GmLecRlk, a Lectin Receptor-like Protein Kinase, Contributes to Salt Stress Tolerance by Regulating Salt-Responsive Genes in Soybean
Abstract
:1. Introduction
2. Results
2.1. Expression Pattern of GmLecRlk
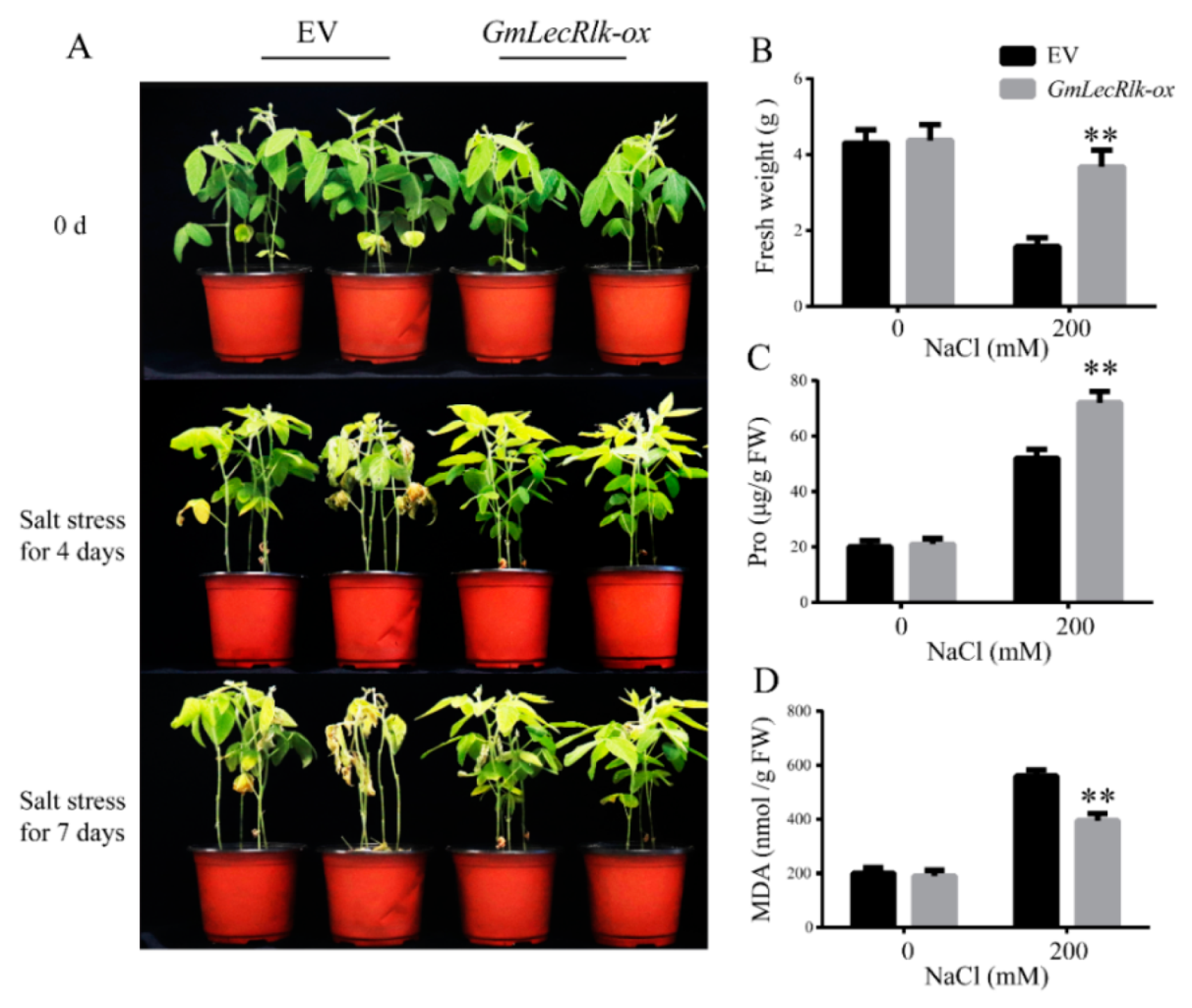
2.2. GmLecRlk Improves Salt Tolerance in Soybean
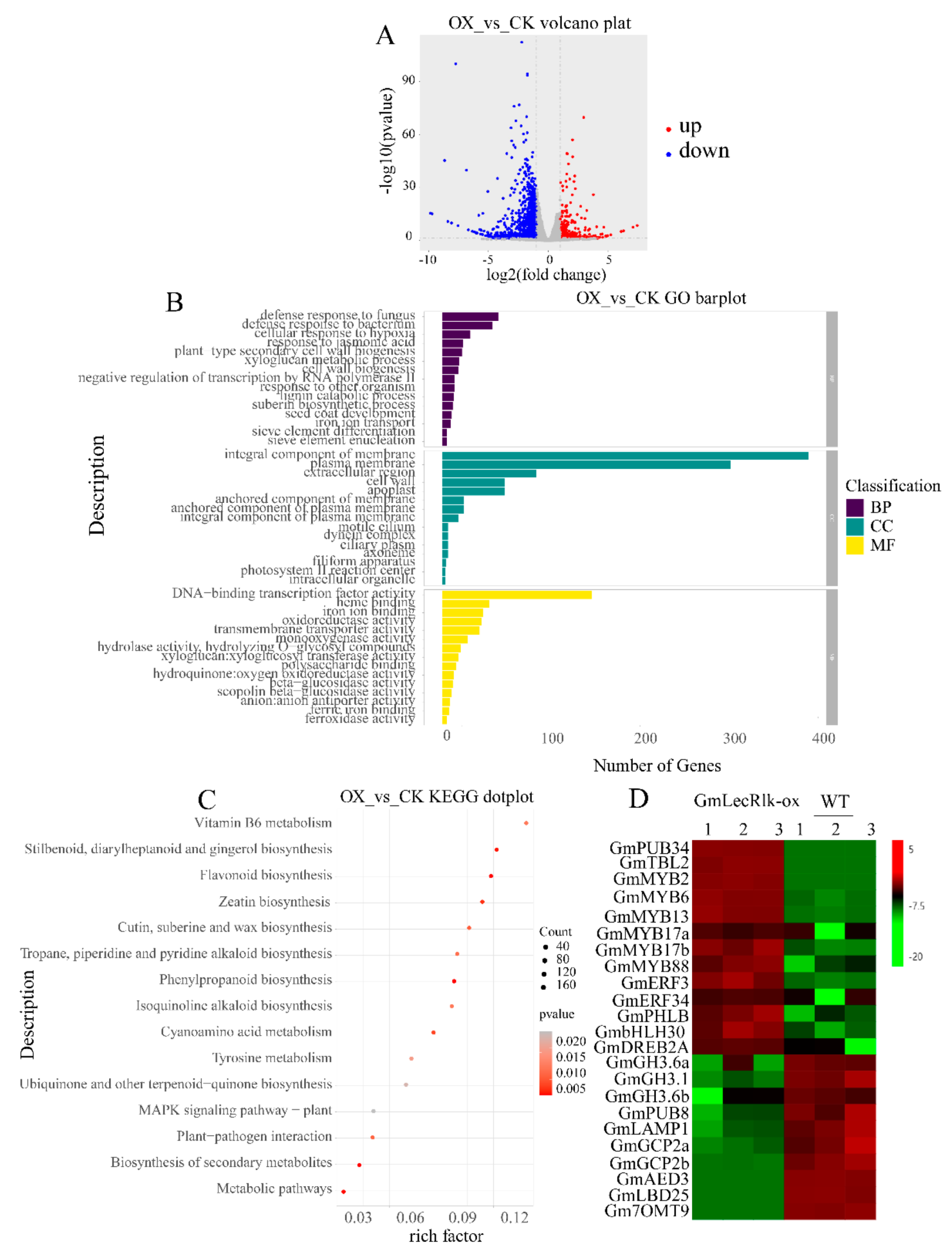
2.3. Transcriptome Sequencing Analysis
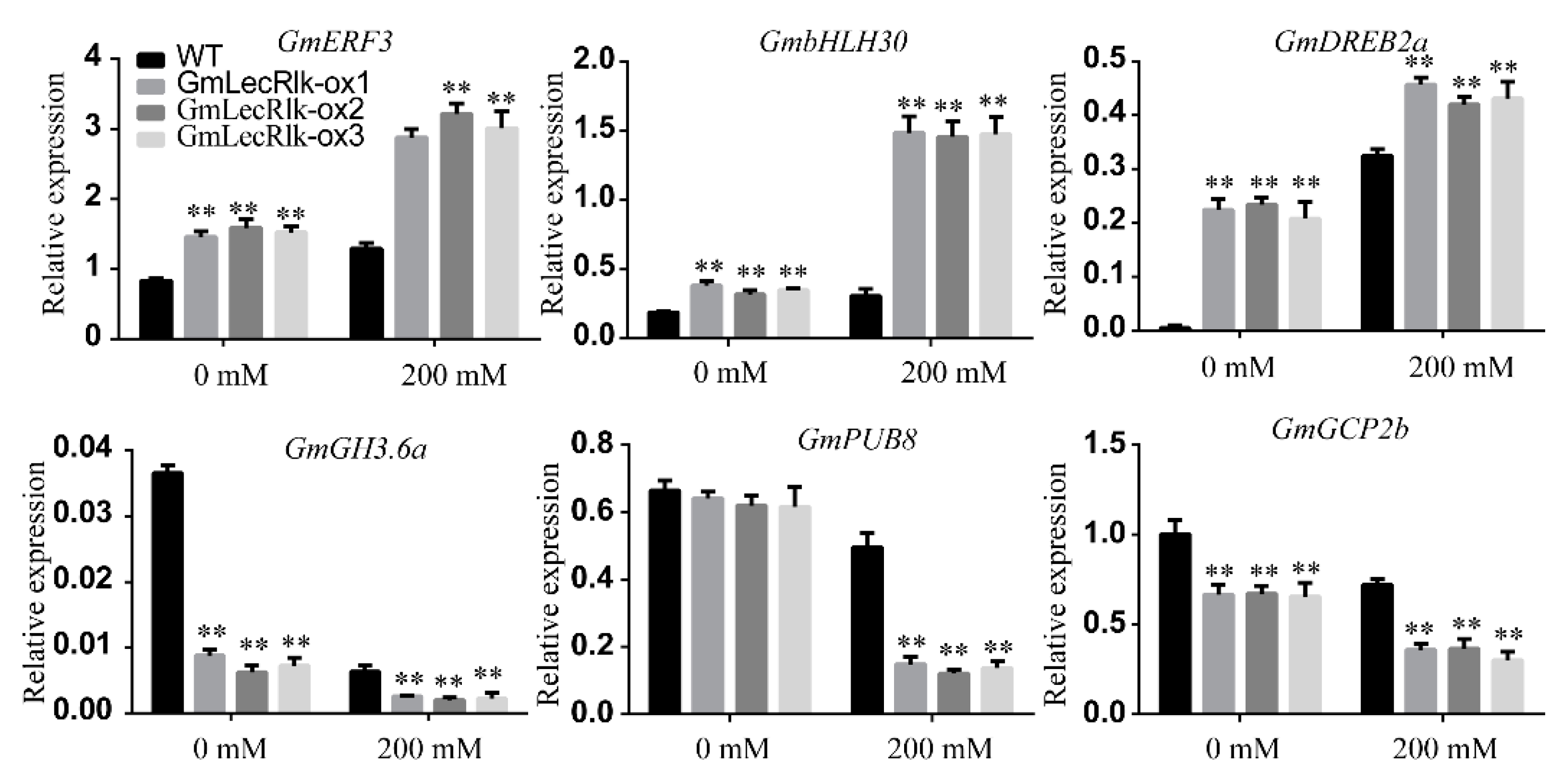
2.4. GmLecRlk Improves Salt Tolerance by Positively Regulating Salt Stress Response Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and NaCl Stress Treatment
4.2. Plasmid Construction and Plant Transformation
4.3. Chlorophyll Fluorescence Measurement
4.4. Measurements of Proline, Malondialdehyde, and H2O2 Contents
4.5. Quantitative PCR Analysis
4.6. Transcriptomic Analysis
4.7. Statistics of Agronomic Characters
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, G.J.; Boerma, H.R.; Villagarcia, M.R.; Zhou, X.; Carter, T.E.; Li, Z.; Gibbs, M.O. A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Appl. Genet. 2004, 109, 1610–1619. [Google Scholar] [CrossRef] [PubMed]
- Kersten, B.; Agrawal, G.K.; Iwahashi, H.; Rakwal, R. Plant phosphoproteomics: A long road ahead. Proteomics 2006, 6, 5517–5528. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, J.B.; Nasrallah, M.E. Pollen[mdash]Stigma Signaling in the Sporophytic Self-Incompatibility Response. Plant Cell Online 1993, 5, 1325–1335. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Becraft, P.W.; Stinard, P.S.; McCarty, D.R. CRINKLY4: A TNFR-Like Receptor Kinase Involved in Maize Epidermal Differentiation. Science 1996, 273, 1406–1409. [Google Scholar] [CrossRef] [PubMed]
- Hervé, C.; Dabos, P.; Galaud, J.-P.; Rougé, P.; Lescure, B. Characterization of an Arabidopsis thaliana gene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. J. Mol. Biol. 1996, 258, 778–788. [Google Scholar] [CrossRef]
- Wang, G.-L.; Ruan, D.-L.; Song, W.-Y.; Sideris, S.; Chen, L.; Pi, L.-Y.; Zhang, S.; Zhang, Z.; Fauquet, C.; Gaut, B.S.; et al. Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell Online 1998, 10, 765–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lease, K.; Ingham, E.; Walker, J.C. Challenges in understanding RLK function. Curr. Opin. Plant Biol. 1998, 1, 388–392. [Google Scholar] [CrossRef]
- Barre, A.; Hervé, C.; Lescure, B.; Rougé, P. Lectin Receptor Kinases in Plants. Crit. Rev. Plant Sci. 2002, 21, 379–399. [Google Scholar] [CrossRef]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in Action: Lectin Receptor-Like Kinases in Plant Development and Stress Responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Patel, A.; Mathieu, M.; Kim, S.-Y.; Xu, D.; Stacey, G. A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol. Biol. 2008, 67, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Pandey, P.K.; Tuteja, N. Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol. Biol. 2012, 80, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Tian, Y.; Liu, R.; Wang, H.; Zhang, L.; Yuan, M. Genome-wide Analysis of Lectin-like Receptor Kinases(LecRLKs) Family in Soybean. Mol. Plant Breed. 2019, 17, 3137–3134. [Google Scholar]
- Desclos-Theveniau, M.; Arnaud, D.; Huang, T.-Y.; Lin, G.J.-C.; Chen, W.-Y.; Lin, Y.-C.; Zimmerli, L. The Arabidopsis Lectin Receptor Kinase LecRK-V.5 Represses Stomatal Immunity Induced by Pseudomonas syringae pv. tomato DC3000. PLOS Pathog. 2012, 8, e1002513. [Google Scholar] [CrossRef]
- Joshi, A.; Dang, H.Q.; Vaid, N.; Tuteja, N. Pea lectin receptor-like kinase promotes high salinity stress tolerance in bacteria and expresses in response to stress in planta. Glycoconj. J. 2010, 27, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Wang, G.; Zhao, J.L.; Zhang, L.Q.; Ai, L.F.; Han, Y.F.; Sun, D.Y.; Zhang, S.W.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.-L.; Yu, Q.-Y.; Tang, L.-L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.-F.; Ding, X.-D.; Zhu, Y.-M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef]
- Li, W.; Bo, L.; Nisa, Z.; Li, Y.; Mallano, A.I.; Wang, T. Soybean GmNFYB1 transcription factor confers abiotic stress tolerance in transgenic Arabidopsis. Can. J. Plant Sci. 2016, 97, 501–515. [Google Scholar] [CrossRef]
- Marutani, Y.; Yamauchi, Y.; Kimura, Y.; Mizutani, M.; Sugimoto, Y. Damage to photosystem II due to heat stress without light-driven electron flow: Involvement of enhanced introduction of reducing power into thylakoid membranes. Planta 2012, 236, 753–761. [Google Scholar] [CrossRef]
- Sun, M.; Qian, X.; Chen, C.; Cheng, S.; Jia, B.; Zhu, Y.; Sun, X. Ectopic Expression of GsSRK in Medicago sativa Reveals Its Involvement in Plant Architecture and Salt Stress Responses. Front. Plant Sci. 2018, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Q.; Yu, T.-F.; Sun, G.-Z.; Zheng, J.-C.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Wei, W.-L.; Xu, Z.-S. Genome-Wide Analysis of the Catharanthus roseus RLK1-Like in Soybean and GmCrRLK1L20 Responds to Drought and Salt Stresses. Front. Plant Sci. 2021, 12, 614909. [Google Scholar] [CrossRef]
- Song, Q.; Qian, S.F.; Chen, X.Q.; Chen, L.M.; Li, K.Z. Study on the Function of Transcription Factor GmbHLH30 on Aluminum Tolerance Preliminary in Tampa Black Soybean. Life Sci. Res. 2014, 18, 332–337. [Google Scholar]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; He, J. Arabidopsis AMP1 Negatively Modulates Plant Responses to Salt Stress. Biotechnol. Bull. 2015, 31, 76–82. [Google Scholar]
- Wang, N.; Liu, Y.; Cong, Y.; Wang, T.; Zhong, X.; Yang, S.; Li, Y.; Gai, J. Genome-Wide Identification of Soybean U-Box E3 Ubiquitin Ligases and Roles of GmPUB8 in Negative Regulation of Drought Stress Response in Arabidopsis. Plant Cell Physiol. 2016, 57, 1189–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wang, R.; Jiao, B.; Dai, P.; Li, Y. Indole Acetic Acid-Amido Synthetase GH3-6 Negatively Regulates Response to Drought and Salt in Arabidopsis. Chin. Bull. Bot. 2016, 51, 586–593. [Google Scholar]
- Zhang, W.; Wang, N.; Yang, J.; Guo, H.; Liu, Z.; Zheng, X.; Li, S.; Xiang, F. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2019, 291, 110326. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Dong, Y.; Yang, X.; Guo, D.; Qian, X.; Yan, F.; Wang, Y.; Li, J.; Wang, Q. Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.). Genome 2020, 63, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-J.; Wang, Y.-Y.; Zhang, Y.-X.; Guo, W.; Jiao, Y.-Q.; Zhou, X.-A. Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3958. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Zou, H.-F.; Wang, H.-W.; Zhang, W.-K.; Ma, B.; Zhang, J.-S.; Chen, S.-Y. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008, 18, 1047. [Google Scholar] [CrossRef] [Green Version]
- Bian, S.; Jin, D.; Sun, G.; Shan, B.; Zhou, H.; Wang, J.; Zhai, L.; Li, X. Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene 2020, 753, 144803. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ren, X.; Zhang, F.; Qi, M.; Zhao, H.; Chen, X.; Ye, Y.; Yang, J.; Li, S.; Zhang, Y.; et al. A R2R3-type MYB transcription factor gene from soybean, GmMYB12, is involved in flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis. Plant Biotechnol. Rep. 2019, 13, 219–233. [Google Scholar] [CrossRef]
- Zhao, L.; Li, M.; Xu, C.; Yang, X.; Li, D.; Zhao, X.; Wang, K.; Li, Y.; Zhang, X.; Liu, L.; et al. Natural variation in GmGBP1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J. 2018, 96, 147–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Chen, H.W.; Li, Q.T.; Wei, W.; Li, W.; Zhang, W.K.; Ma, B.; Bi, Y.D.; Lai, Y.C.; Liu, X.L.; et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015, 83, 224–236. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Fang, Q.; Zheng, J.; Li, Z.; Li, Y.; Feng, Y.; Han, Y.; Li, Y. GmLecRlk, a Lectin Receptor-like Protein Kinase, Contributes to Salt Stress Tolerance by Regulating Salt-Responsive Genes in Soybean. Int. J. Mol. Sci. 2022, 23, 1030. https://doi.org/10.3390/ijms23031030
Zhang Y, Fang Q, Zheng J, Li Z, Li Y, Feng Y, Han Y, Li Y. GmLecRlk, a Lectin Receptor-like Protein Kinase, Contributes to Salt Stress Tolerance by Regulating Salt-Responsive Genes in Soybean. International Journal of Molecular Sciences. 2022; 23(3):1030. https://doi.org/10.3390/ijms23031030
Chicago/Turabian StyleZhang, Yanzheng, Qingwei Fang, Jiqiang Zheng, Zeyang Li, Yue Li, Yuan Feng, Yingpeng Han, and Yongguang Li. 2022. "GmLecRlk, a Lectin Receptor-like Protein Kinase, Contributes to Salt Stress Tolerance by Regulating Salt-Responsive Genes in Soybean" International Journal of Molecular Sciences 23, no. 3: 1030. https://doi.org/10.3390/ijms23031030





