Lipophilic Extracts of Portulaca oleracea L.: Analysis of Bioactive Fatty Acids Targeting Microbial and Cancer Pathways
Abstract
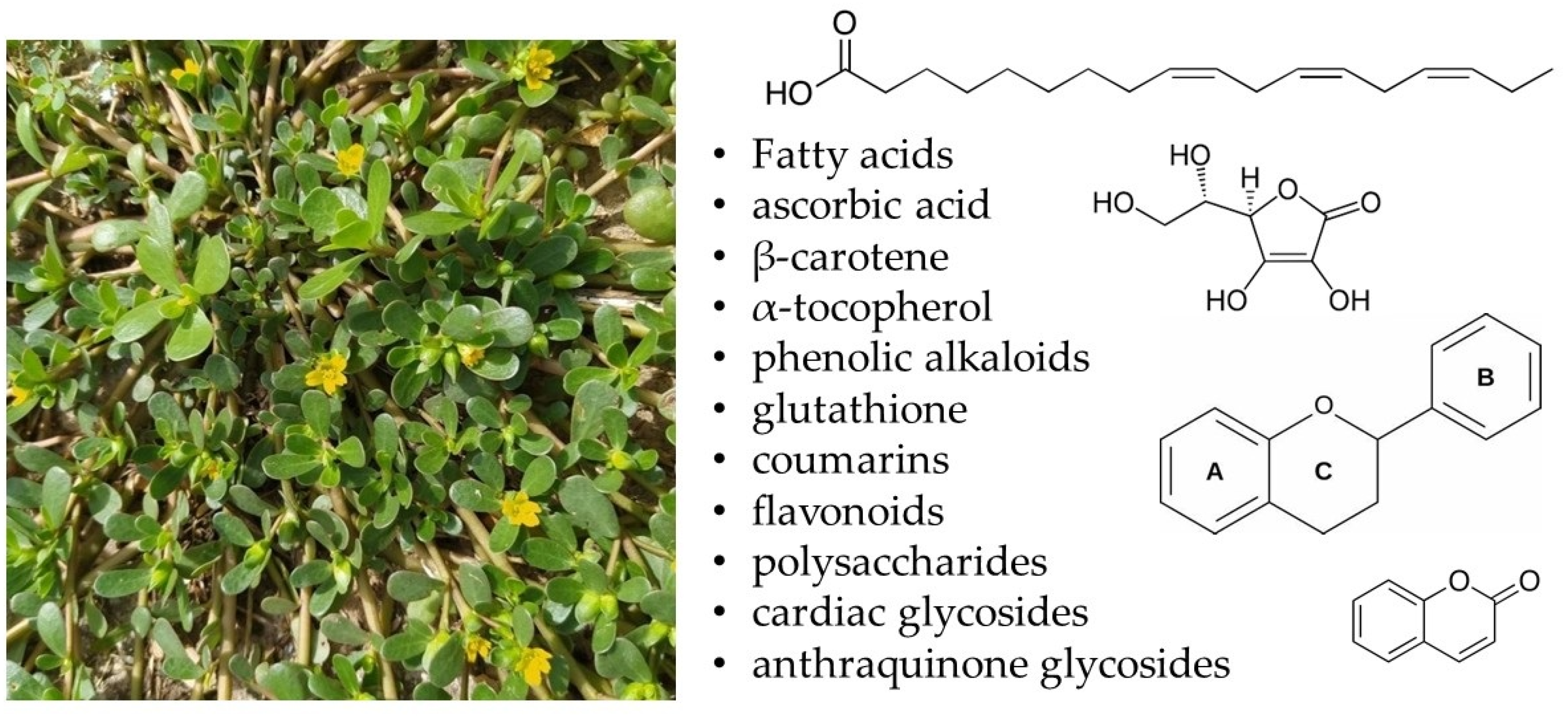
:1. Introduction
2. Results and Discussion
2.1. Fatty Acid Composition
2.2. Antimicrobial Activity
2.3. Antibiofilm Activity
2.4. Cytotoxicity
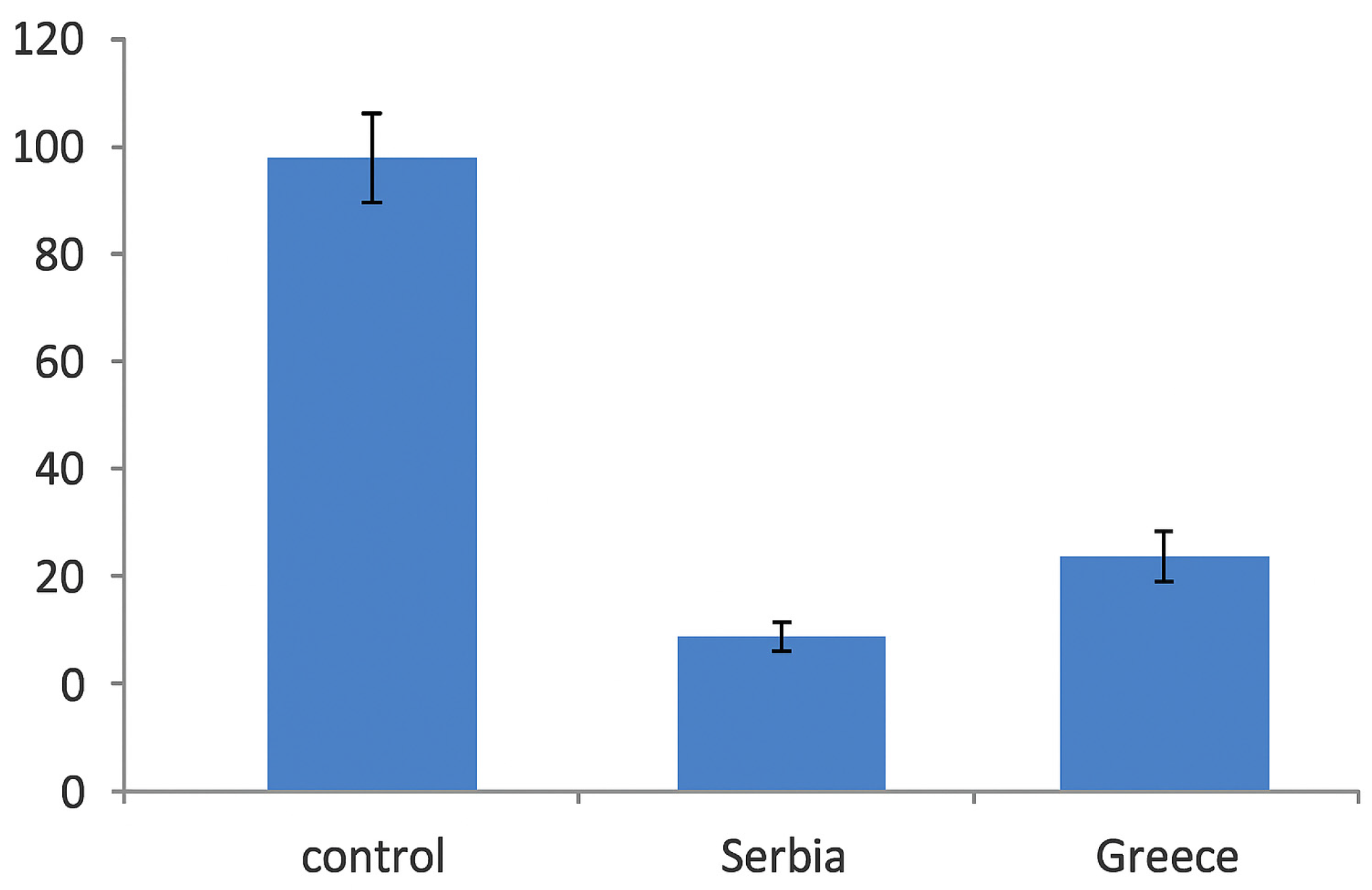
2.5. Bacterial Invasion
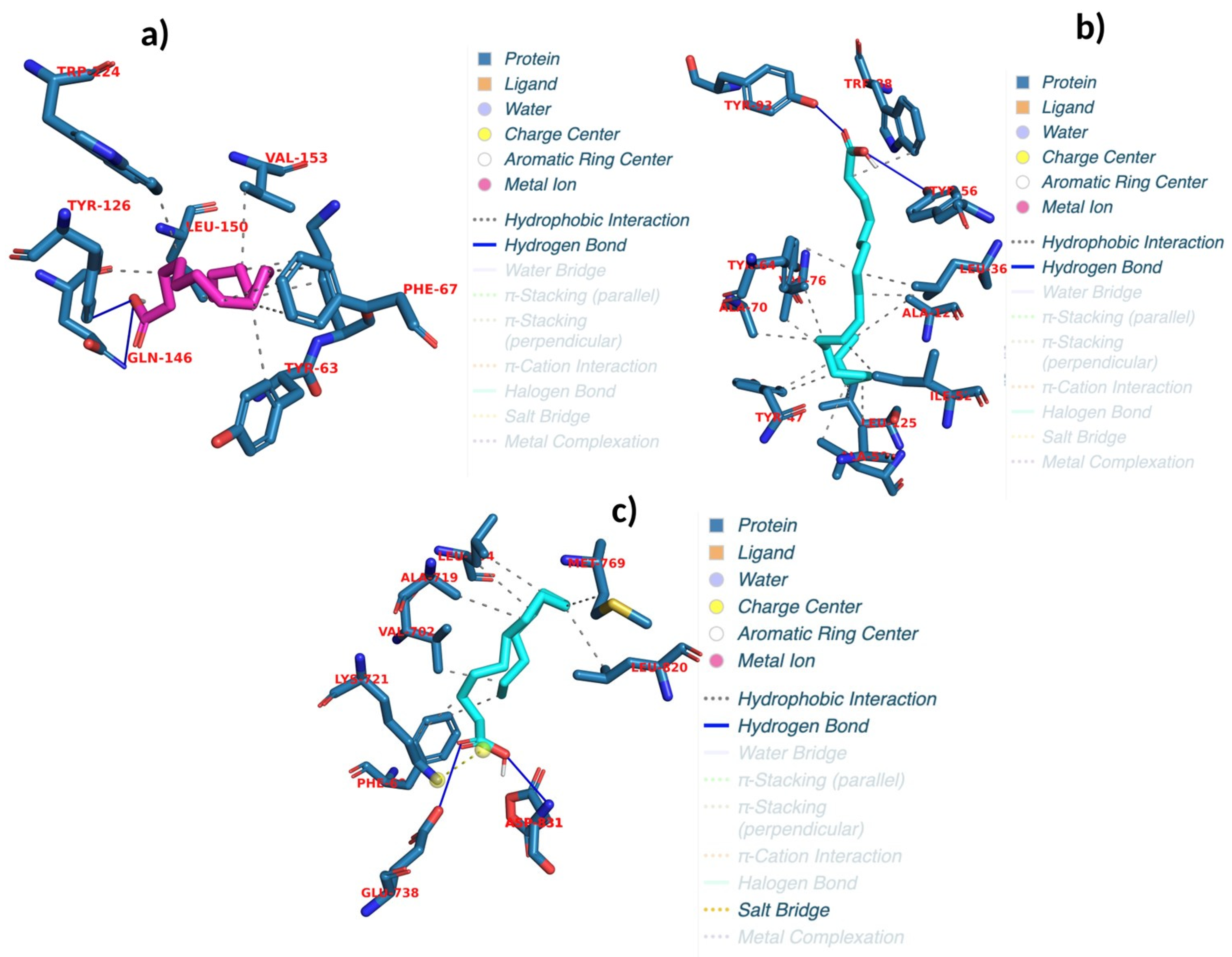
2.6. Evaluating Docking Outcomes: Ligand Binding Energies and Interaction Profiles
2.7. Binding Free Energy Analysis: MM/PBSA Results and Implications for Ligand Efficacy
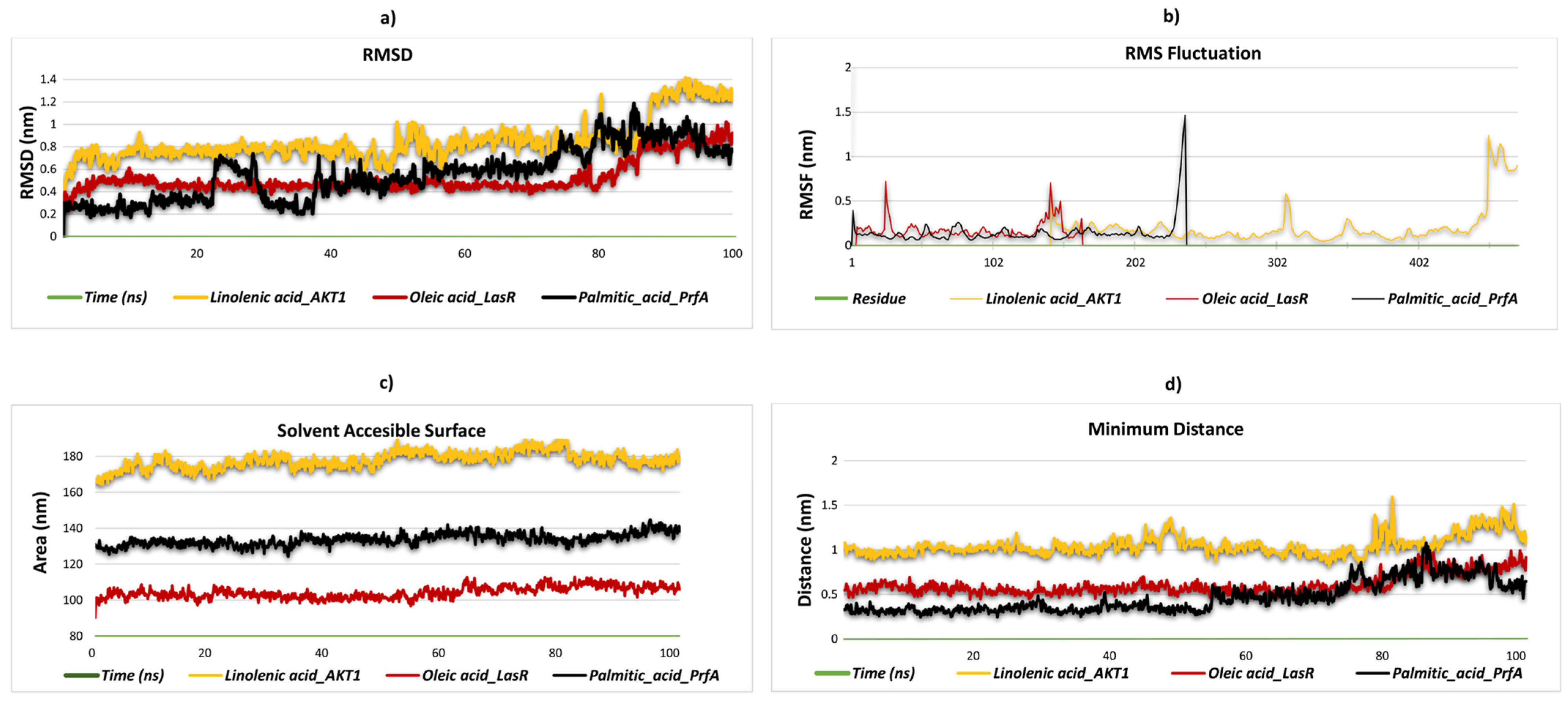
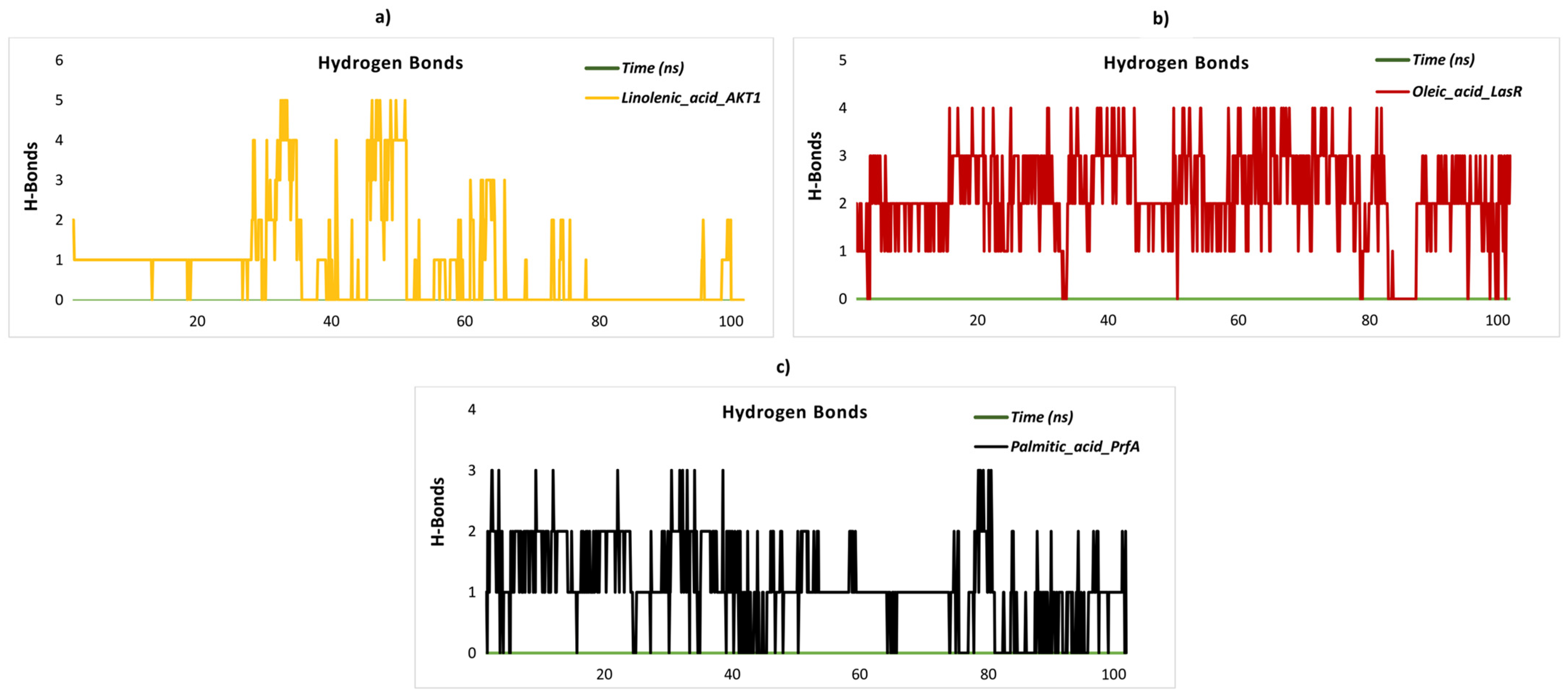
2.8. Stability and Flexibility in Molecular Dynamics Simulation
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Collection of the Samples
3.3. Extraction of the Samples
3.4. Fatty Acids
3.5. Antimicrobial Activity
3.6. Antibiofilm Activity
3.7. Congo Red Test
3.8. eDNA Test
3.9. Analysis of Cytotoxicity of the Extract
3.10. Effects of the Extract on the Invasion Capacities of S. aureus in HaCaT Cells
3.11. Molecular Docking
3.12. Calculation of MM/PBSA Free Energy to Determine Ligand-Binding Affinity
3.13. Molecular Dynamics Simulation Setup and Analysis Parameters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sultana, A.; Raheman, K. Portulaca oleracea Linn: A global panacea with ethnomedicinal and pharmacological potential. Int. J. Pharm. Pharm. Sci. 2013, 5, 33–39. [Google Scholar]
- Iranshahy, M.; Javadi, B.; Iranshahi, M.; Jahanbakhsh, S.P.; Mahyari, S.; Hassani, F.V.; Karimi, G. A review of traditional uses, phytochemistry and pharmacology of Portulaca oleracea L. J. Ethnopharmacol. 2017, 205, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.G.; Plucknett, D.L.; Pancho, J.V.; Herberger, J.P. The World’s Worst Weeds. Distribution and Biology; University Press: Honolulu, HI, USA, 1977. [Google Scholar]
- Uddin, M.K.; Juraimi, A.S.; Anwar, F.; Hossain, M.A.; Alam, M.A. Effect of salinity on proximate mineral composition of purslane (‘Portulca oleracea’ L.). Aust. J. Crop Sci. 2012, 6, 1732–1736. [Google Scholar]
- Ocampo, G.; Columbus, J.T. Molecular phylogenetics, historical biogeography, and chromosome number evolution of Portulaca (Portulacaceae). Mol. Phylogenetics Evol. 2012, 63, 97–112. [Google Scholar] [CrossRef]
- Samir, D.; Sara, C.; Widad, A. The effects of aqueous leaf extract of Portulaca oleracea on haemato-biochemical and histopathological changes induced by sub-chronic aluminium toxicity in male wistar rats. Pharmacol. Res.-Mod. Chin. Med. 2022, 4, 100101. [Google Scholar] [CrossRef]
- Abd El-Hack, M.E.; Alabdali, A.Y.; Aldhalmi, A.K.; Reda, F.M.; Bassiony, S.S.; Selim, S.; El-Saadony, M.T.; Alagawany, M. Impacts of Purslane (Portulaca oleracea) extract supplementation on growing Japanese quails’ growth, carcass traits, blood indices, nutrients digestibility and gut microbiota. Poult. Sci. 2022, 101, 102166. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-X.; Xin, H.-L.; Rahman, K.; Wang, S.-J.; Peng, C.; Zhang, H. Portulaca oleracea L.: A review of phytochemistry and pharmacological effects. BioMed Res. Int. 2015, 2015, 925631. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.I.; Hussein, A.S. Chemical composition of purslane (Portulaca oleracea). Plant Foods Hum. Nutr. 1994, 45, 1–9. [Google Scholar] [CrossRef]
- Naeem, F.; Khan, S.H. Purslane (Portulaca oleracea L.) as phytogenic substance—A review. J. Herbs Spices Med. Plants 2013, 19, 216–232. [Google Scholar] [CrossRef]
- Zouine, N.; Ghachtouli, N.E.; Abed, S.E.; Koraichi, S.I. A comprehensive review on medicinal plant extracts as antibacterial agents: Factors, mechanism insights and future prospects. Sci. Afr. 2024, 26, e02395. [Google Scholar] [CrossRef]
- Puca, V.; Marulli, R.Z.; Grande, R.; Vitale, I.; Niro, A.; Molinaro, G.; Prezioso, S.; Muraro, R.; Di Giovanni, P. Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study. Antibiotics 2021, 10, 1162. [Google Scholar] [CrossRef]
- Yang, S.; Qiao, J.; Zhang, M.; Kwok, L.-Y.; Matijašić, B.B.; Zhang, H.; Zhang, W. Prevention and treatment of antibiotics-associated adverse effects through the use of probiotics: A review. J. Adv. Res. 2024, in press. [Google Scholar] [CrossRef] [PubMed]
- Zasheva, D.; Mladenov, P.; Zapryanova, S.; Gospodinova, Z.; Georgieva, M.; Alexandar, I.; Velinov, V.; Djilianov, D.; Moyankova, D.; Simova-Stoilova, L. Cytotoxic Effects of Plant Secondary Metabolites and Naturally Occurring Bioactive Peptides on Breast Cancer Model Systems: Molecular Mechanisms. Molecules 2024, 29, 5275. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P.; Norman, H.A.; Gillaspy, J.E.; Duke, J.A. Common purslane: A source of omega-3 fatty acids and antioxidants. J. Am. Coll. Nutr. 1992, 11, 374–382. [Google Scholar] [CrossRef]
- Oliveira, I.; Valentão, P.; Lopes, R.; Andrade, P.B.; Bento, A.; Pereira, J.A. Phytochemical characterization and radical scavenging activity of Portulaca oleraceae L. leaves and stems. Microchem. J. 2009, 92, 129–134. [Google Scholar] [CrossRef]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef]
- Othman, A.S. Bactericidal efficacy of omega-3 fatty acids and esters present in Moringa oleifera and Portulaca oleracea fixed oils against oral and gastro enteric bacteria. Int. J. Pharmacol. 2017, 13, 44–53. [Google Scholar]
- Tleubayeva, M.I.; Datkhayev, U.M.; Alimzhanova, M.; Ishmuratova, M.Y.; Korotetskaya, N.V.; Abdullabekova, R.M.; Flisyuk, E.V.; Gemejiyeva, N.G. Component Composition and Antimicrobial Activity of CO2 Extract of Portulaca oleracea, Growing in the Territory of Kazakhstan. Sci. World J. 2021, 2021, 5434525. [Google Scholar] [CrossRef]
- Banerjee, G.; Mukherjee, A. Biological activity of a common weed-Portulaca oleracea L. II. Antifungal activity. Acta Bot. Hung. 2002, 44, 205–208. [Google Scholar] [CrossRef]
- Zheng, C.J.; Yoo, J.-S.; Lee, T.-G.; Cho, H.-Y.; Kim, Y.-H.; Kim, W.-G. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett. 2005, 579, 5157–5162. [Google Scholar] [CrossRef]
- Kelsey, J.; Bayles, K.W.; Shafii, B.; McGuire, M. Fatty acids and monoacylglycerols inhibit growth of Staphylococcus aureus. Lipids 2006, 41, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Fung, K.; Han, Q.; Ip, M.; Yang, X.; Lau, C.B.; Chan, B.C. Synergists from Portulaca oleracea with macrolides against methicillin-resistant Staphylococcus aureus and related mechanism. Hong Kong Med. J. 2017, 23, 38–42. [Google Scholar] [PubMed]
- Nguyen, M.T.; Hanzelmann, D.; Härtner, T.; Peschel, A.; Götz, F. Skin-specific unsaturated fatty acids boost the Staphylococcus aureus innate immune response. Infect. Immun. 2016, 84, 205–215. [Google Scholar] [CrossRef]
- Cho, J.-A.; Roh, Y.J.; Son, H.R.; Choi, H.; Lee, J.-W.; Kim, S.J.; Lee, C.-H. Assessment of the biofilm-forming ability on solid surfaces of periprosthetic infection-associated pathogens. Sci. Rep. 2022, 12, 18669. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Echeverz, M.; Lasa, I. σ(B) Inhibits Poly-N-Acetylglucosamine Exopolysaccharide Synthesis and Biofilm Formation in Staphylococcus aureus. J. Bacteriol. 2019, 201, 10-1128. [Google Scholar] [CrossRef]
- Ma, S.; Liu, J.; Feng, L.; Wang, C.; Ren, J.; Yang, C.; Dai, Y.; Cui, G.; Zhang, K.; Li, C. Study on the antimicrobial effects of aqueous extracts from Portulaca oleracea L. J. Pure Appl. Microbiol. 2013, 7, 2767–2772. [Google Scholar]
- Yuyama, K.T.; Rohde, M.; Molinari, G.; Stadler, M.; Abraham, W.-R. Unsaturated fatty acids control biofilm formation of Staphylococcus aureus and other gram-positive bacteria. Antibiotics 2020, 9, 788. [Google Scholar] [CrossRef]
- Kim, H.-S.; Ham, S.-Y.; Jang, Y.; Sun, P.-F.; Park, J.-H.; Hoon Lee, J.; Park, H.-D. Linoleic acid, a plant fatty acid, controls membrane biofouling via inhibition of biofilm formation. Fuel 2019, 253, 754–761. [Google Scholar] [CrossRef]
- Samrot, A.V.; Abubakar Mohamed, A.; Faradjeva, E.; Si Jie, L.; Hooi Sze, C.; Arif, A.; Chuan Sean, T.; Norbert Michael, E.; Yeok Mun, C.; Xiao Qi, N. Mechanisms and impact of biofilms and targeting of biofilms using bioactive compounds—A review. Medicina 2021, 57, 839. [Google Scholar] [CrossRef]
- Mombeshora, M.; Chi, G.F.; Mukanganyama, S. Antibiofilm Activity of Extract and a Compound Isolated from Triumfetta welwitschii against Pseudomonas aeruginosa. Biochem. Res. Int. 2021, 2021, 9946183. [Google Scholar] [CrossRef]
- Okshevsky, M.; Regina, V.R.; Meyer, R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, S.; Sato, F.; Miyakawa, R.; Chiba, A.; Onodera, S.; Hori, S.; Mizunoe, Y. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and-sensitive strains of Staphylococcus aureus. Sci. Rep. 2018, 8, 2254. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Wang, P.; Li, T.; Wang, Q.; Su, M.; Gu, L.; Wang, S. Antimicrobial and antibiofilm effects of essential fatty acids against clinically isolated vancomycin-resistant Enterococcus faecium. Front. Cell. Infect. Microbiol. 2023, 13, 1266674. [Google Scholar] [CrossRef]
- Shah, S.; Zubair, M. Membrane Disruption and Biofilm Inhibition by Fatty Acids in Pseudomonas aeruginosa. J. Microbiol. 2019, 54, 423–431. [Google Scholar]
- Knap, K.; Kwiecień, K.; Ochońska, D.; Reczyńska-Kolman, K.; Pamuła, E.; Brzychczy-Włoch, M. Synergistic effect of antibiotics, α-linolenic acid and solvent type against Staphylococcus aureus biofilm formation. Pharmacol. Rep. 2024, 76, 1456–1469. [Google Scholar] [CrossRef]
- Al-Sheddi, E.S.; Farshori, N.N.; Al-Oqail, M.M.; Musarrat, J.; Al-Khedhairy, A.A.; Siddiqui, M.A. Portulaca oleracea Seed Oil Exerts Cytotoxic Effects on Human Liver Cancer (HepG2) and Human Lung Cancer (A-549) Cell Lines. Asian Pac. J. Cancer Prev. 2015, 16, 3383–3387. [Google Scholar] [CrossRef]
- Asif, M. Health effects of omega-3,6,9 fatty acids: Perilla frutescens is a good example of plant oils. Orient. Pharm. Exp. Med. 2011, 11, 51–59. [Google Scholar] [CrossRef]
- Petropoulos, S.A.; Fernandes, Â.; Dias, M.I.; Vasilakoglou, I.B.; Petrotos, K.; Barros, L.; Ferreira, I.C.F.R. Nutritional Value, Chemical Composition and Cytotoxic Properties of Common Purslane (Portulaca oleracea L.) in Relation to Harvesting Stage and Plant Part. Antioxidants 2019, 8, 293. [Google Scholar] [CrossRef]
- Scheim, D.E. Cytotoxicity of unsaturated fatty acids in fresh human tumor explants: Concentration thresholds and implications for clinical efficacy. Lipids Health Dis. 2009, 8, 54. [Google Scholar] [CrossRef]
- Banfi, C.; Risé, P.; Mussoni, L.; Galli, C.; Tremoli, E. Linoleic acid enhances the secretion of plasminogen activator inhibitor type 1 by HepG2 cells. J. Lipid Res. 1997, 38, 860–869. [Google Scholar] [CrossRef]
- Nuzzo, I.; Sanges, M.; Folgore, A.; Carratelli, C.R. Apoptosis of human keratinocytes after bacterial invasion. FEMS Immunol. Med. Microbiol. 2000, 27, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Kenny, J.G.; Ward, D.; Josefsson, E.; Jonsson, I.-M.; Hinds, J.; Rees, H.H.; Lindsay, J.A.; Tarkowski, A.; Horsburgh, M.J. The Staphylococcus aureus Response to Unsaturated Long Chain Free Fatty Acids: Survival Mechanisms and Virulence Implications. PLoS ONE 2009, 4, e4344. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Barros, L.; Carvalho, A.M.; Ferreira, I.C. Use of UFLC-PDA for the analysis of organic acids in thirty-five species of food and medicinal plants. Food Anal. Methods 2013, 6, 1337–1344. [Google Scholar] [CrossRef]
- Soković, M.; Glamočlija, J.; Marin, P.D.; Brkić, D.; Van Griensven, L.J. Antibacterial effects of the essential oils of commonly consumed medicinal herbs using an in vitro model. Molecules 2010, 15, 7532–7546. [Google Scholar] [CrossRef] [PubMed]
- Soković, M.; Van Griensven, L.J. Antimicrobial activity of essential oils and their components against the three major pathogens of the cultivated button mushroom, Agaricus bisporus. Eur. J. Plant Pathol. 2006, 116, 211–224. [Google Scholar] [CrossRef]
- Smiljković, M.; Dias, M.I.; Stojković, D.; Barros, L.; Bukvički, D.; Ferreira, I.C.; Soković, M. Characterization of phenolic compounds in tincture of edible Nepeta nuda: Development of antimicrobial mouthwash. Food Funct. 2018, 9, 5417–5425. [Google Scholar] [CrossRef]
- Ivanov, M.; Kostić, M.; Stojković, D.; Soković, M. Rosmarinic acid–modes of antimicrobial and antibiofilm activities of a common plant polyphenol. S. Afr. J. Bot. 2022, 146, 521–527. [Google Scholar] [CrossRef]
- Carević, T.; Kolarević, S.; Kolarević, M.K.; Nestorović, N.; Novović, K.; Nikolić, B.; Ivanov, M. Citrus flavonoids diosmin, myricetin and neohesperidin as inhibitors of Pseudomonas aeruginosa: Evidence from antibiofilm, gene expression and in vivo analysis. Biomed. Pharmacother. 2024, 181, 117642. [Google Scholar] [CrossRef]
- Stojković, D.; Gašić, U.; Drakulić, D.; Zengin, G.; Stevanović, M.; Rajčević, N.; Soković, M. Chemical profiling, antimicrobial, anti-enzymatic, and cytotoxic properties of Phlomis fruticosa L. J. Pharm. Biomed. Anal. 2021, 195, 113884. [Google Scholar] [CrossRef]
- Ahmed, G.F.; Elkhatib, W.F.; Noreddin, A.M. Inhibition of Pseudomonas aeruginosa PAO1 adhesion to and invasion of A549 lung epithelial cells by natural extracts. J. Infect. Public Health 2014, 7, 436–444. [Google Scholar] [CrossRef]
- Deepasree, K.; Subhashree, V. Molecular docking and dynamic simulation studies of terpenoid compounds against phosphatidylinositol-specific phospholipase C from Listeria monocytogenes. Inform. Med. Unlocked 2023, 39, 101252. [Google Scholar] [CrossRef]
- Venugopal, S. Molecular docking and molecular dynamic simulation studies to identify potential terpenes against Internalin A protein of Listeria monocytogenes. Front. Bioinform. 2024, 4, 1463750. [Google Scholar]
- Yu, J.; Zhou, Y.; Tanaka, I.; Yao, M. Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 2010, 26, 46–52. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiao, P.; Li, S.; Feng, X.; Bian, L. Molecular recognition and binding of beta-lactamase II from Bacillus cereus with penicillin V and sulbactam by spectroscopic analysis in combination with docking simulation. Luminescence 2017, 32, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Baburam, S.; Ramasamy, S.; Shanmugam, G.; Mathanmohun, M. Quorum sensing inhibitory potential and molecular docking studies of Phyllanthus emblica phytochemicals against Pseudomonas aeruginosa. Appl. Biochem. Biotechnol. 2022, 194, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Ramaiah, S. Molecular docking and molecular dynamics studies to identify potential OXA-10 extended spectrum β-lactamase non-hydrolysing inhibitors for Pseudomonas aeruginosa. Cell Biochem. Biophys. 2016, 74, 141–155. [Google Scholar] [CrossRef]
- Zuo, K.; Liang, L.; Du, W.; Sun, X.; Liu, W.; Gou, X.; Wan, H.; Hu, J. 3D-QSAR, molecular docking and molecular dynamics simulation of Pseudomonas aeruginosa LpxC inhibitors. Int. J. Mol. Sci. 2017, 18, 761. [Google Scholar] [CrossRef]
- Hetmann, M.; Langner, C.; Durmaz, V.; Cespugli, M.; Köchl, K.; Krassnigg, A.; Blaschitz, K.; Groiss, S.; Loibner, M.; Ruau, D. Identification and validation of fusidic acid and flufenamic acid as inhibitors of SARS-CoV-2 replication using DrugSolver CavitomiX. Sci. Rep. 2023, 13, 11783. [Google Scholar] [CrossRef]
- Saqallah, F.G.; Hamed, W.M.; Talib, W.H.; Dianita, R.; Wahab, H.A. Antimicrobial activity and molecular docking screening of bioactive components of Antirrhinum majus (snapdragon) aerial parts. Heliyon 2022, 8, e10391. [Google Scholar] [CrossRef]
- Sun, R.; Zhu, J.; Sun, K.; Gao, L.; Zheng, B.; Shi, J. Strontium Ranelate Ameliorates Intervertebral Disc Degeneration via Regulating TGF-β1/NF-κB Axis. Int. J. Med. Sci. 2023, 20, 1679. [Google Scholar] [CrossRef]
- Cetiz, M.V.; Isah, M.; Ak, G.; Bakar, K.; Himidi, A.A.; Mohamed, A.; Glamočlija, J.; Nikolić, F.; Gašic, U.; Cespedes-Acuna, C.L.; et al. Exploring of Chemical Profile and Biological Activities of Three Ocimum Species from Comoros Islands: A Combination of In Vitro and In Silico Insights. Cell Biochem. Funct. 2024, 42, e70000. [Google Scholar] [CrossRef] [PubMed]
- Cusumano, G.; Flores, G.A.; Cetiz, M.V.; Kurt, U.; Ak, G.; Saka, E.; Aly, S.H.; Eldahshan, O.A.; Singab, A.N.; Zengin, G.; et al. Small Steps to the Big Picture for Health-Promoting Applications Through the Use of Chickweed (Stellaria media): In Vitro, In Silico, and Pharmacological Network Approaches. Food Sci. Nutr. 2024, 12, 9295–9313. [Google Scholar] [CrossRef] [PubMed]
- Duran, T.; Peron, G.; Zancato, M.; Zengin, G.; Cetiz, M.V.; Bouyahya, A.; Ahmed, S.; Yildiztugay, E.; Dall’Acqua, S.; Kljakić, A.C.; et al. Harnessing the chemical composition and anti-oxidant, anti-enzymatic, and anti-cancer activities of two Corydalis species (C. erdelii and C. solida) by using in vitro and in silico analysis. Food Biosci. 2024, 61, 104762. [Google Scholar] [CrossRef]
- Kurt-Celep, I.; Nilofar; Cetiz, M.V.; Zheleva-Dimitrova, D.; Gevrenova, R.; Celep, E.; Sinan, K.I.; Yildiztugay, E.; Ferrante, C.; Zengin, G. From small-scale studies to an encompassing view: Inhibiting inflammation and clinically relevant enzymes with various extracts of Primula vulgaris using in vitro and in silico techniques. Food Front. 2025, 6, 329–359. [Google Scholar] [CrossRef]
- Yagi, S.; Zengin, G.; Eldahshan, O.A.; Singab, A.N.B.; Selvi, S.; Cetiz, M.V.; Rodrigues, M.J.; Custodio, L.; Dall’Acqua, S.; Elhawary, E.A. Functional constituents of Colchicum lingulatum Boiss. & Spruner subsp. Rigescens K. Perss. Extracts and their biological activities with different perspectives. Food Biosci. 2024, 60, 104496. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Miller, B.R., III; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA. py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Karthick, K.; Abishek, K.; Angel Jemima, E. In Silico Study, Protein Kinase Inhibition and Molecular Docking Study of Benzimidazole Derivatives. Bioinform. Biol. Insights 2024, 18, 11779322241247635. [Google Scholar] [CrossRef] [PubMed]
| Content (%) | ||
|---|---|---|
| Sample from Serbia | Sample from Greece | |
| C6:0 | 0.17 ± 0.00 | 0.21 ± 0.00 |
| C8:0 | 0.25 ± 0.00 | 0.23 ± 0.00 |
| C10:0 | 5.43 ± 0.00 | 6.21 ± 0.00 |
| C12:0 | 0.01 ± 0.00 | 0.04 ± 0.00 |
| C13:0 | 0.08 ± 0.00 | 0.06 ± 0.00 |
| C14:0 | 0.22 ± 0.01 | 0.21 ± 0.01 |
| C15:0 | 0.18 ± 0.00 | 0.16 ± 0.00 |
| C16:0 | 13.87 ± 0.23 | 14.07 ± 0.07 |
| C16:1 | 6.89 ± 0.01 | 0.64 ± 0.01 |
| C17:0 | 0.37 ± 0.01 | 0.37 ± 0.03 |
| C18:0 | 4.41 ± 0.02 | 7.43 ± 0.03 |
| C18:1n9 | 9.72 ± 0.04 | 10.50 ± 0.04 |
| C18:2n6 | 31.42 ±0.07 | 34.52 ± 0.08 |
| C18:3n3 | 23.34 ± 0.02 | 23.22 ± 0.01 |
| C20:0 | 0.22 ± 0.00 | 0.12 ± 0.00 |
| C20:1 | 0.38 ± 0.01 | 0.33 ± 0.01 |
| C20:2 | 0.14 ± 0.00 | 0.12 ± 0.00 |
| C20:3n3 + C21:0 | 0.36 ± 0.00 | 0.03 ± 0.00 |
| C20:5n3 | 0.25 ± 0.01 | 0.01 ± 0.01 |
| C22:0 | 1.01 ± 0.03 | 1.02 ± 0.03 |
| C22:1n9 | 0.11 ± 0.00 | 0.11 ± 0.00 |
| C23:0 | 0.19 ± 0.03 | 0.10 ± 0.03 |
| C24:0 | 0.89 ± 0.09 | 0.21 ± 0.03 |
| C24:1 | 0.09 ± 0.03 | 0.08 ± 0.03 |
| Total SFA (% of total FA) | 27.30 ± 0.07 | 30.44 ± 0.08 |
| Total MUFA (% of total FA) | 17.19 ± 0.01 | 11.66 ± 0.01 |
| Total PUFA (% of total FA) | 55.51 ± 0.07 | 57.90± 0.07 |
| S.a. | B.c. | L.m. | M.f. | P.a. | E.c. | S.t. | En.cl. | ||
|---|---|---|---|---|---|---|---|---|---|
| Greece | MIC | 0.59 | 1.18 | 4.74 | 2.38 | 0.59 | 2.38 | 2.38 | 1.18 |
| MBC | 1.18 | 2.38 | 9.49 | 4.74 | 1.18 | 4.74 | 4.74 | 2.38 | |
| Serbia | MIC | 1.18 | 2.38 | 9.49 | 2.38 | 1.18 | 4.74 | 4.74 | 1.18 |
| MBC | 1.18 | 4.74 | 9.49 | 4.74 | 1.18 | 4.74 | 4.74 | 2.38 | |
| Streptomycin | MIC | 0.25 | 0.05 | 0.15 | 0.125 | 0.05 | 0.05 | 0.05 | 0.05 |
| MBC | 0.50 | 0.10 | 0.30 | 0.25 | 0.10 | 0.10 | 0.10 | 0.10 | |
| Ampicillin | MIC | 0.10 | 0.10 | 0.15 | 0.10 | 0.10 | 0.30 | 0.15 | 0.15 |
| MBC | 0.15 | 0.15 | 0.30 | 0.15 | 0.20 | 0.50 | 0.20 | 0.20 | |
| A.f. | A.v. | A.o. | A.n. | T.v. | P.f. | P.o. | P.c. | ||
| Greece | MIC | 2.38 | 0.025 | 1.19 | 2.38 | 2.38 | 2.38 | 1.19 | 2.38 |
| MFC | 18.96 | 0.95 | 4.74 | 9.49 | 2.38 | 4.74 | 4.74 | 4.47 | |
| Serbia | MIC | 9.49 | 0.95 | 2.38 | 4.74 | 2.38 | 2.38 | 1.19 | 2.38 |
| MBC | 18.96 | 1.19 | 4.74 | 9.49 | 4.74 | 4.74 | 4.74 | 9.49 | |
| Ketoconazole | MIC | 0.20 | 0.20 | 0.15 | 0.20 | 0.20 | 2.50 | 0.20 | 0.25 |
| MFC | 0.50 | 0.50 | 0.20 | 0.50 | 0.30 | 3.50 | 0.50 | 0.50 | |
| Bifonazole | MIC | 0.15 | 0.10 | 0.15 | 0.15 | 0.10 | 0.20 | 0.20 | 0.25 |
| MFC | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.25 | 0.25 | 0.50 |
| Sample | MIC | ½ MIC | ¼ MIC |
|---|---|---|---|
| Greece | 87.91 ± 4.38 | 75.42 ± 5.58 | 41.22 ± 6.44 |
| Serbia | 89.54 ± 7.66 | 81.21 ± 9.11 | 51.47 ± 2.99 |
| Sample | ½ MIC |
|---|---|
| Greece | 56.11 ± 2.93 |
| Serbia | 41.71 ± 7.17 |
| Sample | ½ MIC |
|---|---|
| Greece | 76.61 ± 4.24 |
| Serbia | 62.33 ± 5.65 |
| Cell Lines | Greece IC50 (µg/mL) | Serbia IC50 (µg/mL) | Ellipticine IC50 (µg/mL) |
|---|---|---|---|
| HaCaT | >400 | >400 | >400 |
| MCF7 | 397.31 ± 26.41 | 374.33 ± 18.39 | 1.21 ± 0.01 |
| HeLa | 354.79 ± 20.57 | 361.96 ± 18.36 | 1.02 ± 0.02 |
| CaCo2 | 198.44 ± 11.15 | 178.17 ± 9.33 | 1.42 ± 0.02 |
| HepG2 | 292.37 ± 21.33 | 272.36 ± 17.29 | 1.05 ± 0.01 |
| Compound | Protein Name | PDB ID | Binding Energy | RMSD | Interaction | Binding Site | |
|---|---|---|---|---|---|---|---|
| Type | Number | ||||||
| Linolenic acid | TubR | 6AHT | −5 | nd | Hbond | 2 | ALA A:70; THR A:75 |
| Linolenic acid | Transpeptidase | 5TW8 | −6 | nd | Hbond | 2 | GLY A:261; SER A:262 |
| Linoleic acid | Transpeptidase | 5TW8 | −5 | 13.4 | Hbond | 3 | ASN A:72; GLY A:181; SER A:263 |
| Linolenic acid | PrfA | 6EXL | −7 | 8.1 | Hbond | 2 | TYR A:126; TYR A:126 |
| Palmitic acid | PrfA | 6EXL | −7 | 1.7 | Hbond | 2 | TYR A:126; GLN A:146 |
| Oleic acid | PrfA | 6EXL | −7 | 5.3 | Hbond | 2 | LYS A:64; LYS A:64 |
| Palmitoleic acid | PrfA | 6EXL | −7 | 5.6 | Hbond | 1 | LYS A:64 |
| Linoleic acid | PrfA | 6EXL | −7 | 1.9 | Hbond | 1 | TYR A:126 |
| Linolenic acid | PI3K | 4XE0 | −7 | 7.1 | Hbond | 3 | GLN A:610; LEU A:613; GLN A:792 |
| Palmitic acid | PI3K | 4XE0 | −6 | 8.3 | Hbond | 3 | LEU A:612; LEU A:613; GLN A:792 |
| Oleic acid | PI3K | 4XE0 | −6 | 8.6 | Hbond | 1 | PRO A:812 |
| Linoleic acid | PI3K | 4XE0 | −6 | 6.6 | Hbond | 1 | CYS A:815 |
| Oleic acid | MurE | 4C13 | −5 | 10.0 | Hbond | 2 | TYR A:351; TYR A:351 |
| Linoleic acid | MurE | 4C13 | −5 | nd | Hbond | 5 | GLY A:113; LYS A:114; THR A:115; THR A:115; THR A:137 |
| Linolenic acid | MurE | 1E8C | −5 | nd | Hbond | 1 | GLY B:304 |
| Linolenic acid | MDM2 | 4WT2 | −6 | 1.1 | Hbond | 1 | THR A:16 |
| Palmitic acid | MDM2 | 4WT2 | −5 | 6.3 | Hbond | 1 | VAL A:14 |
| Oleic acid | MDM2 | 4WT2 | −6 | 9.4 | Hbond | 1 | TYR A:100 |
| Palmitoleic acid | MDM2 | 4WT2 | −6 | 7.1 | Hbond | 1 | VAL A:93 |
| Linoleic acid | MDM2 | 4WT2 | −7 | 8.6 | Hbond | 1 | GLN A:59 |
| Oleic acid | LasR | 2UV0 | −8 | 1.7 | Hbond | 2 | TYR E:56; TYR E:93 |
| Linolenic acid | InlA | 1O6T | −6 | 41.9 | Hbond | 4 | SER A:429; SER A:429; SER A:429; SER A:429 |
| Oleic acid | InlA | 1O6T | −5 | nd | Hbond | 5 | ASP A:265; ILE A:266; ASN A:287; ASN A:287; ASN A:287 |
| Linoleic acid | InlA | 1O6T | −5 | 29.6 | Hbond | 3 | GLU A:326; GLN A:328; GLN A:328 |
| Linolenic acid | EGFR | 1M17 | −5 | 8.9 | Hbond | 2 | MET A:769; MET A:769 |
| Oleic acid | EGFR | 1M17 | −5 | nd | Hbond | 2 | GLU A:738; THR A:766 |
| Palmitoleic acid | EGFR | 1M17 | −5 | 1.9 | Hbond | 4 | GLU A:738; ASP A:831; ASP A:831; ASP A:831 |
| Linoleic acid | EGFR | 1M17 | −5 | 1.6 | Hbond | 1 | THR A:766 |
| Palmitoleic acid | DNA Gyrase B | 1KZN | −5 | nd | Hbond | 2 | THR A:165; VAL A:167 |
| Oleic acid | Cyclin D1 | 2W99 | −5 | 1.9 | Hbond | 1 | GLN A:183 |
| Linolenic acid | CDKN1A | 5E0U | −5 | nd | Hbond | 2 | GLN A:131; TYR A:133 |
| Palmitic acid | CDKN1A | 5E0U | −5 | nd | Hbond | 1 | HIS A:44 |
| Oleic acid | CDKN1A | 5E0U | −5 | nd | Hbond | 2 | GLN A:131; TYR A:133 |
| Linolenic acid | CDK2 | 6GUE | −6 | 3.2 | Hbond | 1 | VAL A:163 |
| Palmitic acid | CDK2 | 6GUE | −6 | 3.6 | Hbond | 1 | VAL A:163 |
| Oleic acid | CDK2 | 6GUE | −6 | 3.2 | Hbond | 1 | TYR A:180 |
| Palmitoleic acid | CDK2 | 6GUE | −6 | 7.8 | Hbond | 1 | TYR A:180 |
| Linoleic acid | CDK2 | 6GUE | −6 | 2.4 | Hbond | 1 | VAL A:163 |
| Linolenic acid | AKT-1 | 4VG1 | −5 | 1.7 | Hbond | 2 | PHE A:293; GLY A:294 |
| Linoleic acid | AKT-1 | 4VG1 | −6 | 6.0 | Hbond | 1 | GLU A:191 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stojković, D.; Živković, J.; Bolevich, S.; Zengin, G.; Cetiz, M.V.; Bolevich, S.; Soković, M. Lipophilic Extracts of Portulaca oleracea L.: Analysis of Bioactive Fatty Acids Targeting Microbial and Cancer Pathways. Pharmaceuticals 2025, 18, 587. https://doi.org/10.3390/ph18040587
Stojković D, Živković J, Bolevich S, Zengin G, Cetiz MV, Bolevich S, Soković M. Lipophilic Extracts of Portulaca oleracea L.: Analysis of Bioactive Fatty Acids Targeting Microbial and Cancer Pathways. Pharmaceuticals. 2025; 18(4):587. https://doi.org/10.3390/ph18040587
Chicago/Turabian StyleStojković, Dejan, Jelena Živković, Stefani Bolevich, Gokhan Zengin, Mehmet Veysi Cetiz, Sergey Bolevich, and Marina Soković. 2025. "Lipophilic Extracts of Portulaca oleracea L.: Analysis of Bioactive Fatty Acids Targeting Microbial and Cancer Pathways" Pharmaceuticals 18, no. 4: 587. https://doi.org/10.3390/ph18040587
APA StyleStojković, D., Živković, J., Bolevich, S., Zengin, G., Cetiz, M. V., Bolevich, S., & Soković, M. (2025). Lipophilic Extracts of Portulaca oleracea L.: Analysis of Bioactive Fatty Acids Targeting Microbial and Cancer Pathways. Pharmaceuticals, 18(4), 587. https://doi.org/10.3390/ph18040587










