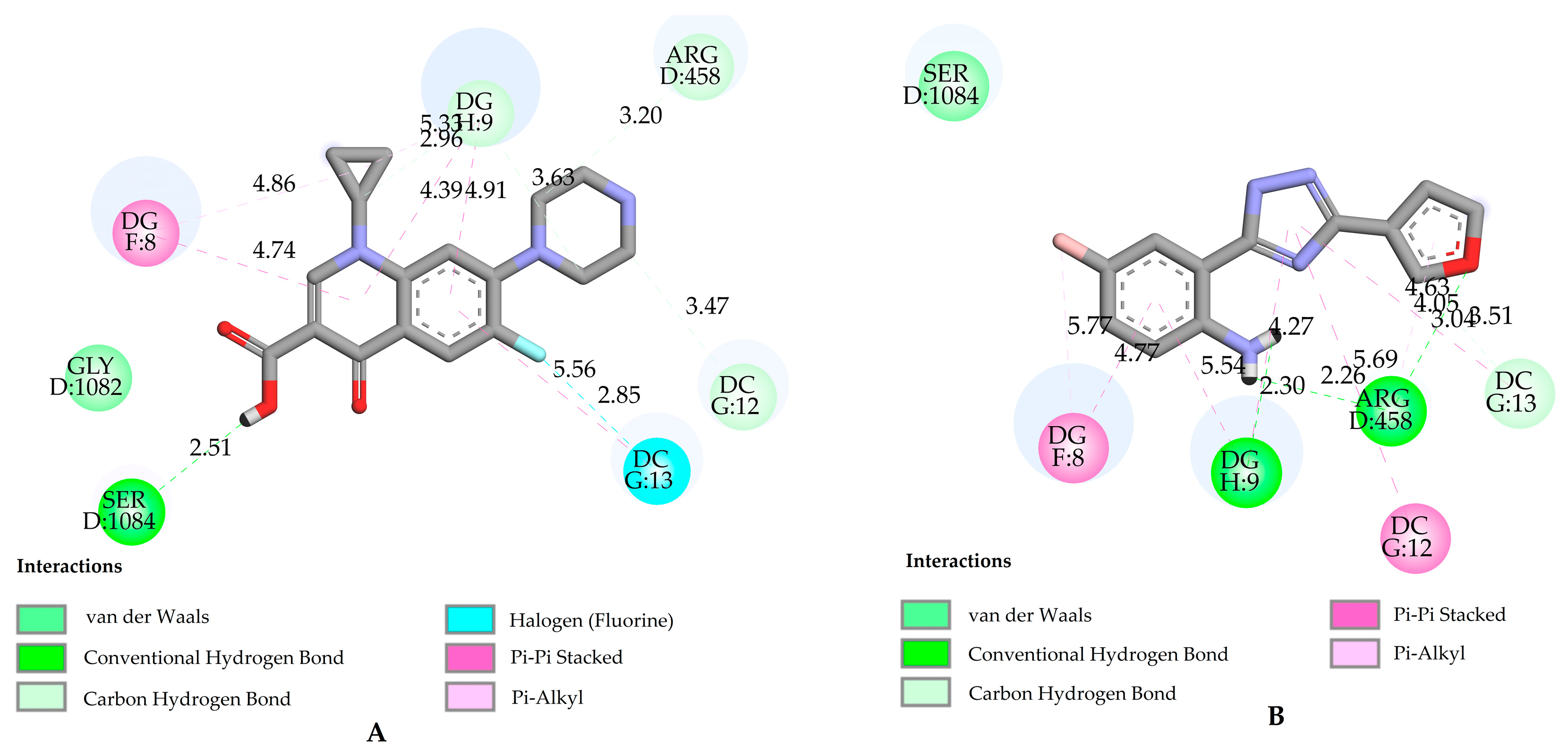
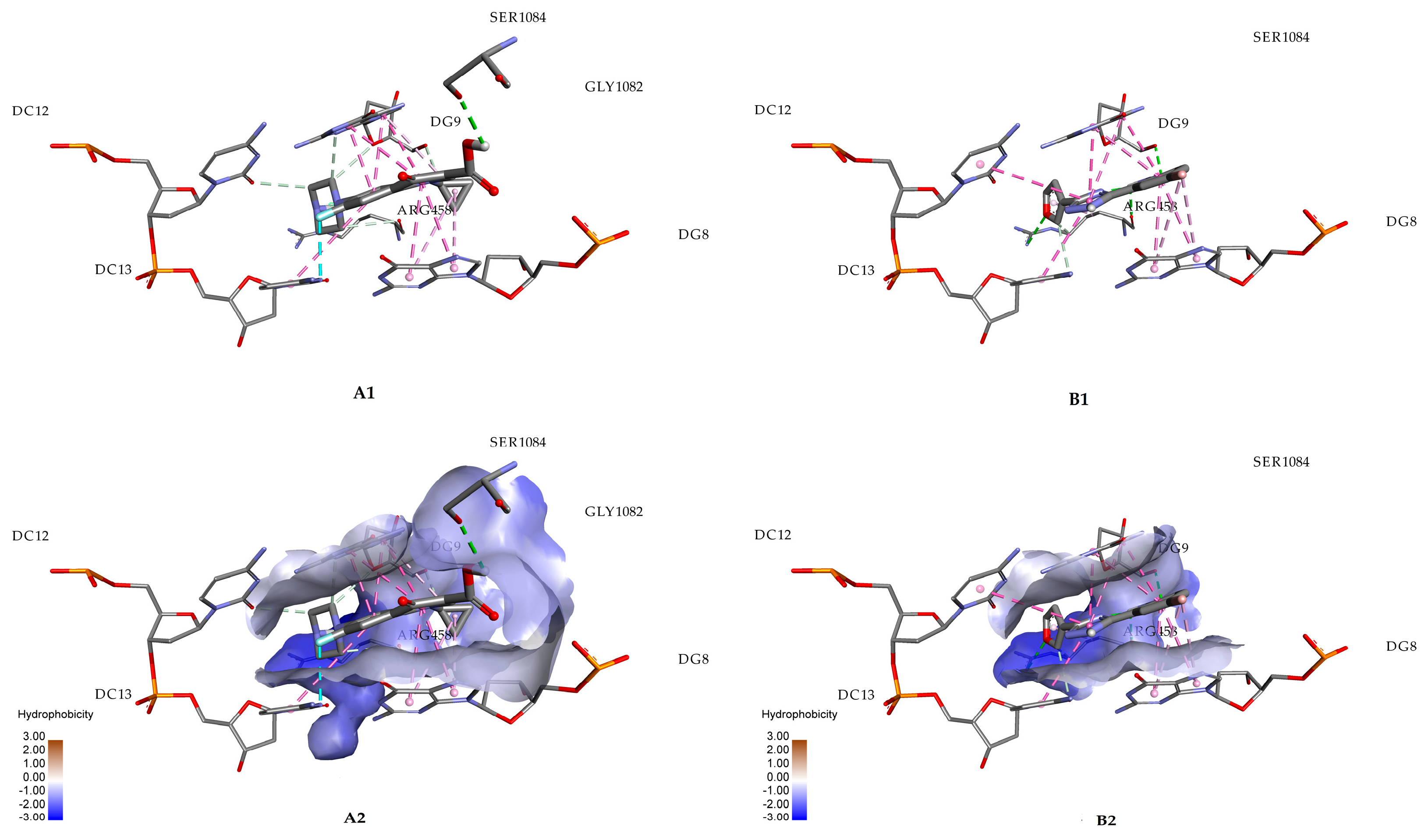
2.2. Molecular Docking Studies
2.3. Antistaphylococcal Activity of Synthesized Compounds
2.4. SAR Analysis
- The introduction of the cyclopropane fragment to the third position of the triazole fragment of 2-(1H-1,2,4-triazol-5-yl)aniline leads to the appearance of an antibacterial effect against S. aureus. The extension of the aliphatic cycle by one or more homologous units increases the antibacterial effect, and the presence of the classic “pharmacophoric” fragment of adamantane in the molecule leads to a high antistaphylococcal effect. Conversely, the modification of the aniline moiety of the molecule through the introduction of halogens results in a loss of antibacterial activity in nearly all instances;
- Replacing the cycloalkyl fragment at the third position of the triazole cycle with the phenyl fragment does not lead to a loss of antistaphylococcal activity, whereas the introduction of a halogen to the phenyl fragment in the third position leads to its reduction, and the relocation of fluorine to the ortho position results in a significant increase thereof;
- The introduction of five- or six-membered heterocyclic fragments to the third position of the triazole cycle, which are electron donors due to the heteroatom (O, N, S), unambiguously leads to high antistaphylococcal activity. The aforementioned phenomenon is associated with an increase in π–electron interactions with nucleotides and, consequently, a greater similarity of contents in the active site of the enzyme. The introduction of the methyl group to the aniline moiety leads to an enhancement of activity. Unlike 2-(3-cycloalkyl-1H-1,2,4-triazol-5-yl)anilines, the introduction of halogens to the aniline moiety of 2-(3-hetheryl-1H-1,2,4-triazol-5-yl)anilines leads to an enhancement of activity.
2.5. SwissADME Analysis
3. Materials and Methods
3.1. Synthetic Section
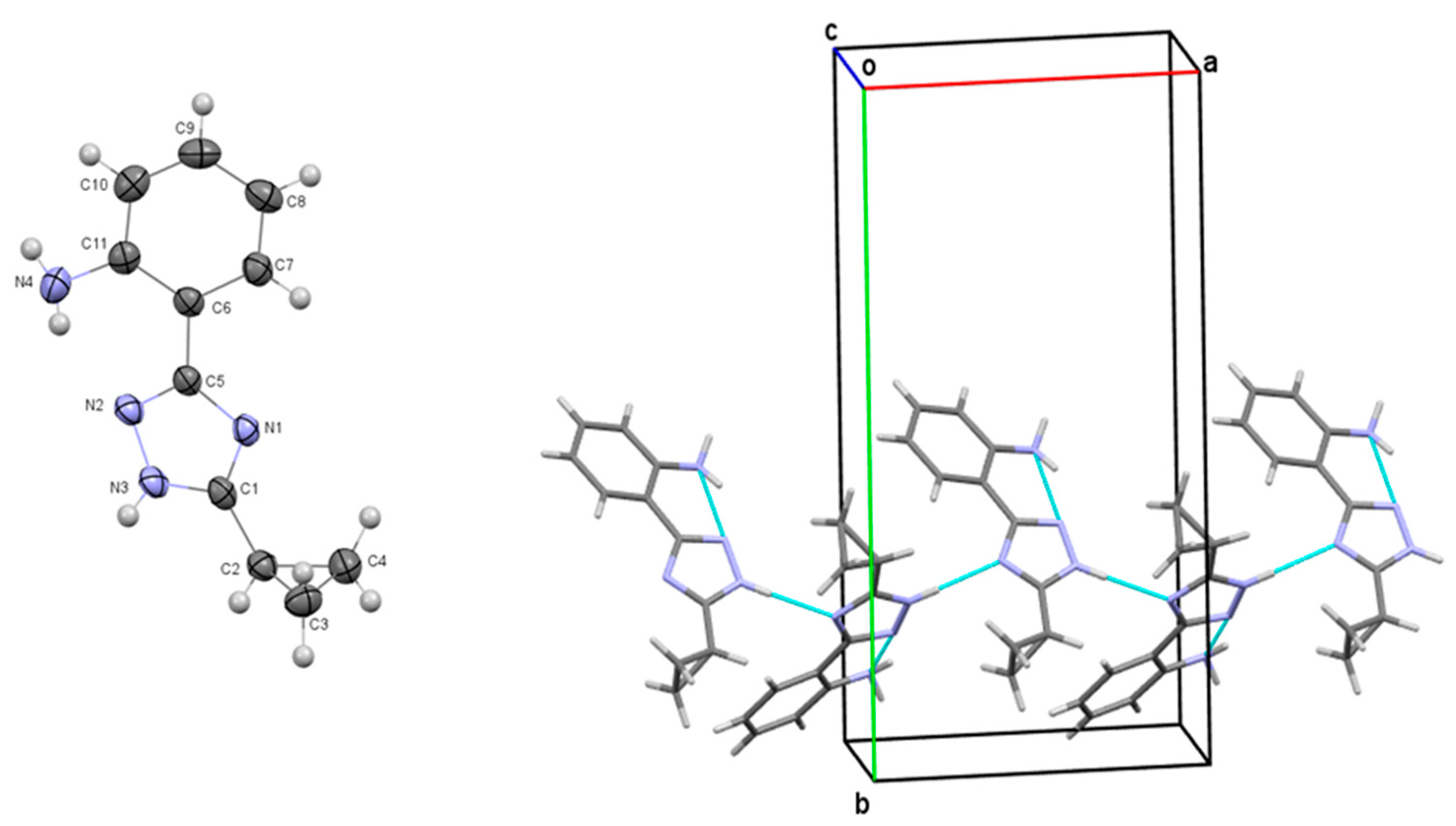
3.2. X-Ray Crystallographic Study of 2-(3-Cyclopropyl-1H-1,2,4-triazol-5-yl)aniline (2.1)
3.3. Molecular Docking
3.3.1. Ligand Preparation
3.3.2. Protein Preparation
3.4. Antimicrobial Activity
3.5. SwissADME Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Aleam, R.H.A.; George, R.F.; Georgey, H.H.; Abdel-Rahman, H.M. Bacterial virulence factors: A target for heterocyclic compounds to combat bacterial resistance. RSC Adv. 2021, 11, 36459–36482. [Google Scholar] [CrossRef] [PubMed]
- Koulenti, D.; Xu, E.; Mok, I.Y.S.; Song, A.; Karageorgopoulos, D.E.; Armaganidis, A.; Lipman, J.; Tsiodras, S. Novel Antibiotics for Multidrug-Resistant Gram-Positive Microorganisms. Microorganisms 2019, 7, 270. [Google Scholar] [CrossRef]
- Theuretzbacher, U. Resistance drives antibacterial drug development. Curr. Opin. Pharmacol. 2011, 11, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Vimalah, V.; Getha, K.; Mohamad, Z.N.; Mazlyzam, A.L. A Review on Antistaphylococcal Secondary Metabolites from Basidiomycetes. Molecules 2020, 25, 5848. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Wright, G. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Blasi, F.; Curtis, N.; Kaplan, S.; Lazzarotto, T.; Meschiari, M.; Mussini, C.; Peghin, M.; Rodrigo, C.; Vena, A.; et al. New Antibiotics for Staphylococcus aureus Infection: An Update from the World Association of Infectious Diseases and Immunological Disorders (WAidid) and the Italian Society of Anti-Infective Therapy (SITA). Antibiotics 2023, 12, 742. [Google Scholar] [CrossRef]
- Wright, P.M.; Seiple, I.B.; Myers, A.G. The evolving role of chemical synthesis in antibacterial drug discovery. Angew. Chem. Int. Ed. 2014, 53, 8840–8869. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I. Drug Design-Past, Present, Future. Molecules 2022, 27, 1496. [Google Scholar] [CrossRef]
- Anstead, G.M.; Cadena, J.; Javeri, H. Treatment of infections due to resistant Staphylococcus aureus. Methods Mol. Biol. 2014, 1085, 259–309. [Google Scholar] [CrossRef] [PubMed]
- Bisacchi, G.S.; Manchester, J.I. A New-Class Antibacterial–Almost. Lessons in Drug Discovery and Development: A Critical Analysis of More than 50 Years of Effort toward ATPase Inhibitors of DNA Gyrase and Topoisomerase IV. ACS Infect. Dis. 2015, 1, 4–41. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Durcik, M.; Tomašič, T.; Zidar, N.; Zega, A.; Kikelj, D.; Mašič, L.P.; Ilaš, J. ATP-competitive DNA gyrase and topoisomerase IV inhibitors as antibacterial agents. Expert Opin. Ther. Pat. 2019, 29, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Dighe, S.N.; Collet, T.A. Recent advances in DNA gyrase-targeted antimicrobial agents. Eur. J. Med. Chem. 2020, 199, 112326. [Google Scholar] [CrossRef]
- Man, R.-J.; Zhang, X.-P.; Yang, Y.-S.; Jiang, A.-Q.; Zhu, H.-L. Recent Progress in Small Molecular Inhibitors of DNA Gyrase. Curr. Med. Chem. 2021, 28, 5808–5830. [Google Scholar] [CrossRef] [PubMed]
- Poonam, P.; Ajay, K.; Akanksha, K.; Tamanna, V.; Vritti, P. Recent Development of DNA Gyrase Inhibitors: An Update. Mini-Rev. Med. Chem. 2024, 24, 1001–1030. [Google Scholar] [CrossRef]
- Ashley, R.E.; Dittmore, A.; McPherson, S.A.; Turnbough, C.L., Jr.; Neuman, K.C.; Osheroff, N. Activities of gyrase and topoisomerase IV on positively supercoiled DNA. Nucleic Acids Res. 2017, 45, 9611–9624. [Google Scholar] [CrossRef] [PubMed]
- Hiasa, H. DNA Topoisomerases as Targets for Antibacterial Agents. Methods Mol. Biol. 2018, 1703, 47–62. [Google Scholar] [CrossRef]
- Watkins, R.R.; Thapaliya, D.; Lemonovich, T.L.; Bonomo, R.A. Gepotidacin: A novel, oral, ‘first-in-class’ triazaacenaphthylene antibiotic for the treatment of uncomplicated urinary tract infections and urogenital gonorrhoea. J. Antimicrob. Chemother. 2023, 78, 1137–1142. [Google Scholar] [CrossRef]
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Badshah, S.L.; Muska, M.; Ahmad, N.; Khan, K. The Current Case of Quinolones: Synthetic Approaches and Antibacterial Activity. Molecules 2016, 21, 268. [Google Scholar] [CrossRef]
- Millanao, A.R.; Mora, A.Y.; Villagra, N.A.; Bucarey, S.A.; Hidalgo, A.A. Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents. Molecules 2021, 26, 7153. [Google Scholar] [CrossRef]
- Fesatidou, M.; Anthi, P.; Geronikaki, A. Heterocycle Compounds with Antimicrobial Activity. Curr. Pharm. Des. 2020, 26, 867–904. [Google Scholar] [CrossRef] [PubMed]
- Murugaiyan, J.; Kumar, P.A.; Rao, G.S.; Iskandar, K.; Hawser, S.; Hays, J.P.; Mohsen, Y.; Adukkadukkam, S.; Awuah, W.A.; Jose, R.A.M.; et al. Progress in Alternative Strategies to Combat Antimicrobial Resistance: Focus on Antibiotics. Antibiotics 2022, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Boparai, J.K.; Sharma, P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2020, 27, 4–16. [Google Scholar] [CrossRef]
- Nasiri Sovari, S.; Zobi, F. Recent Studies on the Antimicrobial Activity of Transition Metal Complexes of Groups 6–12. Chemistry 2020, 2, 418–452. [Google Scholar] [CrossRef]
- Feng, G.; Tengfei, W.; Jiaqi, X.; Gang, H. Antibacterial activity study of 1,2,4-triazole derivatives. Eur. J. Med. Chem. 2019, 173, 274–281. [Google Scholar] [CrossRef]
- Strzelecka, M.; Świątek, P. 1,2,4-Triazoles as Important Antibacterial Agents. Pharmaceuticals 2021, 14, 224. [Google Scholar] [CrossRef] [PubMed]
- Kazeminejad, Z.; Marzi, M.; Shiroudi, A.; Kouhpayeh, S.A.; Farjam, M.; Zarenezhad, E. Novel 1,2,4-Triazoles as Antifungal Agents. Biomed. Res. Int. 2022, 22, 4584846. [Google Scholar] [CrossRef]
- Xuemei, G.; Zhi, X. 1,2,4-Triazole hybrids with potential antibacterial activity against methicillin-resistant Staphylococcus aureus. Arch. Pharm. 2020, 354, e2000223. [Google Scholar] [CrossRef]
- Jie, L.; Junwei, Z. The Antibacterial Activity of 1,2,3-triazole- and 1,2,4-Triazole-containing Hybrids against Staphylococcus aureus: An Updated Review (2020–Present). Curr. Top. Med. Chem. 2022, 22, 41–63. [Google Scholar] [CrossRef]
- Verdirosa, F.; Gavara, L.; Sevaille, L.; Tassone, G.; Corsica, G.; Legru, A.; Feller, G.; Chelini, G.; Mercuri, P.S.; Tanfoni, S.; et al. 1,2,4-Triazole-3-Thione Analogues with a 2-Ethylbenzoic Acid at Position 4 as VIM-type Metallo-β-Lactamase Inhibitors. ChemMedChem 2022, 17, e202100699. [Google Scholar] [CrossRef] [PubMed]
- Sergeieva, T.; Bilichenko, M.; Kholodnyak, S.; Monaykina, Y.; Okovytyy, S.; Kovalenko, S.; Voronkov, E.; Leszczynski, J. Origin of Substituent Effect on Tautomeric Behavior of 1,2,4-Triazole Derivatives. Combined Spectroscopic and Theoretical Study. J. Phys. Chem. A 2016, 120, 10116–10122. [Google Scholar] [CrossRef] [PubMed]
- Pylypenko, O.O.; Okovytyy, S.I.; Sviatenko, L.K.; Voronkov, E.O.; Shabelnyk, K.P.; Kovalenko, S.I. Tautomeric behavior of 1,2,4-triazole derivatives: Combined spectroscopic and theoretical study. Struct. Chem. 2023, 34, 181–192. [Google Scholar] [CrossRef]
- Francis, J.E.; Cash, W.D.; Psychoyos, S.; Ghai, G.; Wenk, P.; Friedmann, R.C.; Atkins, C.; Warren, V.; Furness, P.; Hyun, J.L.; et al. Structure-activity profile of a series of novel triazoloquinazoline adenosine antagonists. J. Med. Chem. 1988, 31, 1014–1020. [Google Scholar] [CrossRef]
- Balo, C.; López, C.; Brea, J.M.; Fernánde, F.; Caamaño, O. Synthesis and Evaluation of Adenosine Antagonist Activity of a Series of [1,2,4]Triazolo[1,5-c]quinazolines. Chem. Pharm. Bull. 2007, 55, 372–375. [Google Scholar] [CrossRef]
- Khan, G.; Sreenivasa, S.; Govindaiah, S.; Chandramohan, V.; Shetty, P.R. Synthesis, biological screening, in silico study and fingerprint applications of novel 1,2,4-triazole derivatives. J. Het. Chem. 2020, 57, 2010–2023. [Google Scholar] [CrossRef]
- Kholodnyak, S.V.; Schabelnyk, K.P.; Zhernova, G.O.; Sergeieva, T.Y.; Ivchuk, V.V.; Voskoboynik, O.Y.; Kovalenko, S.I.; Trzhetsinskii, S.D.; Okovytyy, S.I.; Shishkina, S.V. Hydrolytic cleavage of pyrimidine ring in 2-aryl-[1,2,4]triazolo[1,5-c]-quinazolines: Physico-chemical properties and hypoglycemia activity of the synthesized compounds. News Pharm. 2015, 3, 9–17. [Google Scholar] [CrossRef]
- Pylypenko, O.O.; Sviatenko, L.K.; Shabelnyk, K.P.; Kovalenko, S.I.; Okovytyy, S.I. Synthesis and hydrolytic decomposition of 2-hetaryl[1,2,4]triazolo[1,5-c]quinazolines: DFT Study. Struct. Chem. 2024, 35, 97–104. [Google Scholar] [CrossRef]
- Kovalenko, S.I.; Antypenko, L.M.; Bilyi, A.K.; Kholodnyak, S.V.; Karpenko, O.V.; Antypenko, O.M.; Mykhaylova, N.S.; Los, T.I.; Kolomoets, O.S. Synthesis and Anticancer Activity of 2-(Alkyl-, Alkaryl-, Aryl-, Hetaryl-)-[1,2,4]triazolo[1,5-c]quinazolines. Sci. Pharm. 2013, 81, 359–391. [Google Scholar] [CrossRef]
- Breitmaier, E. Structure Elucidation by NMR in Organic Chemistry: A Practical Guide, 3rd ed.; Wiley: Hoboken, NJ, USA, 2002; 270p, ISBN 978-0-470-85007-7. [Google Scholar]
- Zefirov, Y.V. Reduced intermolecular contacts and specific interactions in molecular crystals. Crystallogr. Rep. 1997, 42, 865–886. Available online: https://inis.iaea.org/search/searchsinglerecord.aspx?recordsFor=SingleRecord&RN=30003683 (accessed on 10 March 2024).
- Tao, L.; Zhang, P.; Qin, C.; Chen, S.Y.; Zhang, C.; Chen, Z.; Zhu, F.; Yang, S.Y.; Wei, Y.Q.; Chen, Y.Z. Recent progresses in the exploration of machine learning methods as in-silico ADME prediction tools. Adv. Drug Deliv. Rev. 2015, 86, 83–100. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Combin. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Martin, Y.C. A bioavailability score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, L.; Xu, H.; Li, X.; Zhao, L.; Wang, W.; Li, B.; Zhang, X. 6,7-Dimorpholinoalkoxy quinazoline derivatives as potent EGFR inhibitors with enhanced antiproliferative activities against tumor cells. Eur. J. Med. Chem. 2018, 147, 77–89. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar] [CrossRef]
- Kumar, H.S.S.; Kumar, S.R.; Kumar, N.N.; Ajith, S. Molecular docking studies of gyrase inhibitors: Weighing earlier screening bedrock. In Silico Pharmacol. 2021, 9, 2. [Google Scholar] [CrossRef]
- MarvinSketch version 20.21.0, ChemAxon. Available online: http://www.chemaxon.com (accessed on 20 December 2023).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Discovery Studio Visualizer v21.1.0.20298. Accelrys Software Inc. Available online: https://www.3dsbiovia.com (accessed on 20 December 2023).
- Baber, J.C.; Thompson, D.C.; Cross, J.B.; Humblet, C. GARD: A Generally Applicable Replacement for RMSD. J. Chem. Inf. Model. 2009, 49, 1889–1900. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Andrews, C.W.; Capelli, A.-M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2005, 49, 5912–5931. [Google Scholar] [CrossRef]
- DockRMSD. Docking Pose Distance Calculation. 2022. Available online: https://zhanggroup.org/DockRMSD/ (accessed on 23 January 2024).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Wayne, P.A., Ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2020; ISBN 978-1-68440-066-9 [Print]; ISBN 978-1-68440-067-6 [Electronic]. [Google Scholar]
- SwissADME. Available online: http://www.swissadme.ch/index.php# (accessed on 27 October 2023).
- Bilyi, A.K.; Antypenko, L.M.; Ivchuk, V.V.; Kamyshnyi, O.M.; Polishchuk, N.M.; Kovalenko, S.I. 2-Heteroaryl-[1,2,4]triazolo[1,5-c]quinazoline-5(6 H)-thiones and Their S-Substituted Derivatives: Synthesis, Spectroscopic Data, and Biological Activity. ChemPlusChem 2015, 80, 980–989. [Google Scholar] [CrossRef] [PubMed]
- Nosulenko, I.S.; Voskoboynik, O.Y.; Berest, G.G.; Safronyuk, S.L.; Kovalenko, S.I.; Kamyshnyi, O.M.; Polishchuk, N.M.; Sinyak, R.S.; Katsev, A.V. Synthesis and Antimicrobial Activity of 6-Thioxo-6,7-dihydro-2H-[1,2,4]triazino[2,3-c]-quinazolin-2-one Derivatives. Sci. Pharm. 2014, 82, 483–500. [Google Scholar] [CrossRef]

| Compound | Affinity (kcal/mol) | Compound | Affinity (kcal/mol) | Compound | Affinity (kcal/mol) |
|---|---|---|---|---|---|
| TA * | −6.3 | 2.17 | −5.5 | 2.34 | −8.1 |
| 2.1 | −6.7 | 2.18 | −5.6 | 2.35 | −7.9 |
| 2.2 | −7.4 | 2.19 | −6.1 | 2.36 | −8.3 |
| 2.3 | −8.4 | 2.20 | −6.3 | 2.37 | −8.3 |
| 2.4 | −7.1 | 2.21 | −6.0 | 2.38 | −8.9 |
| 2.5 | −7.1 | 2.22 | −7.9 | 2.39 | −8.5 |
| 2.6 | −7.6 | 2.23 | −8.3 | 2.40 | −7.9 |
| 2.7 | −7.4 | 2.24 | −8.4 | 2.41 | −9.2 |
| 2.8 | −7.6 | 2.25 | −8.0 | 2.42 | −8.1 |
| 2.9 | −7.5 | 2.26 | −8.5 | 2.43 | −8.6 |
| 2.10 | −8.6 | 2.27 | −8.2 | 2.44 | −8.9 |
| 2.11 | −8.4 | 2.28 | −8.6 | 2.45 | −8.5 |
| 2.12 | −7.8 | 2.29 | −8.9 | 2.46 | −8.0 |
| 2.13 | −8.1 | 2.30 | −8.6 | 2.47 | −7.8 |
| 2.14 | −8.3 | 2.31 | −8.7 | 2.48 | −8.7 |
| 2.15 | −7.8 | 2.32 | −7.5 | Ciprofloxacin | −6.7 |
| 2.16 | −8.1 | 2.33 | −7.8 | - | - |
| Compounds | R | R1 | MIC *, μM | MBC **, μM | MBC/MIC |
|---|---|---|---|---|---|
| 2.1 | cyclopropyl | H | 62.4 | 124.8 | 2 |
| 2.2 | cyclopropyl | 6-Me | 933.4 | 933.4 | 1 |
| 2.3 | cyclopropyl | 5-F | 458.2 | 916.5 | 2 |
| 2.4 | cyclopropyl | 4-Cl | 852.2 | 852.2 | 1 |
| 2.5 | cyclobutyl | H | 14.6 | 23.3 | 1.5 |
| 2.6 | cyclobutyl | 6-Me | 876.1 | 876.1 | 1 |
| 2.7 | cyclobutyl | 5-F | 430.6 | 861.2 | 2 |
| 2.8 | cyclobutyl | 4-Cl | 402.1 | 804.2 | 2 |
| 2.9 | cyclopentyl | H | 27.4 | 438.0 | 16 |
| 2.10 | cyclopentyl | 6-Me | 825.4 | 825.4 | 1 |
| 2.11 | cyclopentyl | 5-F | 203.0 | 406.0 | 2 |
| 2.12 | cyclopentyl | 4-Cl | 47.6 | 47.6 | 1 |
| 2.13 | cyclohexyl | H | 26.8 | 206.3 | 7.7 |
| 2.14 | cyclohexyl | 6-Me | 390.1 | 390.1 | 1 |
| 2.15 | cyclohexyl | 5-F | 192.1 | 768.3 | 4 |
| 2.16 | cyclohexyl | 4-Cl | 361.3 | 723.6 | 2 |
| 2.17 | adamantyl-1 | H | 10.6 | 21.2 | 2 |
| 2.18 | adamantyl-1 | 6-Me | 10.1 | 20.2 | 2 |
| 2.19 | adamantyl-1 | 5-F | 320.1 | 640.2 | 2 |
| 2.20 | adamantyl-1 | 4-Cl | 304.1 | 608.2 | 2 |
| 2.21 | adamantyl-1 | 4-Br | 267.9 | 535.7 | 2 |
| 2.22 | Ph | H | 26.4 | 211.6 | 8 |
| 2.23 | 4-FC6H4 | H | 196.6 | 786.6 | 4 |
| 2.24 | 4-ClC6H4 | H | 92.3 | 184.6 | 2 |
| 2.25 | 4-BrC6H4 | H | 317.3 | 634.6 | 2 |
| 2.26 | 2-FC6H4 | H | 12.4 | 24.8 | 2 |
| 2.27 | furan-2-yl | H | 221.0 | 442.0 | 2 |
| 2.28 | furan-3-yl | 6-Me | 13.0 | 52.0 | 4 |
| 2.29 | furan-3-yl | 5-F | 25.6 | 51.2 | 2 |
| 2.30 | furan-3-yl | 4-Cl | 11.9 | 23.8 | 2 |
| 2.31 | furan-3-yl | 4-Br | 5.2 | 10.4 | 2 |
| 2.32 | thiophen-2-yl | H | 103.2 | 206.4 | 2 |
| 2.33 | thiophen-2-yl | 5-F | 24.0 | 48.0 | 2 |
| 2.34 | thiophen-3-yl | 6-Me | 12.2 | 12.2 | 1 |
| 2.35 | thiophen-3-yl | 5-F | 6.1 | 48.0 | 8 |
| 2.36 | thiophen-3-yl | 4-Cl | 45.2 | 180.8 | 4 |
| 2.37 | thiophen-3-yl | 4-Br | 77.8 | 311.3 | 4 |
| 2.38 | benzofuran-2-yl | H | 180.9 | 361.8 | 2 |
| 2.39 | benzofuran-2-yl | 6-Me | 10.7 | 21.4 | 2 |
| 2.40 | benzofuran-2-yl | 5-F | 42.5 | 84.9 | 2 |
| 2.41 | benzofuran-2-yl | 4-Cl | 20.1 | 40.2 | 2 |
| 2.42 | benzofuran-2-yl | 4-Br | 140.7 | 140.7 | 1 |
| 2.43 | benzothiophen-2-yl | H | 171.0 | 342.0 | 2 |
| 2.44 | indol-2-yl | H | 181.6 | 363.2 | 2 |
| 2.45 | pyridin-2-yl | H | 105.3 | 210.6 | 2 |
| 2.46 | pyridin-3-yl | H | 13.2 | 52.8 | 4 |
| 2.47 | pyridin-4-yl | H | 105.3 | 210.6 | 2 |
| 2.48 | pyridin-4-yl | Br | 79.1 | 158.2 | 2 |
| Ciprofloxacin | 4.7 | 9.6 | 2 |
| Physicochemical Descriptors and Predicted Pharmacokinetic Properties * | Compounds | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.17 | 2.18 | 2.26 | 2.28 | 2.30 | 2.31 | 2.34 | 2.35 | 2.39 | 2.46 | CF ** | |
| MW (Da) (<500) | 294.39 | 308.42 | 252.27 | 240.26 | 260.68 | 305.13 | 256.33 | 260.29 | 290.32 | 237.26 | 331.34 |
| n-ROTB (<10) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 |
| n-HBA (<10) | 2 | 2 | 2 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 5 |
| n-HBD (≤5) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 |
| TPSA (<140, Å2) | 67.59 | 67.59 | 87.82 | 80.73 | 80.73 | 80.73 | 95.83 | 95.83 | 80.73 | 80.48 | 74.57 |
| logP (≤5) | 3.21 | 3.54 | 2.05 | 2.06 | 2.30 | 2.34 | 2.74 | 2.69 | 3.07 | 1.66 | 1.10 |
| Molar refractivity | 88.11 | 93.08 | 73.68 | 68.89 | 68.94 | 71.63 | 74.50 | 69.49 | 86.40 | 69.45 | 95.25 |
| Gastrointestinal absorption | high | high | high | high | high | high | high | high | high | high | high |
| Drug-likeness | |||||||||||
| Lipinski (Pfizer) filter [43] | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Veber (GSK) filter [44] | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Muegge (Bayer) filter [45] | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Ghose filter [46] | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Egan filter [47] | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Bioavailability score [48] | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| Lead-likeness | no | no | yes | no | yes | yes | yes | yes | yes | no | yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shabelnyk, K.; Fominichenko, A.; Antypenko, O.; Gaponov, O.; Koptieva, S.; Shyshkina, S.; Voskoboinik, O.; Okovytyy, S.; Kovalenko, S.; Oksenych, V.; et al. Antistaphylococcal Triazole-Based Molecular Hybrids: Design, Synthesis and Activity. Pharmaceuticals 2025, 18, 83. https://doi.org/10.3390/ph18010083
Shabelnyk K, Fominichenko A, Antypenko O, Gaponov O, Koptieva S, Shyshkina S, Voskoboinik O, Okovytyy S, Kovalenko S, Oksenych V, et al. Antistaphylococcal Triazole-Based Molecular Hybrids: Design, Synthesis and Activity. Pharmaceuticals. 2025; 18(1):83. https://doi.org/10.3390/ph18010083
Chicago/Turabian StyleShabelnyk, Kostiantyn, Alina Fominichenko, Oleksii Antypenko, Olexandr Gaponov, Svitlana Koptieva, Svitlana Shyshkina, Oleksii Voskoboinik, Sergiy Okovytyy, Serhii Kovalenko, Valentyn Oksenych, and et al. 2025. "Antistaphylococcal Triazole-Based Molecular Hybrids: Design, Synthesis and Activity" Pharmaceuticals 18, no. 1: 83. https://doi.org/10.3390/ph18010083
APA StyleShabelnyk, K., Fominichenko, A., Antypenko, O., Gaponov, O., Koptieva, S., Shyshkina, S., Voskoboinik, O., Okovytyy, S., Kovalenko, S., Oksenych, V., & Kamyshnyi, O. (2025). Antistaphylococcal Triazole-Based Molecular Hybrids: Design, Synthesis and Activity. Pharmaceuticals, 18(1), 83. https://doi.org/10.3390/ph18010083










