Elucidating Drug-Like Compounds and Potential Mechanisms of Corn Silk (Stigma Maydis) against Obesity: A Network Pharmacology Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Extracts Preparation
2.2. GC-MS Analysis Condition
2.3. GC-MS Compounds in CS and Screening of DLCs
2.4. Identification of Target Proteins Associated with Bioactives or Obesity
2.5. PPI Construction of Final Target Proteins and Identification of Rich Factor
2.6. The Construction of STB Network
2.7. Bioactives and Target Proteins Preparation for MDT
2.8. MDT of Bioactives on Target Proteins Related to Two Key Signaling Pathways
3. Results
3.1. Physicochemical Properties of Chemical Compounds from Corn Silk (CS)
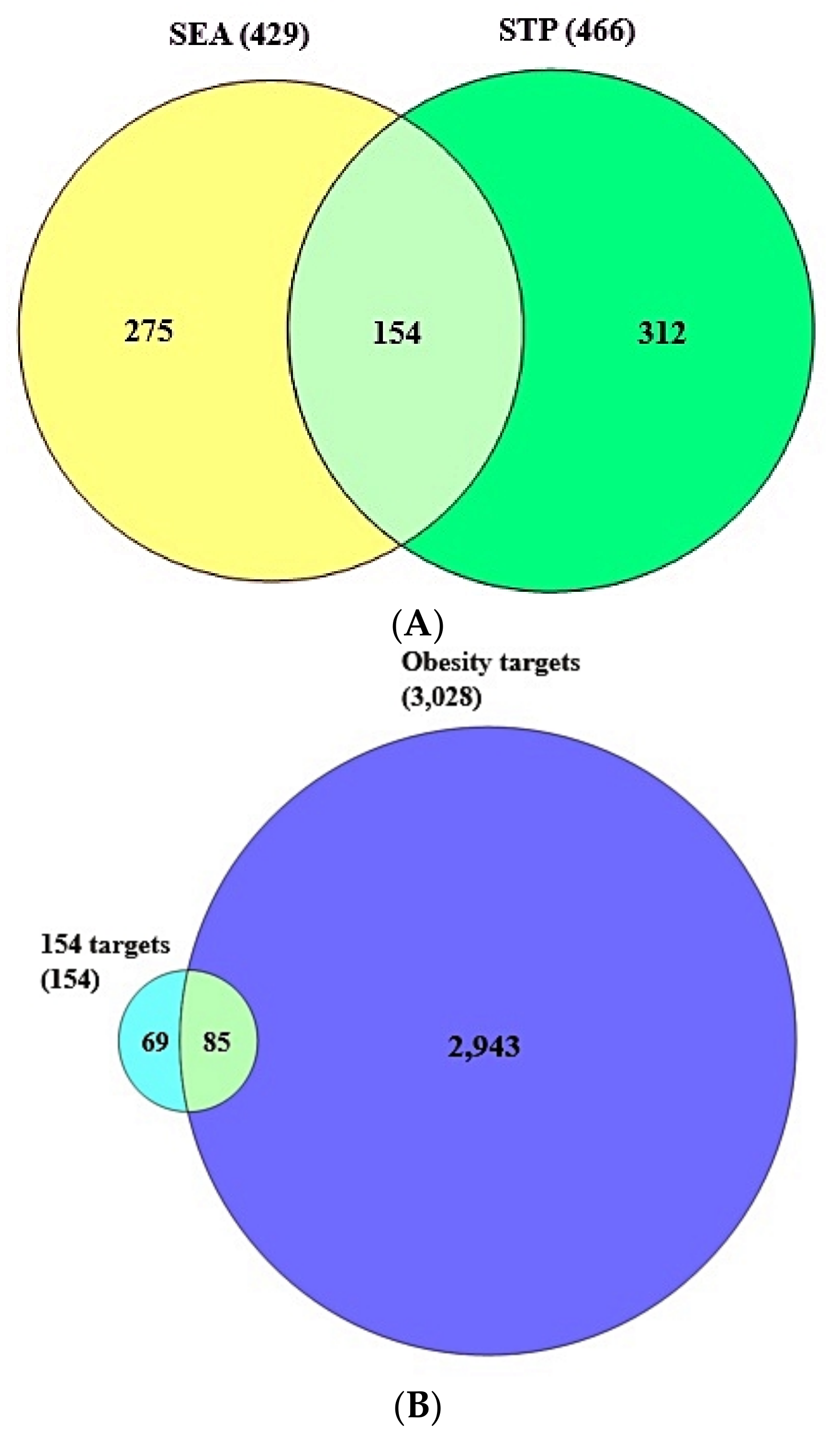
3.2. Identification of Overlapping Target Proteins between SEA and STP Linked to 36 Compounds
3.3. The Final Overlapping Target Proteins between Obesity-Related Target Proteins and the 154 Overlapping Target Proteins
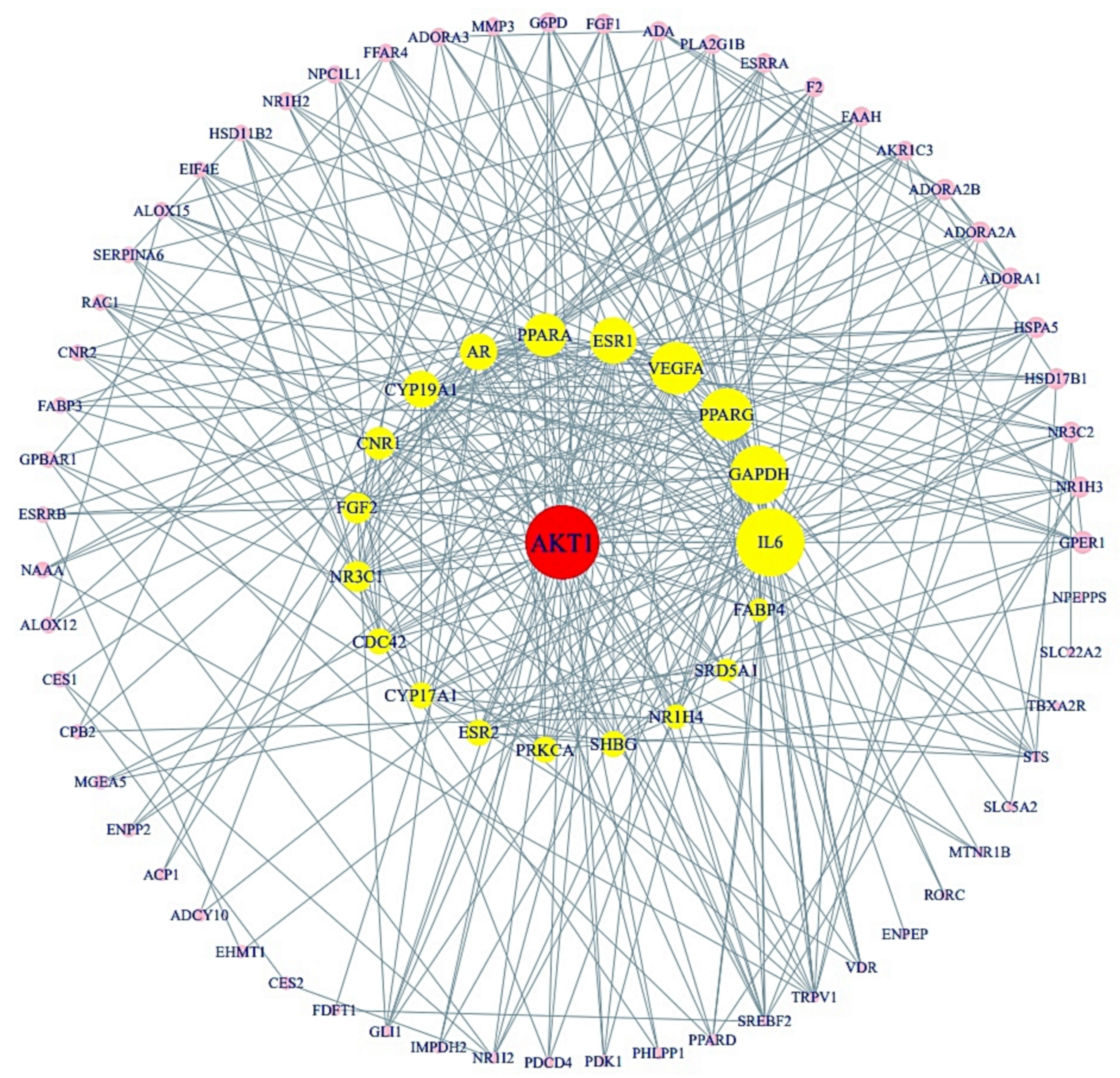
3.4. Protein-Protein Interaction (PPI) from Final 85 Target Proteins
3.5. The 12 Signaling Pathways and Identification of Two Key Pathways of CS against Obesity
3.6. The Construction of a Signaling Pathway-Target Protein-Bioactive (STB) Networks
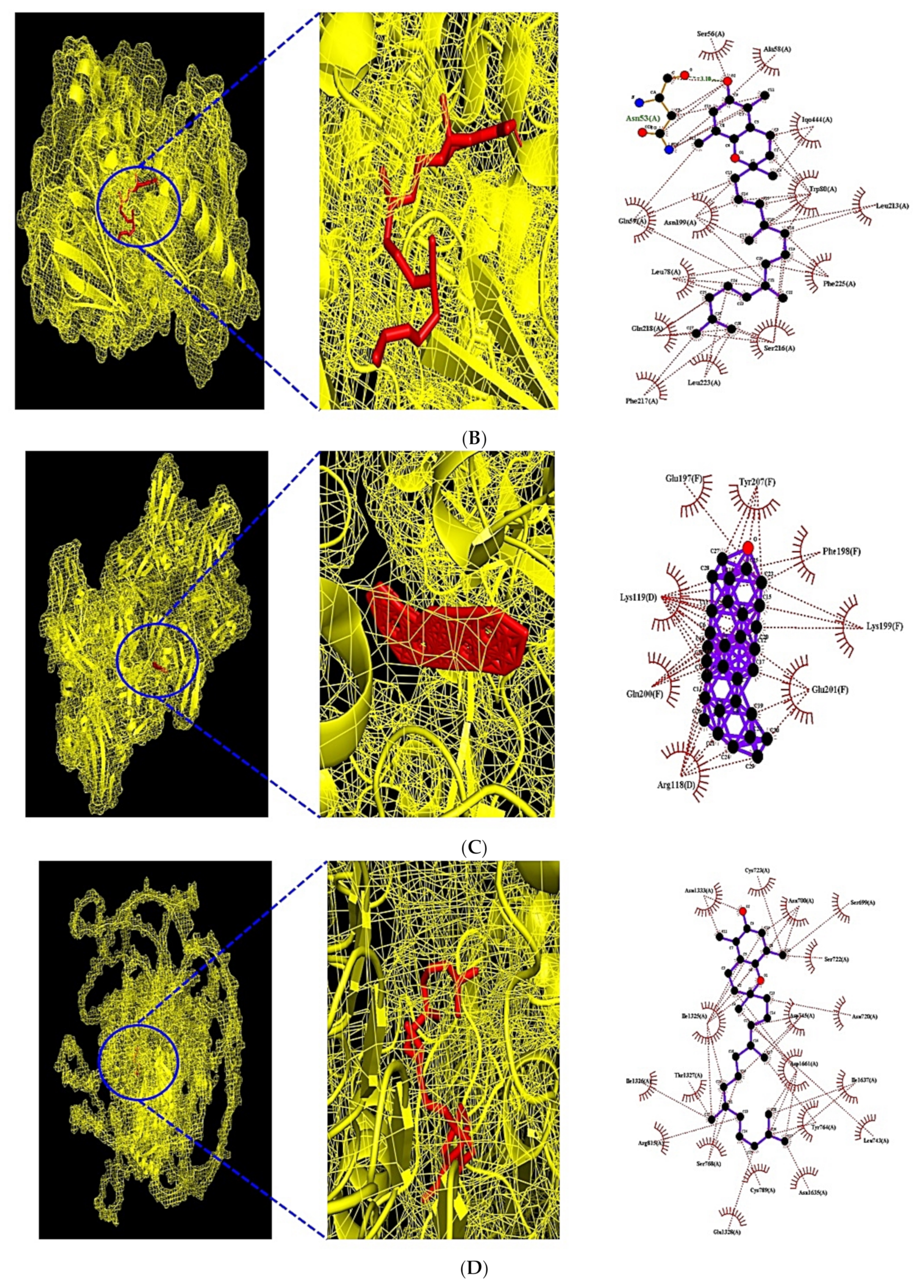
3.7. MDT of 6 Target Proteins, 2 Key Bioactives, and 9 Positive Controls on PPAR Signaling Pathway
3.8. MDT of 7 Target Proteins, 3 Key Bioactives, and 15 Positive Controls on PI3K-Akt1 Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
International Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| cAMP | cyclic Adenosine MonoPhosphate; |
| CS | Corn Silk; |
| DLCs | Drug Like Compounds (DLCs); |
| DM | Diabetes Mellitus; |
| FGF1 | Fibroblast Growth Factor 1; |
| FGF2 | Fibroblast Growth Factor 2; |
| GC-MS | Gas Chromatography Mass Spectrometry; |
| GLP-1 | Glucagon-Like Peptide-1; |
| HIF-1 | Hypoxia Inducible Factor-1; |
| HFWD | High-Fat Western-style fat Diet; |
| MAPK | Mitogen-Activated Protein Kinase; |
| MDT | Molecular Docking Test; |
| PI3K-Akt | Phosphoinositide 3-Kinase–Protein Kinase B; |
| PPAR | Peroxisome Proliferator-Activated Receptor; |
| PPARA | Peroxisome Proliferator-Activated Receptor Alpha; |
| PPARD | Peroxisome Proliferator-Activated Receptor Delta; |
| PPARG | Peroxisome Proliferator-Activated Receptor Gamma; |
| PPI | Protein Protein Interaction (PPI); |
| Rap1 | Repressor activator protein 1; |
| RAS | Renin Angiotensin System; |
| SEA | Similarity Ensemble Approach; |
| STB | Signaling pathways-Targets-Bioactives; |
| STP | SwissTargetPrediction; |
| TPSA | Topological Polar Surface Area; |
| VEGFA | Vascular Endothelial Growth Factor A. |
References
- González Jiménez, E. Obesity: Etiologic and pathophysiological analysis. Endocrinología y Nutrición 2013, 60, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.K.; Adnan, M.; Ju, I.; Cho, D.H. A network pharmacology study on main chemical compounds from Hibiscus cannabinus L. leaves. RSC Adv. 2021, 11, 11062–11082. [Google Scholar] [CrossRef]
- Racette, S.B.; Deusinger, S.S.; Deusinger, R.H. Obesity: Overview of Prevalence, Etiology, and Treatment. Phys. Ther. 2003, 83, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Lancha, L.O.; Campos-Ferraz, P.L.; Lancha, A.H. Junior Obesity: Considerations about etiology, metabolism, and the use of experimental models. Diabetes Metab. Syndr. Obes. Targets Ther. 2012, 5, 75. [Google Scholar] [CrossRef]
- Kuczmarski, R.J.; Flegal, K.M. Criteria for definition of overweight in transition: Background and recommendations for the United States. Am. J. Clin. Nutr. 2000, 72, 1074–1081. [Google Scholar] [CrossRef]
- Widjaja, N.A.; Prihaningtyas, R.A. Determinants of Food Choice in Obesity. Indones. J. Public Health 2020, 15, 122–132. [Google Scholar] [CrossRef]
- Kelly, T.; Yang, W.; Chen, C.S.; Reynolds, K.; He, J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. 2008, 32, 1431–1437. [Google Scholar] [CrossRef]
- Barja-Fernandez, S.; Leis, R.; Casanueva, F.; Seoane, L. Drug development strategies for the treatment of obesity: How to ensure efficacy, safety, and sustainable weight loss. Drug Des. Dev. Ther. 2014, 8, 2391. [Google Scholar] [CrossRef]
- Rodgers, R.J.; Tschöp, M.H.; Wilding, J.P.H. Anti-obesity drugs: Past, present and future. Dis. Models Mech. 2012, 5, 621. [Google Scholar] [CrossRef]
- Heck, D.A.M.; Yanovski, D.J.A.; Calis, D.K.A. Orlistat, a New Lipase Inhibitor for the Management of Obesity. Pharmacotherapy 2000, 20, 270. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cheung, B.M.Y. Pharmacotherapy for obesity. Br. J. Clin.Pharmacol. 2009, 68, 804. [Google Scholar] [CrossRef]
- Derosa, G.; Maffioli, P. Anti-obesity drugs: A review about their effects and their safety. Expert Opin. Drug Saf. 2012, 11, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Onakpoya, I.J.; Posadzki, P.; Eddouks, M. The safety of herbal medicine: From prejudice to evidence. Evid. Based Complementary Altern. Med. 2015, 2015, 316706. [Google Scholar] [CrossRef]
- Watanabe, T. Therapeutic properties of the new phytochemical osmotin for preventing atherosclerosis. Vessel Plus 2020, 4, 4. [Google Scholar] [CrossRef]
- Jo, M.G.; Kim, M.W.; Jo, M.H.; bin Abid, N.; Kim, M.O. Adiponectin homolog osmotin, a potential anti-obesity compound, suppresses abdominal fat accumulation in C57BL/6 mice on high-fat diet and in 3T3-L1 adipocytes. Int. J. Obes. 2019, 43, 2422–2433. [Google Scholar] [CrossRef]
- Shaji, J.; Patole, V. Protein and Peptide Drug Delivery: Oral Approaches. Indian J. Pharm. Sci. 2008, 70, 269. [Google Scholar] [CrossRef]
- Pérez, M.; Pérez, D.I.; Martínez, A.; Castro, A.; Gómez, G.; Fall, Y. The first enantioselective synthesis of palinurin. Chem. Commun. 2009, 22, 3252–3254. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Guo, Y.; Jiang, H.; Shen, X. A sesquiterpene quinone, dysidine, from the sponge Dysidea villosa, activates the insulin pathway through inhibition of PTPases. Acta Pharmacol. Sin. 2009, 30, 333–345. [Google Scholar] [CrossRef]
- Noinart, J.; Buttachon, S.; Dethoup, T.; Gales, L.; Pereira, J.A.; Urbatzka, R.; Freitas, S.; Lee, M.; Silva, A.M.S.; Pinto, M.M.M.; et al. A New Ergosterol Analog, a New Bis-Anthraquinone and Anti-Obesity Activity of Anthraquinones from the Marine Sponge-Associated Fungus Talaromyces stipitatus KUFA 0207. Mar. Drugs 2017, 15, 139. [Google Scholar] [CrossRef]
- Seo, Y.J.; Lee, K.T.; Rho, J.R.; Choi, J.H. Phorbaketal A, Isolated from the Marine Sponge Phorbas sp., Exerts Its Anti-Inflammatory Effects via NF-κB Inhibition and Heme Oxygenase-1 Activation in Lipopolysaccharide-Stimulated Macrophages. Mar. Drugs 2015, 13, 7005–7019. [Google Scholar] [CrossRef] [PubMed]
- Karri, S.; Sharma, S.; Hatware, K.; Patil, K. Natural anti-obesity agents and their therapeutic role in management of obesity: A future trend perspective. Biomed. Pharmacother. 2019, 110, 224–238. [Google Scholar] [CrossRef]
- Chaiittianan, R.; Chayopas, P.; Rattanathongkom, A.; Tippayawat, P.; Sutthanut, K. Anti-obesity potential of corn silks: Relationships of phytochemicals and antioxidation, anti-pre-adipocyte proliferation, anti-adipogenesis, and lipolysis induction. J. Funct. Foods 2016, 23, 497–510. [Google Scholar] [CrossRef]
- Cosme, P.; Rodríguez, A.B.; Espino, J.; Garrido, M. Plant Phenolics: Bioavailability as a Key Determinant of Their Potential Health-Promoting Applications. Antioxidants 2020, 9, 1263. [Google Scholar] [CrossRef]
- Chen, Y.; Bi, F.; Sun, Z. A network pharmacology approach to determine the underlying mechanisms of action of Yishen Tongluo formula for the treatment of oligoasthenozoospermia. PLoS ONE 2021, 16, e0252906. [Google Scholar] [CrossRef]
- Li, W.; Yuan, G.; Pan, Y.; Wang, C.; Chen, H. Network Pharmacology Studies on the Bioactive Compounds and Action Mechanisms of Natural Products for the Treatment of Diabetes Mellitus: A Review. Front. Pharmacol. 2017, 8, 74. [Google Scholar] [CrossRef]
- Shi, S.; Cai, Y.; Cai, X.; Zheng, X.; Cao, D.; Ye, F.; Xiang, Z. A Network Pharmacology Approach to Understanding the Mechanisms of Action of Traditional Medicine: Bushenhuoxue Formula for Treatment of Chronic Kidney Disease. PLoS ONE 2014, 9, e89123. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.K.; Adnan, M.; Cho, D.H. A network pharmacology analysis on drug-like compounds from Ganoderma lucidum for alleviation of atherosclerosis. J. Food Biochem. 2021, 45, e13906. [Google Scholar] [CrossRef]
- Matsson, P.; Kihlberg, J. How Big Is Too Big for Cell Permeability? J. Med. Chem. 2017, 60, 1662–1664. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Schoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Chaput, L.; Villoutreix, B.O. Virtual screening web servers: Designing chemical probes and drug candidates in the cyberspace. Brief. Bioinform. 2021, 22, 1790–1818. [Google Scholar] [CrossRef]
- Soo, H.-C.; Chung, F.F.-L.; Lim, K.-H.; Yap, V.A.; Bradshaw, T.D.; Hii, L.-W.; Tan, S.-H.; See, S.-J.; Tan, Y.-F.; Leong, C.-O.; et al. Cudraflavone C Induces Tumor-Specific Apoptosis in Colorectal Cancer Cells through Inhibition of the Phosphoinositide 3-Kinase (PI3K)-AKT Pathway. PLoS ONE 2017, 12, e0170551. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Khanal, P.; Patil, B.M.; Chand, J.; Naaz, Y. Anthraquinone Derivatives as an Immune Booster and their Therapeutic Option Against COVID-19. Nat. Prod. Bioprospect. 2020, 10, 325. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Modeling 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Shityakov, S.; Förster, C. In silico predictive model to determine vector-mediated transport properties for the blood–brain barrier choline transporter. Adv. Appl. Bioinform. Chem. AABC 2014, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- De Melo, K.M.; de Oliveira, F.T.B.; Costa Silva, R.A.; Gomes Quinderé, A.L.; Marinho Filho, J.D.B.; Araújo, A.J.; Barros Pereira, E.D.; Carvalho, A.A.; Chaves, M.H.; Rao, V.S.; et al. α, β-Amyrin, a pentacyclic triterpenoid from Protium heptaphyllum suppresses adipocyte differentiation accompanied by down regulation of PPARγ and C/EBPα in 3T3-L1 cells. Biomed. Pharmacother. 2019, 109, 1860–1866. [Google Scholar] [CrossRef]
- Santos, F.A.; Frota, J.T.; Arruda, B.R.; de Melo, T.S.; de Castro Brito, G.A.; Chaves, M.H.; Rao, V.S. Antihyperglycemic and hypolipidemic effects of α, β-amyrin, a triterpenoid mixture from Protium heptaphyllum in mice. Lipids Health Dis. 2012, 11, 1–8. [Google Scholar] [CrossRef]
- Feng, S.; Dai, Z.; Liu, A.B.; Huang, J.; Narsipur, N.; Guo, G.; Kong, B.; Reuhl, K.; Lu, W.; Luo, Z.; et al. Intake of stigmasterol and β-sitosterol alters lipid metabolism and alleviates NAFLD in mice fed a high-fat western-style diet. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Moller, D.E.; Berger, J.P. Role of PPARs in the regulation of obesity-related insulin sensitivity and inflammation. Int. J. Obes. 2003, 27 (Suppl. S3), S17–S21. [Google Scholar] [CrossRef]
- Laurencikiene, J.; Rydén, M. Liver X receptors and fat cell metabolism. Int. J. Obes. 2012, 36, 1494. [Google Scholar] [CrossRef]
- Wan, M.; Easton, R.M.; Gleason, C.E.; Monks, B.R.; Ueki, K.; Kahn, C.R.; Birnbaum, M.J. Loss of Akt1 in mice increases energy expenditure and protects against diet-induced obesity. Mol. Cell. Biol. 2012, 32, 96–106. [Google Scholar] [CrossRef]
- Buzzi, F.; Xu, L.; Zuellig, R.A.; Boller, S.B.; Spinas, G.A.; Hynx, D.; Chang, Z.; Yang, Z.; Hemmings, B.A.; Tschopp, O.; et al. Differential Effects of Protein Kinase B/Akt Isoforms on Glucose Homeostasis and Islet Mass. Mol. Cell. Biol. 2010, 30, 601. [Google Scholar] [CrossRef]
- Nijhawans, P.; Behl, T.; Bhardwaj, S. Angiogenesis in obesity. Biomed. Pharmacother. 2020, 126, 110103. [Google Scholar] [CrossRef] [PubMed]
- Ngo, D.T.; Farb, M.G.; Kikuchi, R.; Karki, S.; Tiwari, S.; Bigornia, S.J.; Bates, D.O.; LaValley, M.P.; Hamburg, N.M.; Vita, J.A.; et al. Anti-Angiogenic Actions of VEGF-A165b, an Inhibitory Isoform of VEGF-A, in Human Obesity. Circulation 2014, 130, 1072. [Google Scholar] [CrossRef] [PubMed]
- Boothby-Shoemaker, W.; Benham, V.; Paithankar, S.; Shankar, R.; Chen, B.; Bernard, J.J. The Relationship between Leptin, the Leptin Receptor and FGFR1 in Primary Human Breast Tumors. Cells 2020, 9, 2224. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, X.; Huang, C.; Liu, H.; Liu, S.; Zhang, Q.; Dong, M.; Hou, M.; Liu, Y.; Lin, H. FGF2 disruption enhances thermogenesis in brown and beige fat to protect against obesity and hepatic steatosis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kay, A.M.; Simpson, C.L.; Stewart, J.A. The Role of AGE/RAGE Signaling in Diabetes-Mediated Vascular Calcification. J. Diabetes Res. 2016, 2016, 6809703. [Google Scholar] [CrossRef]
- Sellegounder, D.; Zafari, P.; Rajabinejad, M.; Taghadosi, M.; Kapahi, P. Advanced glycation end products (AGEs) and its receptor, RAGE, modulate age-dependent COVID-19 morbidity and mortality. A review and hypothesis. Int. Immunopharmacol. 2021, 98, 107806. [Google Scholar] [CrossRef]
- Bandurska-Stankiewicz, E. Thyroid hormones—Obesity and metabolic syndrome. Thyroid Res. 2013, 6, A5. [Google Scholar] [CrossRef][Green Version]
- Bernard, V.; Young, J.; Binart, N. Prolactin—A pleiotropic factor in health and disease. Nat. Rev. Endocrinol. 2019, 15, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Grantham, J.P.; Henneberg, M. The Estrogen Hypothesis of Obesity. PLoS ONE 2014, 9, e99776. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.E.; Meoli, C.C.; Mangiafico, S.P.; Fazakerley, D.J.; Cogger, V.C.; Mohamad, M.; Pant, H.; Kang, M.-J.; Powter, E.; Burchfield, J.G.; et al. Systemic VEGF-A Neutralization Ameliorates Diet-Induced Metabolic Dysfunction. Diabetes 2014, 63, 2656–2667. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Molina, A.; Lopez-Guadamillas, E.; De Cabo, R.; Serrano, M. Pharmacological Inhibition of PI3K Reduces Adiposity and Metabolic Syndrome in Obese Mice and Rhesus Monkeys. Cell Metab. 2015, 21, 558–570. [Google Scholar] [CrossRef]
- Li, N.; Chen, K.; Dong, H.; Yang, J.; Yoshizawa, M.; Kagami, H.; Li, X. Alliin inhibits adipocyte differentiation by downregulating Akt expression: Implications for metabolic disease. Exp. Ther. Med. 2021, 21, 1–8. [Google Scholar] [CrossRef]
- Lee, Y.S.; Riopel, M.; Cabrales, P.; Bandyopadhyay, G.K. Hepatocyte-specific HIF-1 ablation improves obesity-induced glucose intolerance by reducing first-pass GLP-1 degradation. Sci. Adv. 2019, 5, eaaw4176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, H.; Deng, R.; Wang, N.; Zhang, Y.; Wang, Y.; Liu, Y.; Li, F.; Wang, X.; Zhou, L. Berberine Suppresses Adipocyte Differentiation via Decreasing CREB Transcriptional Activity. PLoS ONE 2015, 10, e0125667. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K.; Xu, P.; Cordonier, E.L.; Chen, S.S.; Ng, A.; Xu, Y.; Morozov, A.; Fukuda, M. Neuronal Rap1 Regulates Energy Balance, Glucose Homeostasis, and Leptin Actions. Cell Rep. 2016, 16, 3003–3015. [Google Scholar] [CrossRef]
- Chae, S.Y.; Seo, S.G.; Yang, H.; Yu, J.G.; Suk, S.J.; Jung, E.S.; Ji, H.; Kwon, J.Y.; Lee, H.J.; Lee, K.W. Anti-adipogenic effect of erucin in early stage of adipogenesis by regulating Ras activity in 3T3-L1 preadipocytes. J. Funct. Foods 2015, 19, 700–709. [Google Scholar] [CrossRef]
- Oh, J.H.; Karadeniz, F.; Lee, J.I.; Seo, Y.; Kong, C.-S. Artemisia princeps Inhibits Adipogenic Differentiation of 3T3-L1 Pre-Adipocytes via Downregulation of PPARγ and MAPK Pathways. Prev. Nutr. Food Sci. 2019, 24, 299. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Jang, H.-Y.; Park, E.H.; Kim, J.K.; Kim, J.-M.; Kim, E.-K.; Yea, K.; Kim, Y.-H.; Lee-Kwon, W.; Ryu, S.H.; et al. Wedelolactone inhibits adipogenesis through the ERK pathway in human adipose tissue-derived mesenchymal stem cells. J. Cell. Biochem. 2012, 113, 3436–3445. [Google Scholar] [CrossRef] [PubMed]
- Gross, B.; Pawlak, M.; Lefebvre, P.; Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2016, 13, 36–49. [Google Scholar] [CrossRef] [PubMed]








| No. | Compounds | PubChem ID | RT (mins) | Area (%) | Taxonomic Compound Classification |
|---|---|---|---|---|---|
| 1 | Ethylamine | 6341 | 3.625 | 2.37 | Amines |
| 2 | cis-2,3-Epoxybutane | 92162 | 4.097 | 2.77 | Epoxides |
| 3 | 5-Hydroxymethylfurfural | 237332 | 4.683 | 8.2 | Carbonyl compounds |
| 4 | Mannitan | 10909888 | 5.135 | 0.36 | Tetrahydrofurans |
| 5 | 5-Aminovaleric acid | 138 | 5.164 | 0.67 | Amino acids, peptides, and analogues |
| 6 | Nitrous acid, 1-methylpropyl ester | 13544 | 5.270 | 1.18 | Organic nitroso compounds |
| 7 | Formicin | 69365 | 5.356, 5.481 | 1.76 | Carboxylic acid derivatives |
| 8 | Diethyl acetal | 7765 | 5.818 | 1.39 | Ethers |
| 9 | Xanthosine | 64959 | 6.337 | 7.91 | Purine nucleosides |
| 10 | Cytidine | 6175 | 6.395 | 6.33 | Pyrimidine nucleosides |
| 11 | 2,4,4-Trimethylpentane-1,3-diyl bis(2-methylpropanoate) | 93439 | 6.606 | 1.33 | Dicarboxylic acids and derivatives |
| 12 | α-D-2-deoxyribose | 441475 | 7.116 | 3.4 | Oxanes |
| 13 | 2R,3S-9-[1,3,4-Trihydroxy-2-butoxymethyl]guanine | 135789714 | 7.193 | 3.51 | Purines and purine derivatives |
| 14 | Palmitic acid | 985 | 8.250, 8.616 | 5.2 | Fatty acids and conjugates |
| 15 | Ethyl palmitate | 12366 | 8.318 | 3.35 | Fatty acid esters |
| 16 | Linoleic acid | 5280450 | 8.914 | 1.85 | Lineolic acids and derivatives |
| 17 | Ethyl linoleate | 5282184 | 8.952 | 4.24 | Lineolic acids and derivatives |
| 18 | Ethyl stearate | 8122 | 9.058 | 1.41 | Fatty acid esters |
| 19 | Estradiol, 3-deoxy | 537293 | 9.414 | 0.74 | Estrane steroids |
| 20 | Oleic Acid | 445639 | 9.645 | 0.59 | Fatty acids and conjugates |
| 21 | Ethyl isovalerate | 7945 | 9.731 | 0.76 | Fatty acid esters |
| 22 | Eicosane | 8222 | 10.058, 10.721, 11.529 | 2.71 | Alkanes |
| 23 | (Z)-9-Hexadecenal | 5364643 | 10.164 | 1.61 | Fatty aldehydes |
| 24 | Heneicosanoic acid, 2,4-dimethyl-, methyl ester | 560463 | 10.366 | 1.29 | Fatty acid esters |
| 25 | 7-Pentadecyne | 549063 | 10.789 | 1.32 | Acetylenes |
| 26 | Ethyl heptadecanoate | 26397 | 11.096 | 0.9 | Fatty acid esters |
| 27 | Squalene | 638072 | 11.193 | 0.76 | Triterpenoids |
| 28 | 1,3-Dioxolane, 4-ethyl-5-octyl-2,2-bis(trifluoromethyl)-, trans- | 91694992 | 12.423 | 0.24 | Ethers |
| 29 | Neotocopherol | 86052 | 12.462 | 0.74 | 1-hydroxy-4-unsubstituted benzenoids |
| 30 | N,2-diaminopropane | 7210 | 12.731, 14.356 | 1.02 | 1-hydroxy-4-unsubstituted benzenoids |
| 31 | 24-epicampesterol | 5283637 | 13.895 | 2.85 | Ergostane steroids |
| 32 | β-Stigmasterol | 6432745 | 14.116 | 5.32 | Stigmastanes and derivatives |
| 33 | β-Sitosterol | 222284 | 14.721 | 12.41 | Triterpenoids |
| 34 | β-Amyrone | 612782 | 14.895, 15.385 | 6.95 | Triterpenoids |
| 35 | Sitostenone | 5484202 | 16.087 | 1.64 | Stigmastanes and derivatives |
| 36 | 2-Ethylacridine | 610161 | 18.308 | 0.91 | Benzoquinolines |
| No. | Compounds | Lipinski Rules | Lipinski’s Violations | Bioavailability Score | TPSA(Å2) | |||
|---|---|---|---|---|---|---|---|---|
| MW | HBA | HBD | MLog P | |||||
| <500 | <10 | ≤5 | ≤4.15 | ≤1 | >0.1 | <140 | ||
| 1 | Ethylamine | 45.08 | 1 | 1 | −0.23 | 0 | 0.55 | 26.02 |
| 2 | cis-2,3-Epoxybutane | 72.11 | 1 | 0 | 0.35 | 0 | 0.55 | 12.53 |
| 3 | 5-Hydroxymethylfurfural | 126.11 | 3 | 1 | −1.06 | 0 | 0.55 | 50.44 |
| 4 | Mannitan | 164.16 | 5 | 4 | −2.35 | 0 | 0.55 | 90.15 |
| 5 | 5-Aminovaleric acid | 117.15 | 3 | 2 | 0.01 | 0 | 0.55 | 63.32 |
| 6 | Nitrous acid, 1-methylpropyl ester | 103.12 | 3 | 0 | 0.42 | 0 | 0.55 | 38.66 |
| 7 | Formicin | 89.09 | 2 | 2 | −0.85 | 0 | 0.55 | 49.33 |
| 8 | Diethyl acetal | 118.17 | 2 | 0 | 1.01 | 0 | 0.55 | 18.46 |
| 9 | Xanthosine | 284.23 | 7 | 5 | −2.30 | 0 | 0.55 | 153.46 |
| 10 | Cytidine | 243.22 | 6 | 4 | −2.29 | 0 | 0.55 | 130.83 |
| 11 | 2,4,4-Trimethylpentane-1,3-diyl bis(2-methylpropanoate) | 286.41 | 4 | 0 | 3.17 | 0 | 0.55 | 52.60 |
| 12 | α-D-2-deoxyribose | 134.13 | 4 | 3 | −1.49 | 0 | 0.55 | 69.92 |
| 13 | 2R,3S-9-[1,3,4-Trihydroxy-2-butoxymethyl] guanine | 285.26 | 7 | 5 | −2.76 | 0 | 0.55 | 159.51 |
| 14 | Palmitic acid | 256.42 | 2 | 1 | 4.19 | 1 | 0.85 | 37.30 |
| 15 | Ethyl palmitate | 284.48 | 2 | 0 | 4.67 | 1 | 0.55 | 26.30 |
| 16 | Linoleic acid | 280.45 | 2 | 1 | 4.47 | 1 | 0.85 | 37.30 |
| 17 | Ethyl linoleate | 308.50 | 2 | 0 | 4.93 | 1 | 0.55 | 26.30 |
| 18 | Ethyl stearate | 312.53 | 2 | 0 | 5.13 | 1 | 0.55 | 26.30 |
| 19 | Estradiol, 3-deoxy | 256.38 | 1 | 1 | 4.19 | 1 | 0.55 | 20.23 |
| 20 | Oleic Acid | 282.46 | 2 | 1 | 4.57 | 1 | 0.85 | 37.30 |
| 21 | Ethyl isovalerate | 130.18 | 2 | 0 | 1.63 | 0 | 0.55 | 26.30 |
| 22 | Eicosane | 282.55 | 0 | 0 | 7.38 | 1 | 0.55 | 0.00 |
| 23 | (Z)-9-Hexadecenal | 238.41 | 1 | 0 | 4.20 | 1 | 0.55 | 17.07 |
| 24 | Heneicosanoic acid, 2,4-dimethyl-, methyl ester | 368.64 | 2 | 0 | 6.00 | 1 | 0.55 | 26.30 |
| 25 | 7-Pentadecyne | 208.38 | 0 | 0 | 6.04 | 1 | 0.55 | 0.00 |
| 26 | Ethyl heptadecanoate | 298.50 | 2 | 0 | 4.91 | 1 | 0.55 | 26.30 |
| 27 | Squalene | 410.72 | 0 | 0 | 7.93 | 1 | 0.55 | 0.00 |
| 28 | 1,3-Dioxolane, 4-ethyl-5-octyl-2,2-bis(trifluoromethyl)-, trans- | 350.34 | 8 | 0 | 4.02 | 0 | 0.55 | 18.46 |
| 29 | Neotocopherol | 416.68 | 2 | 1 | 5.94 | 1 | 0.55 | 29.46 |
| 30 | N,2-diaminopropane | 282.34 | 5 | 4 | 1.81 | 0 | 0.55 | 65.18 |
| 31 | 24-epicampesterol | 400.68 | 1 | 1 | 6.54 | 1 | 0.55 | 20.23 |
| 32 | β-Stigmasterol | 412.69 | 1 | 1 | 6.62 | 1 | 0.55 | 20.23 |
| 33 | β-Sitosterol | 414.71 | 1 | 1 | 6.73 | 1 | 0.55 | 20.23 |
| 34 | β-Amyrone | 424.70 | 1 | 0 | 6.82 | 1 | 0.55 | 17.07 |
| 35 | Sitostenone | 412.69 | 1 | 0 | 6.62 | 1 | 0.55 | 17.07 |
| 36 | 2-Ethylacridine | 207.27 | 1 | 0 | 3.58 | 0 | 0.55 | 12.89 |
| No. | Target | Degree of Value | No. | Target | Degree of Value |
|---|---|---|---|---|---|
| 1 | AKT1 | 43 | 41 | NR1I2 | 7 |
| 2 | IL6 | 39 | 42 | ADORA3 | 6 |
| 3 | GAPDH | 33 | 43 | ALOX15 | 6 |
| 4 | PPARG | 29 | 44 | EIF4E | 6 |
| 5 | VEGFA | 29 | 45 | FFAR4 | 6 |
| 6 | ESR1 | 25 | 46 | GLI1 | 6 |
| 7 | PPARA | 23 | 47 | HSD11B2 | 6 |
| 8 | AR | 19 | 48 | NPC1L1 | 6 |
| 9 | CYP19A1 | 19 | 49 | NR1H2 | 6 |
| 10 | CNR1 | 16 | 50 | RAC1 | 6 |
| 11 | FGF2 | 15 | 51 | SERPINA6 | 6 |
| 12 | NR3C1 | 15 | 52 | VDR | 6 |
| 13 | CDC42 | 12 | 53 | PPARD | 6 |
| 14 | CYP17A1 | 12 | 54 | CNR2 | 5 |
| 15 | ESR2 | 12 | 55 | FABP3 | 5 |
| 16 | PRKCA | 12 | 56 | GPBAR1 | 5 |
| 17 | TRPV1 | 12 | 57 | ESRRB | 4 |
| 18 | SREBF2 | 11 | 58 | NAAA | 4 |
| 19 | NR1H4 | 10 | 59 | PDK1 | 4 |
| 20 | SHBG | 10 | 60 | PDCD4 | 4 |
| 21 | SRD5A1 | 10 | 61 | ALOX12 | 3 |
| 22 | FABP4 | 9 | 62 | CES1 | 3 |
| 23 | GPER1 | 9 | 63 | CPB2 | 3 |
| 24 | HSD17B1 | 9 | 64 | ENPP2 | 3 |
| 25 | HSPA5 | 9 | 65 | MGEA5 | 3 |
| 26 | NR1H3 | 9 | 66 | MTNR1B | 3 |
| 27 | NR3C2 | 9 | 67 | IMPDH2 | 3 |
| 28 | STS | 9 | 68 | PHLPP1 | 3 |
| 29 | ADORA1 | 8 | 69 | ACP1 | 2 |
| 30 | ADORA2A | 8 | 70 | CES2 | 2 |
| 31 | ADORA2B | 8 | 71 | EHMT1 | 2 |
| 32 | AKR1C3 | 8 | 72 | FDFT1 | 2 |
| 33 | ESRRA | 8 | 73 | RORC | 2 |
| 34 | F2 | 8 | 74 | SLC5A2 | 2 |
| 35 | FAAH | 8 | 75 | TBXA2R | 2 |
| 36 | PLA2G1B | 8 | 76 | ADCY10 | 1 |
| 37 | ADA | 7 | 77 | ENPEP | 1 |
| 38 | FGF1 | 7 | 78 | NPEPPS | 1 |
| 39 | G6PD | 7 | 79 | SLC22A2 | 1 |
| 40 | MMP3 | 7 |
| KEGG ID | Targets | False Discovery Rate |
|---|---|---|
| hsa03320:PPAR signaling pathway | PPARA, PPARD, PPARG, FABP3, FABP4, NR1H3 | 0.0001200 |
| hsa04370:VEGF signaling pathway | AKT1, VEGFA, PRKCA, CDC42 | 0.0049000 |
| hsa04917:Prolactin signaling pathway | AKT1, ESR1, ESR2, CYP17A1 | 0.0080000 |
| hsa04066:HIF-1 signaling pathway | AKT1, IL6, GAPDH, VEGFA, PRKCA, PDK1 | 0.0006900 |
| hsa04933:AGE-RAGE signaling pathway in diabetic complications | AKT1, IL6, VEGFA, PRKCA, CDC42 | 0.0035000 |
| hsa04015:Rap1 signaling pathway | AKT1, VEGFA, CDC42, PRKCA, FGF1, FGF2, CNR1, ADRA2A, ADORA2B | 0.0000376 |
| hsa04919:Thyroid hormone signaling pathway | AKT1, PRKCA, ESR1, THRA | 0.0337000 |
| hsa04014:Ras signaling pathway | AKT1, VEGFA, CDC42, PRKCA, FGF1, FGF2 | 0.0031000 |
| hsa04915:Estrogen signaling pathway | AKT1, ESR1, ESR2, GPER1 | 0.0480000 |
| hsa04024:cAMP signaling pathway | AKT1, PPARA, ADORA2A, GLI1, ADORA1, ADCY10 | 0.0089000 |
| hsa04010:MAPK signaling pathway | AKT1, VEGFA, CDC42, PRKCA, FGF1, FGF2 | 0.0328000 |
| hsa04151:PI3K-Akt signaling pathway | AKT1, IL6, VEGFA, PRKCA, FGF1, FGF2, PHLPP1 | 0.0194000 |
| No. | Target | Degree of Value | No. | Target | Degree of Value |
|---|---|---|---|---|---|
| 1 | AKT1 | 11 | 15 | PPARG | 1 |
| 2 | PRKCA | 8 | 16 | FABP4 | 1 |
| 3 | VEGFA | 7 | 17 | CYP17A1 | 1 |
| 4 | CDC42 | 5 | 18 | GAPDH | 1 |
| 5 | FGF1 | 4 | 19 | PDK1 | 1 |
| 6 | FGF2 | 4 | 20 | CNR1 | 1 |
| 7 | ESR1 | 3 | 21 | ADORA2B | 1 |
| 8 | IL6 | 3 | 22 | THRA | 1 |
| 9 | PPARA | 2 | 23 | PLA2G1B | 1 |
| 10 | ESR2 | 2 | 24 | GPER1 | 1 |
| 11 | ADORA2A | 2 | 25 | GLI1 | 1 |
| 12 | FABP3 | 1 | 26 | ADORA1 | 1 |
| 13 | NR1H3 | 1 | 27 | ADCY10 | 1 |
| 14 | PPARD | 1 | 28 | PHLPP1 | 1 |
| Grid Box | Hydrogen Bond Interactions | Hydrophobic Interactions | |||||
|---|---|---|---|---|---|---|---|
| Protein | Ligand | PubChem ID | Binding Energy (kcal/mol) | Center | Dimension | Amino Acid Residue | Amino Acid Residue |
| PPARA (PDB ID: 3SP6) | (*) β-Amyrone | 612782 | −16.1 | x = 8.006 | size_x = 40 | N/A | Tyr334, Ala333, Val324 |
| y = −0.459 | size_y = 40 | Met320, Phe218, Met220 | |||||
| z = 23.392 | size_z = 40 | Glu286, Val332, Asn219 | |||||
| Thr279 | |||||||
| Squalene | 638072 | −6.0 | x = 8.006 | size_x = 40 | N/A | Tyr334, Asn336, Ala333 | |
| y = −0.459 | size_y = 40 | Thr279, Leu254, Val332 | |||||
| z = 23.392 | size_z = 40 | Ile241, Glu251, Ala250 | |||||
| Cys275, Cys278, Val255 | |||||||
| Ethyl palmitate | 12366 | −6.0 | x = 8.006 | size_x = 40 | N/A | Met320, Leu321, Val324 | |
| y = −0.459 | size_y = 40 | Leu331, Val332, Ala333 | |||||
| z = 23.392 | size_z = 40 | Thr279, Tyr334, Asn219 | |||||
| Thr283, Ile317 | |||||||
| Heneicosanoic acid, 2,4-dimethyl-, methyl ester | 560463 | −5.8 | x = 8.006 | size_x = 40 | N/A | Lys257, Val255, His274 | |
| y = −0.459 | size_y = 40 | Leu254, Ala250, Glu251 | |||||
| z = 23.392 | size_z = 40 | Ala333, Cys275, Cys278 | |||||
| Leu258 | |||||||
| Oleic Acid | 445639 | −5.3 | x = 8.006 | size_x = 40 | N/A | Leu254, Val255, Ala250 | |
| y = −0.459 | size_y = 40 | Ala333, Asn219, Thr283 | |||||
| z = 23.392 | size_z = 40 | Met320, Leu321, Val324 | |||||
| Ile317, Thr279, Tyr334 | |||||||
| Cys275, Glu251 | |||||||
| Ethyl linoleate | 5282184 | −5.0 | x = 8.006 | size_x = 40 | N/A | Glu282, Tyr334, Thr279 | |
| y = −0.459 | size_y = 40 | Ala333, Glu251, Leu254 | |||||
| z = 23.392 | size_z = 40 | Cys275, Ala250, Val255 | |||||
| Cys278, Val281 | |||||||
| Palmitic acid | 985 | −4.9 | x = 8.006 | size_x = 40 | N/A | Val332, Ile241, Ala333 | |
| y = −0.459 | size_y = 40 | Thr279, Val255, Tyr334 | |||||
| z = 23.392 | size_z = 40 | Leu258, Cys275, Ala250 | |||||
| Leu254, Glu251 | |||||||
| Linoleic acid | 5280450 | −4.9 | x = 8.006 | size_x = 40 | Ser323, Tyr214 | Asn221, Met320, Val324 | |
| y = −0.459 | size_y = 40 | Met320, Asn219, Tyr334 | |||||
| z = 23.392 | size_z = 40 | Thr279, Leu331, Leu321 | |||||
| Thr283, Ile317 | |||||||
| (Z)-9-Hexadecenal | 5364643 | −3.8 | x = 8.006 | size_x = 40 | Thr307 | Asn303, Lys310, Tyr311 | |
| y = −0.459 | size_y = 40 | Pro389, Asp466, Ser688 | |||||
| z = 23.392 | size_z = 40 | Val306 | |||||
| PPARD (PDB ID: 5U3Q) | (*) β-Stigmasterol | 6432745 | −10.8 | x = 39.265 | size_x = 40 | N/A | Ser271, Glu262, Ser271 |
| y = −18.736 | size_y = 40 | Pro268, Ser266, Lys265 | |||||
| z = 119.392 | size_z = 40 | ||||||
| β-Sitosterol | 222284 | −7.1 | x = 39.265 | size_x = 40 | Arg407, Glu288 | Asp439, Tyr284, Pro362 | |
| y = −18.736 | size_y = 40 | Met440, Val410, Thr411 | |||||
| z = 119.392 | size_z = 40 | ||||||
| Heneicosanoic acid, 2,4-dimethyl-, methyl ester | 560463 | −5.9 | x = 39.265 | size_x = 40 | N/A | Met440, Thr411, Val410 | |
| y = −18.736 | size_y = 40 | Tyr441, Pro362, Asp360 | |||||
| z = 119.392 | size_z = 40 | Arg361, Arg407, Tyr284 | |||||
| Ethyl linoleate | 5282184 | −5.6 | x = 39.265 | size_x = 40 | N/A | Met440, Tyr441, Asp439 | |
| y = −18.736 | size_y = 40 | Pro362, Tyr284, Arg361 | |||||
| z = 119.392 | size_z = 40 | Val410, Thr411 | |||||
| Linoleic acid | 5280450 | −5.2 | x = 39.265 | size_x = 40 | N/A | Val410, Arg407, Met440 | |
| y = −18.736 | size_y = 40 | Asp439, Thr411, Tyr411 | |||||
| z = 119.392 | size_z = 40 | Tyr284, Asp360, Pro362 | |||||
| Arg361, Glu288 | |||||||
| Oleic Acid | 445639 | −4.9 | x = 39.265 | size_x = 40 | N/A | Asp360, Pro362, Tyr284 | |
| y = −18.736 | size_y = 40 | Val410, Met440, Tyr441 | |||||
| z = 119.392 | size_z = 40 | Thr411 | |||||
| Palmitic acid | 985 | −4.6 | x = 39.265 | size_x = 40 | N/A | Tyr441, Pro362, Arg361 | |
| y = −18.736 | size_y = 40 | Val410, Tyr284, Glu288 | |||||
| z = 119.392 | size_z = 40 | Met440, Thr411, Ala414 | |||||
| Arg407 | |||||||
| (Z)-9-Hexadecenal | 5364643 | −3.7 | x = 39.265 | size_x = 40 | Arg361 | Thr411, Arg407, Pro362 | |
| y = −18.736 | size_y = 40 | Asp439, Met440 | |||||
| z = 119.392 | size_z = 40 | ||||||
| PPARG (PDB ID: 3E00) | (*) β-Amyrone | 612782 | −14.0 | x = 2.075 | size_x = 40 | N/A | Glu343, Glu295, Ile326 |
| y = 31.910 | size_y = 40 | Ile296, Ala292, Phe226 | |||||
| z = 18.503 | size_z = 40 | Met329, Leu333, Pro227 | |||||
| Leu228 | |||||||
| Ethyl linoleate | 5282184 | −5.9 | x = 2.075 | size_x = 40 | N/A | Ile296, Met329, Ile326 | |
| y = 31.910 | size_y = 40 | Leu228, Leu333, Ala292 | |||||
| z = 18.503 | size_z = 40 | Arg288, Phe226, Glu295 | |||||
| Linoleic acid | 5280450 | −5.3 | x = 2.075 | size_x = 40 | Thr162, Leu167 | Arg202, Tyr192, Asp166 | |
| y = 31.910 | size_y = 40 | Lys336, Glu369, Val372 | |||||
| z = 18.503 | size_z = 40 | Arg350, Glu351, Gln193 | |||||
| Lys354 | |||||||
| Oleic Acid | 445639 | −5.1 | x = 2.075 | size_x = 40 | Asp441, Asn377 | Arg426, Lys381, Asp379 | |
| y = 31.910 | size_y = 40 | Phe370, Lys373, Glu448 | |||||
| z = 18.503 | size_z = 40 | Ile445, Pro366, Glu369 | |||||
| Gln444 | |||||||
| Palmitic acid | 985 | −5.1 | x = 2.075 | size_x = 40 | Glu291, Arg288 | Glu343, Leu333, Leu330 | |
| y = 31.910 | size_y = 40 | Leu228, Met329, Ala292 | |||||
| z = 18.503 | size_z = 40 | Ile326, Glu295 | |||||
| (Z)-9-Hexadecenal | 5364643 | −5.0 | x = 2.075 | size_x = 40 | N/A | Met329, Leu333, Ser332 | |
| y = 31.910 | size_y = 40 | Leu228, Arg288, Glu295 | |||||
| z = 18.503 | size_z = 40 | Glu343, Ala292 | |||||
| FABP3 (PDB ID: 5HZ9) | (*) β-Amyrone | 612782 | −21.5 | x = −1.215 | size_x = 40 | N/A | Phe28, Gln32, Lys32 |
| y = 46.730 | size_y = 40 | Thr57, Ala29 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Heneicosanoic acid, 2,4-dimethyl-, methyl ester | 560463 | −8.8 | x = −1.215 | size_x = 40 | N/A | Glu27, Phe28, Gly25 | |
| y = 46.730 | size_y = 40 | Ala29, Lys22, Gln32 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Eicosane | 8222 | −8.6 | x = −1.215 | size_x = 40 | N/A | Lys22, Thr57, Gln32 | |
| y = 46.730 | size_y = 40 | Gly25, Phe28, Ala29 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Ethyl stearate | 8122 | −8.3 | x = −1.215 | size_x = 40 | N/A | Phe58, Gln32, Gly25 | |
| y = 46.730 | size_y = 40 | Phe28 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Ethyl heptadecanoate | 26397 | −8.3 | x = −1.215 | size_x = 40 | N/A | Gly27, Gln32, Ala29 | |
| y = 46.730 | size_y = 40 | Phe28 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Ethyl linoleate | 5282184 | −8.2 | x = −1.215 | size_x = 40 | N/A | Lys22, Ala29, Phe28 | |
| y = 46.730 | size_y = 40 | Gly25, Gln32 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Ethyl palmitate | 12366 | −8.2 | x = −1.215 | size_x = 40 | N/A | Ala29, Gln32, Phe28 | |
| y = 46.730 | size_y = 40 | Gly25, Gly27, Lys22 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Linoleic acid | 5280450 | −7.4 | x = −1.215 | size_x = 40 | Lys22 | Ala29, Gln32, Phe28 | |
| y = 46.730 | size_y = 40 | Gly25, Gly27 | |||||
| z = −15.099 | size_z = 40 | ||||||
| (Z)-9-Hexadecenal | 5364643 | −6.9 | x = −1.215 | size_x = 40 | N/A | Gly27, Gly25, Gln32 | |
| y = 46.730 | size_y = 40 | Ala29, Phe28, Ala29 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Oleic Acid | 445639 | −6.9 | x = −1.215 | size_x = 40 | N/A | Phe28, Gly27, Ala29 | |
| y = 46.730 | size_y = 40 | Gln32 | |||||
| z = −15.099 | size_z = 40 | ||||||
| Palmitic acid | 985 | −6.5 | x = −1.215 | size_x = 40 | N/A | Val33, Gln32, Ala29 | |
| y = 46.730 | size_y = 40 | Phe58, Lys22, Thr57 | |||||
| z = −15.099 | size_z = 40 | ||||||
| 5-Aminovaleric acid | 138 | −5.1 | x = −1.215 | size_x = 40 | Ala29, Phe28, Val26 | Gly25, Gly27, Lys22 | |
| y = 46.730 | size_y = 40 | Phe28 | |||||
| z = −15.099 | size_z = 40 | ||||||
| FABP4 (PDB ID: 3P6D) | (*) β-Amyrone | 612782 | −13.2 | x = 7.693 | size_x = 40 | N/A | Val90, Lys107, Glu109 |
| y = 9.921 | size_y = 40 | Glu116, Val114, Lys105 | |||||
| z = 14.698 | size_z = 40 | ||||||
| Heneicosanoic acid, 2,4-dimethyl-, methyl ester | 560463 | −5.6 | x = 7.693 | size_x = 40 | Leu86 | Leu66, Ile49, Asp47 | |
| y = 9.921 | size_y = 40 | Cys1, Ser1, Met0 | |||||
| z = 14.698 | size_z = 40 | ||||||
| Ethyl stearate | 8122 | −5.5 | x = 7.693 | size_x = 40 | Gly88 | Asp87, Leu86, Asp47 | |
| y = 9.921 | size_y = 40 | Leu66, Gly46, Ile49 | |||||
| z = 14.698 | size_z = 40 | Val44, Ser1, Cys1 | |||||
| Met0 | |||||||
| Ethyl palmitate | 12366 | −5.4 | x = 7.693 | size_x = 40 | N/A | Ala29, Gln32, Phe28 | |
| y = 9.921 | size_y = 40 | Gly25, Gly27, Lys22 | |||||
| z = 14.698 | size_z = 40 | ||||||
| Ethyl heptadecanoate | 26397 | −5.3 | x = 7.693 | size_x = 40 | Gly88 | Asp87, Leu86, Asp47 | |
| y = 9.921 | size_y = 40 | Ile49, Ser1, Leu66 | |||||
| z = 14.698 | size_z = 40 | Cys1, Met0 | |||||
| Ethyl linoleate | 5282184 | −5.2 | x = 7.693 | size_x = 40 | Gly88 | Asp87, Met0, Ile49 | |
| y = 9.921 | size_y = 40 | Cys1, Gly46, Asp47 | |||||
| z = 14.698 | size_z = 40 | Ser1, Leu66, Leu86 | |||||
| 5-Aminovaleric acid | 138 | −5.0 | x = 7.693 | size_x = 40 | Arg106, Gln96, Glu72 | Thr60, Ala75, Thr74 | |
| y = 9.921 | size_y = 40 | ||||||
| z = 14.698 | size_z = 40 | ||||||
| Oleic Acid | 445639 | −5.0 | x = 7.693 | size_x = 40 | Leu86 | Gly88, Ile49, Val44 | |
| y = 9.921 | size_y = 40 | Gly46, Asp47, Ile65 | |||||
| z = 14.698 | size_z = 40 | Leu66, Asp87 | |||||
| Linoleic acid | 5280450 | −4.9 | x = 7.693 | size_x = 40 | Leu86 | Thr85, Leu66, Asp47 | |
| y = 9.921 | size_y = 40 | Cys1, Gly46, Ser1 | |||||
| z = 14.698 | size_z = 40 | Met0 | |||||
| Palmitic acid | 985 | −4.4 | x = 7.693 | size_x = 40 | Glu72, Val80 | Lys79, Asp71, Val73 | |
| y = 9.921 | size_y = 40 | Glu61, Thr60 | |||||
| z = 14.698 | size_z = 40 | ||||||
| (Z)-9-Hexadecenal | 5364643 | −4.0 | x = 7.693 | size_x = 40 | N/A | Glu109, Val90, Lys105 | |
| y = 9.921 | size_y = 40 | Lys107, Val114, Glu116 | |||||
| z = 14.698 | size_z = 40 | ||||||
| NR1H3 (PDB ID: 2ACL) | (*) β-Amyrone | 612782 | −15.4 | x = 48.735 | size_x = 40 | N/A | Gln330, Ala325, Gly328 |
| y = 39.677 | size_y = 40 | Arg248, Arg245, Lys431 | |||||
| z = 77.096 | size_z = 40 | Gln297, Leu294, Gln429 | |||||
| Val298, Asp295, Leu329 | |||||||
| β-Stigmasterol | 6432745 | −11.1 | x = 48.735 | size_x = 40 | N/A | Gln429, Arg248, Gly328 | |
| y = 39.677 | size_y = 40 | Leu329, Gln330, Glu332 | |||||
| z = 77.096 | size_z = 40 | Ile299, Val331, Arg302 | |||||
| Val298, Asp295, Leu294 | |||||||
| β-Sitosterol | 222284 | −8.1 | x = 48.735 | size_x = 40 | Asn385 | Pro237, Glu388, Glu322 | |
| y = 39.677 | size_y = 40 | Ala391, Lys395, Ala398 | |||||
| z = 77.096 | size_z = 40 | Leu400, Glu394, Pro240 | |||||
| Lys326, Ile238 | |||||||
| Estradiol, 3-deoxy | 537293 | −8.0 | x = 48.735 | size_x = 40 | N/A | Pro240, Leu347, Ala343 | |
| y = 39.677 | size_y = 40 | Asp379, Ser411, Arg404 | |||||
| z = 77.096 | size_z = 40 | Met407, Lys408, Glu390 | |||||
| Glu346 | |||||||
| Sitostenone | 5484202 | −7.7 | x = 48.735 | size_x = 40 | Asn385 | Glu388, Ala391, Lys395 | |
| y = 39.677 | size_y = 40 | Pro240, Glu322, Glu394 | |||||
| z = 77.096 | size_z = 40 | Leu400, Asp241, Trp236 | |||||
| Pro242, Arg251, Lys326 | |||||||
| Ile238, Pro237 | |||||||
| 24-epicampesterol | 5283637 | −7.7 | x = 48.735 | size_x = 40 | Asn385 | Glu388, Ala391, Lys395 | |
| y = 39.677 | size_y = 40 | Glu394, Asp241, Pro242 | |||||
| z = 77.096 | size_z = 40 | Pro240, Lys326, Ile238 | |||||
| Glu322, Pro237 | |||||||
| Ethyl linoleate | 5282184 | −6.0 | x = 48.735 | size_x = 40 | N/A | Gln330, Lys381, Ile299 | |
| y = 39.677 | size_y = 40 | Asp295, Leu294, Lys431 | |||||
| z = 77.096 | size_z = 40 | Arg248, Gln429, Val298 | |||||
| Arg302, Gly382 | |||||||
| Linoleic acid | 5280450 | −5.2 | x = 48.735 | size_x = 40 | Ser411 | Leu347, Arg404, Pro378 | |
| y = 39.677 | size_y = 40 | Asp379, Ala387, Pro386 | |||||
| z = 77.096 | size_z = 40 | Glu390, Pro240, Glu346 | |||||
| Lys408, Ala343 | |||||||
| Oleic Acid | 445639 | −4.9 | x = 48.735 | size_x = 40 | N/A | Arg404, Glu390, Lys408 | |
| y = 39.677 | size_y = 40 | Arg342, Glu339, Pro386 | |||||
| z = 77.096 | size_z = 40 | Pro240, Glu346, Tyr397 | |||||
| Met407 | |||||||
| Grid Box | Hydrogen Bond Interactions | Hydrophobic Interactions | |||||
|---|---|---|---|---|---|---|---|
| Protein | Ligand | PubChem ID | Binding Energy (kcal/mol) | Center | Dimension | Amino Acid Residue | Amino Acid Residue |
| AKT1 (PDB ID: 3O96) | (*) Neotocopherol | 86052 | −6.6 | x = 6.313 | size_x = 40 | Asn53 | Ser56, Ala58, Trp80 |
| y = −7.926 | size_y = 40 | Leu213, Phe225, Ser216 | |||||
| z = 17.198 | size_z = 40 | Leu223, Phe217, Gln218 | |||||
| Leu78, Gln59, Asn199 | |||||||
| IL6 (PDB ID: 4NI9) | (*) Xanthosine | 64959 | −7.4 | x = 11.213 | size_x = 40 | Ser37 | Asp34, Ala38 |
| y = 33.474 | size_y = 40 | ||||||
| z = 11.162 | size_z = 40 | ||||||
| 2-Ethylacridine | 610161 | −6.7 | x = 11.213 | size_x = 40 | N/A | Asp34, Gly35, Tyr31 | |
| y = 33.474 | size_y = 40 | Gln111 | |||||
| z = 11.162 | size_z = 40 | ||||||
| Linoleic acid | 5280450 | −5.0 | x = 11.213 | size_x = 40 | Lys39 | Glu81, Pro80, Ser168 | |
| y = 33.474 | size_y = 40 | Phe83, Glu105, Leu104 | |||||
| z = 11.162 | size_z = 40 | Gln166, Lys103, Glu165 | |||||
| Pro40, Ile106 | |||||||
| VEGFA (PDB ID: 3V2A) | (*) Ethyl palmitate | 12366 | −6.4 | x = 38.009 | size_x = 40 | N/A | Gly196, Lys48, Ile215 |
| y = −10.962 | size_y = 40 | Ile80, Met81, Ile91 | |||||
| z = 12.171 | size_z = 40 | Gln79, His133, Pro49 | |||||
| Tyr165 | |||||||
| Ethyl heptadecanoate | 26397 | −5.1 | x = 38.009 | size_x = 40 | N/A | Pro40, Asp276, Phe36 | |
| y = −10.962 | size_y = 40 | Lys48, Phe47, Ile46 | |||||
| z = 12.171 | size_z = 40 | Lys286, Asp34 | |||||
| Ethyl stearate | 8122 | −5.0 | x = 38.009 | size_x = 40 | N/A | Gln87, Gly88, Tyr137 | |
| y = −10.962 | size_y = 40 | Ile138, Lys144, Val146 | |||||
| z = 12.171 | size_z = 40 | Ser189, Thr45, Thr139 | |||||
| His86 | |||||||
| Ethyl linoleate | 5282184 | −4.9 | x = 38.009 | size_x = 40 | N/A | Pro40, Asp276, Asp34 | |
| y = −10.962 | size_y = 40 | Phe47, Lys48, Asn253 | |||||
| z = 12.171 | size_z = 40 | Ile46, Lys286, Phe36 | |||||
| α-D-2-deoxyribose | 441475 | −4.2 | x = 38.009 | size_x = 40 | Gly255, Ser310, Gly312 | Glu44, Asp257, Lys84 | |
| y = −10.962 | size_y = 40 | Pro85, Ser311 | |||||
| z = 12.171 | size_z = 40 | ||||||
| PRKCA (PDB ID: 3IW4) | (*) Ethyl palmitate | 12366 | −6.4 | x = −14.059 | size_x = 40 | N/A | Gly196, Met197, Ile80 |
| y = 38.224 | size_y = 40 | Met81, Ile91, Gln79 | |||||
| z = 32.319 | size_z = 40 | His133, Pro49, Lys48 | |||||
| Tyr165, Ile215 | |||||||
| Ethyl heptadecanoate | 26397 | −6.2 | x = −14.059 | size_x = 40 | Lys396 | Leu393, Asn660, Gln402 | |
| y = 38.224 | size_y = 40 | Pro666, Ile667, Glu474 | |||||
| z = 32.319 | size_z = 40 | Lys478, Pro398, Val664 | |||||
| Pro397, Arg608 | |||||||
| Ethyl linoleate | 5282184 | −6.2 | x = −14.059 | size_x = 40 | Lys396 | Asn660, Gln402, Pro666 | |
| y = 38.224 | size_y = 40 | Val664, Glu418, His665 | |||||
| z = 32.319 | size_z = 40 | Arg608, Lys478, Pro398 | |||||
| Asp395, Leu393, Leu394 | |||||||
| 2,4,4-Trimethylpentane-1,3-diyl bis(2-methylpropanoate) | 93439 | −6.0 | x = −14.059 | size_x = 40 | Asp472, His476, Arg608 | Ser670, Ile510, Met551 | |
| y = 38.224 | size_y = 40 | Gln548, Glu609, Asp544 | |||||
| z = 32.319 | size_z = 40 | Glu552, Ile667, Asn607 | |||||
| Leu668, Glu474 | |||||||
| Linoleic acid | 5280450 | −5.4 | x = −14.059 | size_x = 40 | Lys396 | Leu393, Pro397, Asn660 | |
| y = 38.224 | size_y = 40 | Leu394, Ser549, Gln662 | |||||
| z = 32.319 | size_z = 40 | Gln548, His553, Glu552 | |||||
| Val664, Pro398, Gln402 | |||||||
| Ethyl stearate | 8122 | −5.0 | x = −14.059 | size_x = 40 | N/A | Arg275, Phe36, Asp34 | |
| y = 38.224 | size_y = 40 | Lys48, Phe47, Ile46 | |||||
| z = 32.319 | size_z = 40 | Lys286, Asp276, Pro40 | |||||
| (Z)-9-Hexadecenal | 5364643 | −4.2 | x = −14.059 | size_x = 40 | Asp395, Lys396 | Gln402, Pro398, Val664 | |
| y = 38.224 | size_y = 40 | Glu552, Gln662, Leu394 | |||||
| z = 32.319 | size_z = 40 | ||||||
| Palmitic acid | 985 | −3.8 | x = −14.059 | size_x = 40 | Phe47 | Phe36, Lys286, Leu252 | |
| y = 38.224 | size_y = 40 | Leu277, Asp276, Ile46 | |||||
| z = 32.319 | size_z = 40 | Asn253, Ser50 | |||||
| Oleic Acid | 445639 | −3.5 | x = −14.059 | size_x = 40 | N/A | Thr145, Val146, Ile138 | |
| y = 38.224 | size_y = 40 | His86, Leu313, Thr139 | |||||
| z = 32.319 | size_z = 40 | Tyr137, Ser189, Lys144 | |||||
| FGF1 (PDB ID: 3OJ2) | (*) Sitostenone | 5484202 | −8.5 | x = 9.051 | size_x = 40 | N/A | Arg203, Ser220, Val222 |
| y = 22.527 | size_y = 40 | Phe172, Ile257, Ser282 | |||||
| z = −0.061 | size_z = 40 | Pro19, Tyr281, Ile204 | |||||
| Ala260 | |||||||
| 24-epicampesterol | 5283637 | −8.3 | x = 9.051 | size_x = 40 | Ile204 | Arg203, Val222, Tyr281 | |
| y = 22.527 | size_y = 40 | Pro19, Ile257, Ser220 | |||||
| z = −0.061 | size_z = 40 | Ala260 | |||||
| Cytidine | 6175 | −6.7 | x = 9.051 | size_x = 40 | Asn350, Arg255, Gln351 | Asn107, Asn173, Phe172 | |
| y = 22.527 | size_y = 40 | Leu258, Ser220, Ala349 | |||||
| z = −0.061 | size_z = 40 | Thr174 | |||||
| α-D-2-deoxyribose | 441475 | −5.2 | x = 9.051 | size_x = 40 | Gln348, Asn350, Thr174 | Ala349, Phe172 | |
| y = 22.527 | size_y = 40 | Asn173, Asn107, Arg255 | |||||
| z = −0.061 | size_z = 40 | ||||||
| FGF2 (PDB ID: 1IIL) | (*) β-Amyrone | 612782 | −14.4 | x = 26.785 | size_x = 40 | N/A | Glu197, Tyr207, Phe198 |
| y = 14.360 | size_y = 40 | Lys199, Glu201, Arg118 | |||||
| z = −1.182 | size_z = 40 | Gln200, Lys119 | |||||
| β-Stigmasterol | 6432745 | −10.9 | x = 26.785 | size_x = 40 | Arg118 | Glu201, Asp99, Gln200 | |
| y = 14.360 | size_y = 40 | Tyr207, Val209, Lys119 | |||||
| z = −1.182 | size_z = 40 | ||||||
| Sitostenone | 5484202 | −7.7 | x = 26.785 | size_x = 40 | His254 | Ala172, Val222, Ile204 | |
| y = 14.360 | size_y = 40 | Ser220, Leu258, Gln259 | |||||
| z = −1.182 | size_z = 40 | Ala260, Ile257 | |||||
| 24-epicampesterol | 5283637 | −7.7 | x = 26.785 | size_x = 40 | Ser137 | Thr139, Trp123, Lys13 | |
| y = 14.360 | size_y = 40 | Leu312, Asp336, Tyr340 | |||||
| z = −1.182 | size_z = 40 | Ile329, Tyr328, Leu327 | |||||
| Ser122, Glu323 | |||||||
| β-Sitosterol | 222284 | −7.2 | x = 26.785 | size_x = 40 | Asp336, Tyr340 | Ile329, Leu327, Ser122 | |
| y = 14.360 | size_y = 40 | Thr319, Lys313, Leu312 | |||||
| z = −1.182 | size_z = 40 | Lys292 | |||||
| Cytidine | 6175 | −6.4 | x = 26.785 | size_x = 40 | Tyr328, Lys313, Thr319 | Pro141, Glu323 | |
| y = 14.360 | size_y = 40 | Asn318, Ser122 | |||||
| z = −1.182 | size_z = 40 | ||||||
| α-D-2-deoxyribose | 441475 | −4.6 | x = 26.785 | size_x = 40 | Arg255, Phe352 | Ala172, Thr174, Ser351 | |
| y = 14.360 | size_y = 40 | Asn173, His353 | |||||
| z = −1.182 | size_z = 40 | ||||||
| PHLPP1 (not available in the PDB) | (*) Neotocopherol | 86052 | −7.2 | x = 26.785 | size_x = 40 | N/A | Asn1333, Cys273, Asn700 |
| y = 14.360 | size_y = 40 | Ser699, Ser722, Asp745 | |||||
| z = −1.182 | size_z = 40 | Asn720, Asp1661, Ile1637 | |||||
| Tyr764, Leu743, Asn1635 | |||||||
| Cys789, Glu1328, Ser768 | |||||||
| Arg815, Ile1326, Thr1327 | |||||||
| Ile1325 | |||||||
| Compounds | PubChem ID | Docking Score (kcal/mol) | |||||
|---|---|---|---|---|---|---|---|
| PPARA (PDB ID: 3SP6) | PPARD (PDB ID: 5U3Q) | PPARG (PDB ID: 3E00) | FABP3 (PDB ID: 5HZ9) | FABP4 (PDB ID: 3P6D) | NR1H3 (PDB ID: 2ACL) | ||
| β-Amyrone | 612782 | −16.1 | |||||
| (1) Clofibrate | 2796 | −6.4 | |||||
| (2) Gemfibrozil | 3463 | −6.3 | |||||
| (3) Ciprofibrate | 2763 | −5.4 | |||||
| (4) Bezafibrate | 39042 | −5.8 | |||||
| (5) Fenofibrate | 3339 | −5.4 | |||||
| β-Stigmasterol | 6432745 | −10.8 | |||||
| (6) Cardarine | 9803963 | −8.5 | |||||
| β-Amyrone | 612782 | −14.0 | |||||
| (7) Pioglitazone | 4829 | −7.7 | |||||
| (8) Rosiglitazone | 77999 | −7.4 | |||||
| (9) Lobeglitazone | 9826451 | −7.3 | |||||
| β-Amyrone | 612782 | −21.5 | |||||
| β-Amyrone | 612782 | −13.2 | |||||
| β-Amyrone | 612782 | −15.4 | |||||
| (10) GW3965 | 447905 | −11.9 | |||||
| (11) T0901317 | 447912 | −8.2 | |||||
| Compounds | PubChem ID | Docking Score (kcal/mol) | ||||||
|---|---|---|---|---|---|---|---|---|
| AKT1 (PDB ID: 3O96) | IL6 (PDB ID: 4NI9) | VEGFA (PDB ID: 3V2A) | PRKCA (PDB ID: 3IW4) | FGF1 (PDB ID: 3OJ2) | FGF2 (PDB ID: 1IIL) | PHLPP1 (N/A in the PDB) | ||
| Neotocopherol | 86052 | −7.5 | ||||||
| (12) AT13148 | 24905401 | −6.9 | ||||||
| (13) Afuresertib | 46843057 | −6.9 | ||||||
| (14) Alliin | 87310 | −4.8 | ||||||
| Xanthosine | 64959 | −7.4 | ||||||
| (15) APX-115 free base | 51036475 | −7.2 | ||||||
| (16) Resatorvid | 11703255 | −7.1 | ||||||
| (17) Myrislignan | 21636106 | −7.1 | ||||||
| (18) Muscone | 10947 | −6.7 | ||||||
| (19) 2′,5′-Dihydroxyacetophenone | 10279 | −6.5 | ||||||
| (20) α-Cyperone | 6452086 | −6.3 | ||||||
| (21) Veratric acid | 7121 | −6.1 | ||||||
| (22) Triolein | 5497163 | −5.5 | ||||||
| (23) Methylthiouracil | 667493 | −5.4 | ||||||
| (24) Falcarindiol | 5281148 | −5.2 | ||||||
| Ethyl palmitate | 12366 | −6.4 | ||||||
| (25) BAW2881 | 16004702 | −7.6 | ||||||
| Ethyl palmitate | 12366 | −6.4 | ||||||
| (26) Midostaurin | 9829523 | −11.0 | ||||||
| Sitostenone | 5484202 | −8.5 | ||||||
| (27) Suramin | 5361 | −15.4 | ||||||
| β-Amyrone | 612782 | −14.4 | ||||||
| (28) PD 166866 | 5328127 | −8.3 | ||||||
| Neotocopherol | 86052 | −7.2 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, K.-K.; Adnan, M.; Cho, D.-H. Elucidating Drug-Like Compounds and Potential Mechanisms of Corn Silk (Stigma Maydis) against Obesity: A Network Pharmacology Study. Curr. Issues Mol. Biol. 2021, 43, 1906-1936. https://doi.org/10.3390/cimb43030133
Oh K-K, Adnan M, Cho D-H. Elucidating Drug-Like Compounds and Potential Mechanisms of Corn Silk (Stigma Maydis) against Obesity: A Network Pharmacology Study. Current Issues in Molecular Biology. 2021; 43(3):1906-1936. https://doi.org/10.3390/cimb43030133
Chicago/Turabian StyleOh, Ki-Kwang, Md. Adnan, and Dong-Ha Cho. 2021. "Elucidating Drug-Like Compounds and Potential Mechanisms of Corn Silk (Stigma Maydis) against Obesity: A Network Pharmacology Study" Current Issues in Molecular Biology 43, no. 3: 1906-1936. https://doi.org/10.3390/cimb43030133
APA StyleOh, K.-K., Adnan, M., & Cho, D.-H. (2021). Elucidating Drug-Like Compounds and Potential Mechanisms of Corn Silk (Stigma Maydis) against Obesity: A Network Pharmacology Study. Current Issues in Molecular Biology, 43(3), 1906-1936. https://doi.org/10.3390/cimb43030133








