Implications of Microorganisms in Alzheimer’s Disease
Abstract
:1. Introduction
2. AD Etiology
2.1. Role of Bacteria
2.2. Role of Viruses
2.3. Role of Fungi
2.4. AD and Periodontis
2.5. Other Factors Responsible for AD
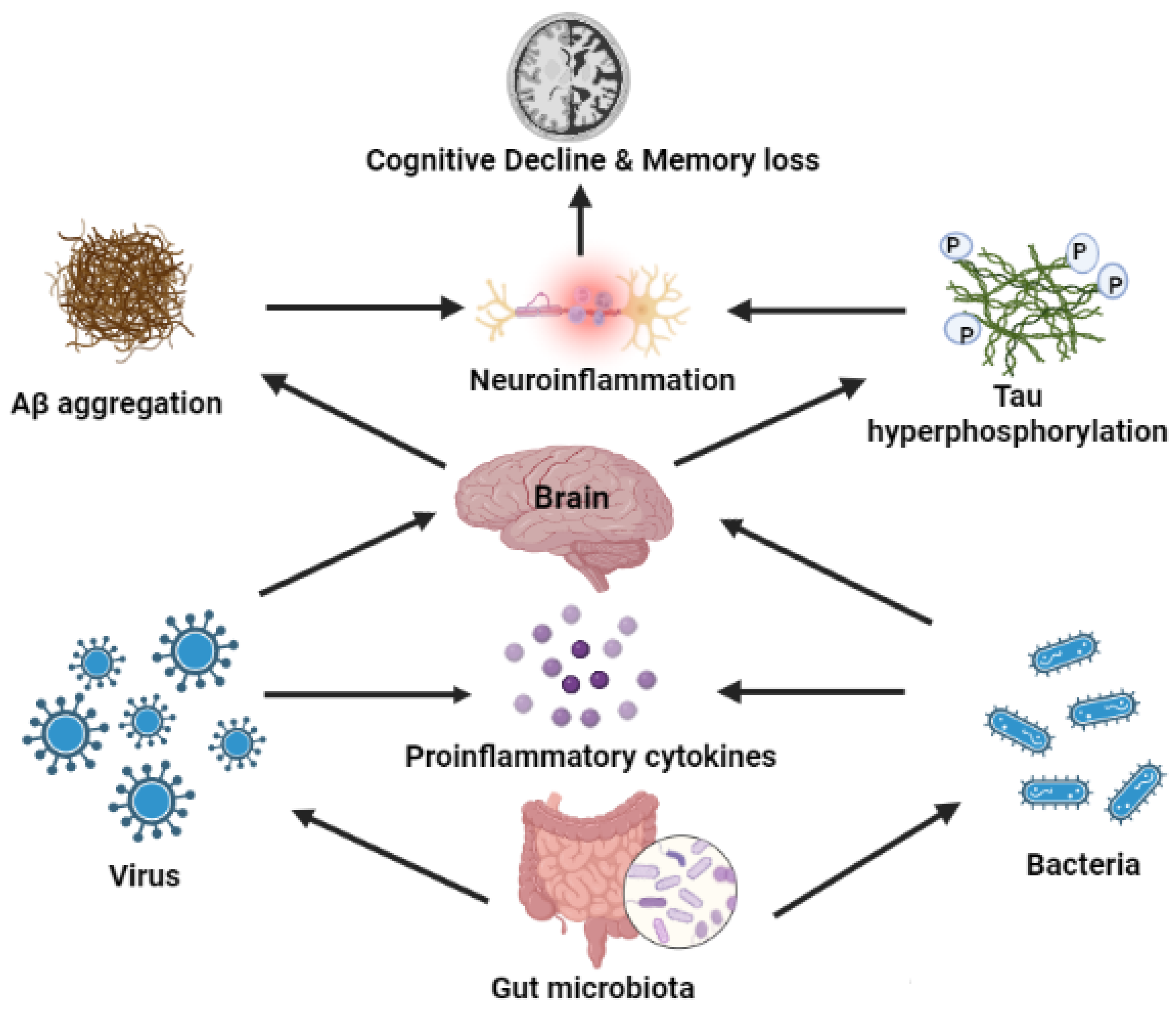
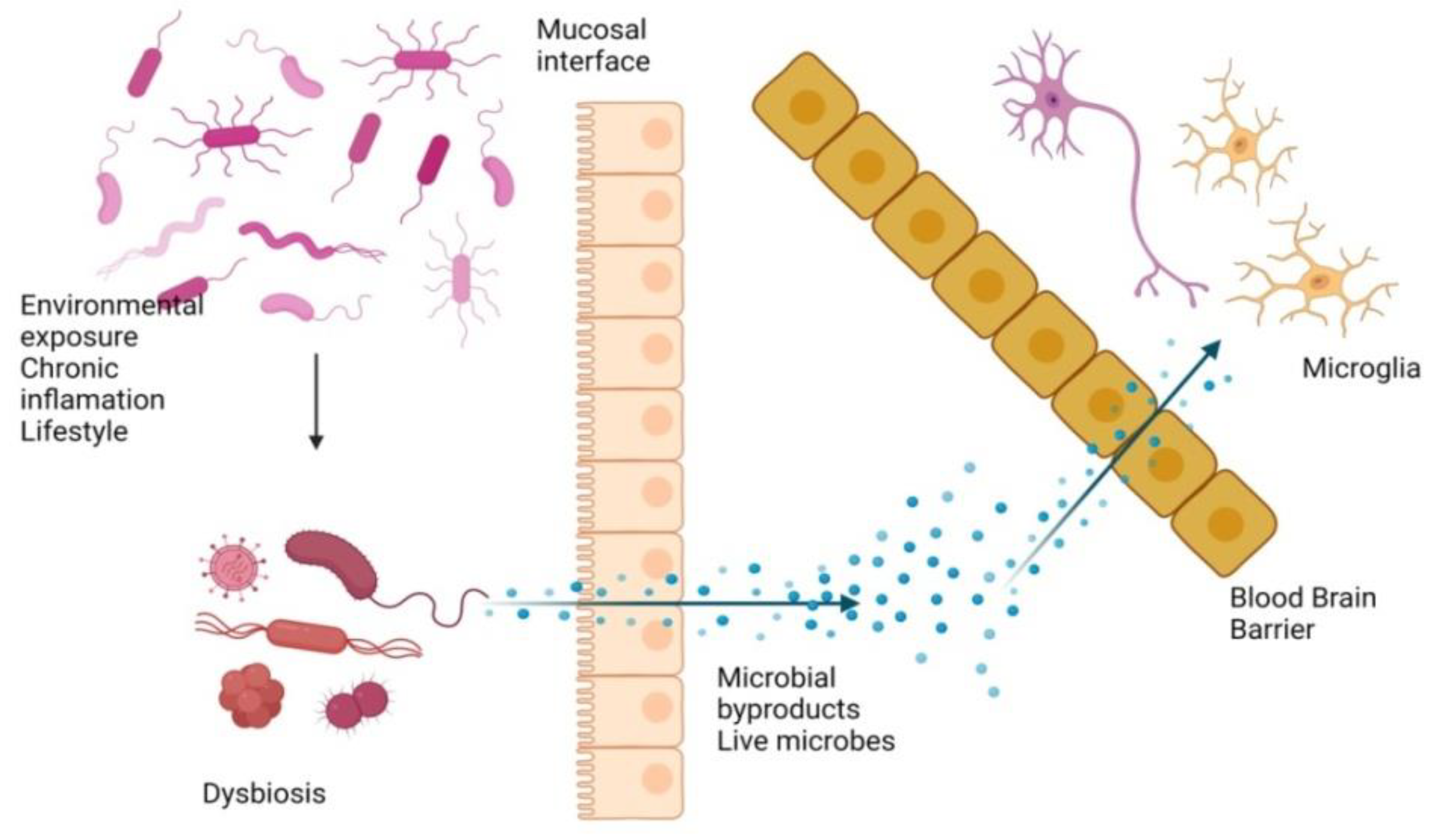
3. Gut Microbiota and AD
4. Effects of Bacterial and Viral Infection on AD
5. Animal Models Used to Identify a Role for Various Microorganisms in AD
5.1. Mouse Models
5.2. Drosophila Models
6. Potential Therapeutic Approaches in AD
6.1. Fecal Microbiota Transplantation
6.2. Beta-Secretase
6.3. Gamma-Secretase
6.4. Tau
6.5. APP Forms in AD
6.6. Drugs Approved for the Treatment of AD
6.7. Other Therapeutic Approaches
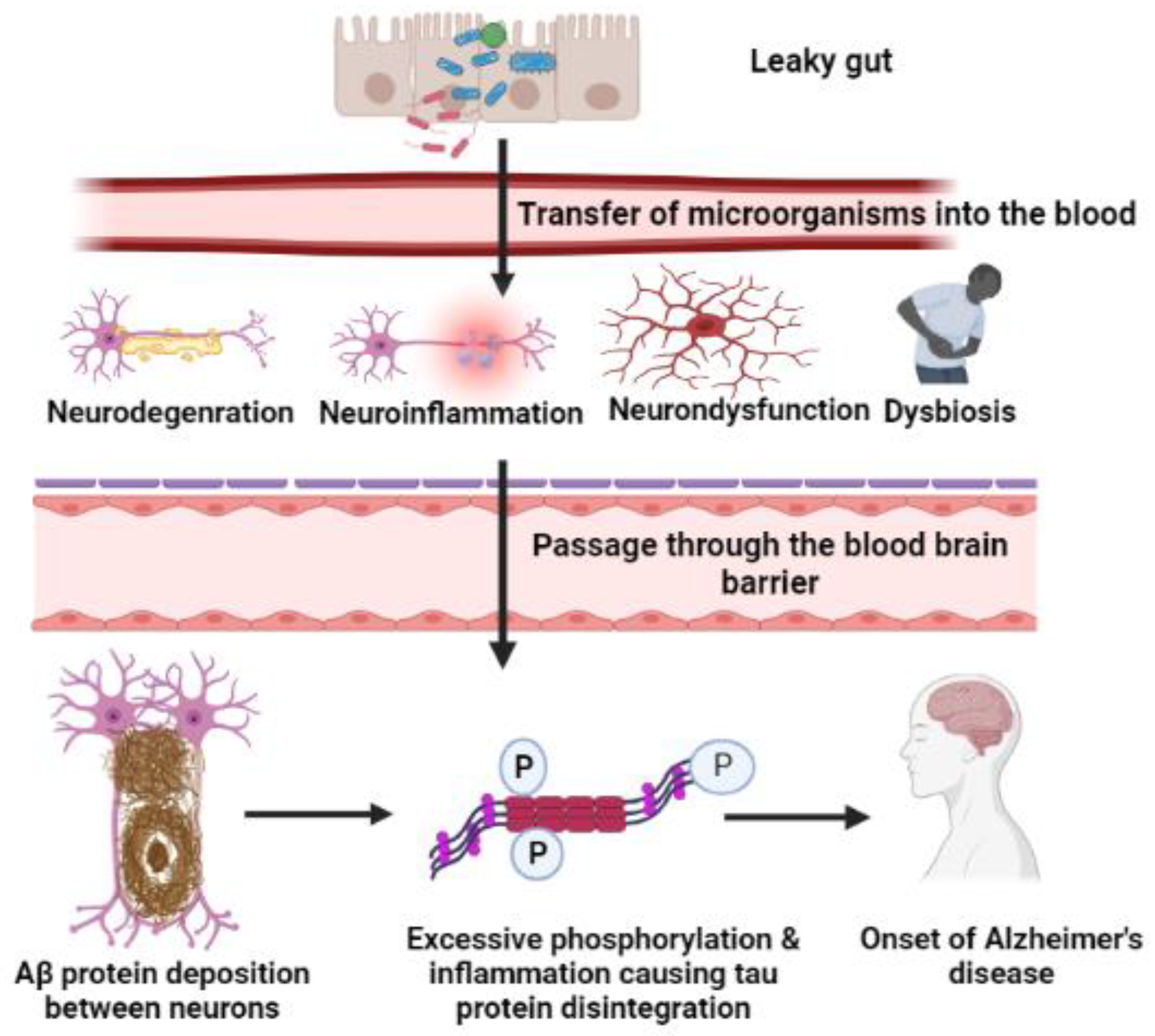
7. Microorganism-Mediated Gut Barrier Dysfunction, BBB Passage, and Activation of Chemokines/Cytokines, Leading to Breakdown of APP
7.1. Gut Inflammation and Dysbiosis
7.2. Amyloid Cascade Hypothesis
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferry, C.; Prince, M.; Brayne, C. Alzheimer’s Disease International Global prevalence of dementia: A Delphi concensus Study. Lancet 2005, 366, 2112–2117. [Google Scholar] [CrossRef]
- Kumar, A.; Sidhu, J.; Goyal, A.; Tsao, J.W. Alzheimer Disease; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Kamer, A.R.; Craig, R.G.; Dasanayake, A.P.; Brys, M.; Glodzik-Sobanska, L.; de Leon, M.J. Inflammation and Alzheimer’s disease: Possible role of periodontal diseases. Alzheimers Dement. 2008, 4, 242–250. [Google Scholar] [CrossRef]
- Sochocka, M.; Donskow-Lysoniewska, K.; Diniz, B.S.; Kurpas, D.; Brzozowska, E.; Leszek, J. The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer’s Disease—A Critical Review. Mol. Neurobiol. 2019, 56, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Landry, R.L.; Embers, M.E. Does Dementia Have a Microbial Cause? NeuroSci 2022, 3, 262–283. [Google Scholar] [CrossRef]
- Sait, A.; Angeli, C.; Doig, A.J.; Day, P.J.R. Viral Involvement in Alzheimer’s Disease. ACS Chem. Neurosci. 2021, 12, 1049–1060. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Mee, A.P.; Itzhaki, R.F. Herpes simplex virus type 1 DNA is located within Alzheimer’s disease amyloid plaques. J. Pathol. 2009, 217, 131–138. [Google Scholar] [CrossRef]
- Miklossy, J. Alzheimer’s disease--a spirochetosis? Neuroreport 1993, 4, 841–848. [Google Scholar] [CrossRef]
- Miklossy, J.; Kis, A.; Radenovic, A.; Miller, L.; Forro, L.; Martins, R.; Reiss, K.; Darbinian, N.; Darekar, P.; Mihaly, L.; et al. Beta-amyloid deposition and Alzheimer’s type changes induced by Borrelia spirochetes. Neurobiol. Aging 2006, 27, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, M.A.; Itzhaki, R.F.; Shipley, S.J.; Dobson, C.B. Herpes simplex virus infection causes cellular beta-amyloid accumulation and secretase upregulation. Neurosci. Lett. 2007, 429, 95–100. [Google Scholar] [CrossRef]
- Ramesh, G.; Alvarez, A.L.; Roberts, E.D.; Dennis, V.A.; Lasater, B.L.; Alvarez, X.; Philipp, M.T. Pathogenesis of Lyme neuroborreliosis: Borrelia burgdorferi lipoproteins induce both proliferation and apoptosis in rhesus monkey astrocytes. Eur. J. Immunol. 2003, 33, 2539–2550. [Google Scholar] [CrossRef] [PubMed]
- Ho, G.J.; Drego, R.; Hakimian, E.; Masliah, E. Mechanisms of cell signaling and inflammation in Alzheimer’s disease. Curr. Drug Targets Inflamm. Allergy 2005, 4, 247–256. [Google Scholar] [CrossRef]
- Wyss-Coray, T.; Rogers, J. Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring Harb. Perspect Med. 2012, 2, a006346. [Google Scholar] [CrossRef]
- Dosunmu, R.; Wu, J.; Basha, M.R.; Zawia, N.H. Environmental and dietary risk factors in Alzheimer’s disease. Expert Rev. Neurother. 2007, 7, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Bulgart, H.R.; Neczypor, E.W.; Wold, L.E.; Mackos, A.R. Microbial involvement in Alzheimer disease development and progression. Mol. Neurodegener. 2020, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Askarova, S.; Umbayev, B.; Masoud, A.R.; Kaiyrlykyzy, A.; Safarova, Y.; Tsoy, A.; Olzhayev, F.; Kushugulova, A. The Links Between the Gut Microbiome, Aging, Modern Lifestyle and Alzheimer’s Disease. Front. Cell Infect. Microbiol. 2020, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Rajmohan, R.; Reddy, P.H. Amyloid-Beta and Phosphorylated Tau Accumulations Cause Abnormalities at Synapses of Alzheimer’s disease Neurons. J. Alzheimers Dis. 2017, 57, 975–999. [Google Scholar] [CrossRef]
- Murphy, M.P.; LeVine, H., 3rd. Alzheimer’s disease and the amyloid-beta peptide. J. Alzheimers Dis. 2010, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, W.D.; Wang, Y.D. beta-Amyloid: The key peptide in the pathogenesis of Alzheimer’s disease. Front. Pharmacol. 2015, 6, 221. [Google Scholar] [CrossRef]
- Guo, J.; Jia, X.; Liu, Y.; Wang, S.; Cao, J.; Zhang, B.; Xiao, G.; Wang, W. Screening of Natural Extracts for Inhibitors against Japanese Encephalitis Virus Infection. Antimicrob. Agents Chemother. 2020, 64, e02373-19. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Alzheimer’s disease is a synaptic failure. Science 2002, 298, 789–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, J.; Yue, Z. Neuronal aggregates: Formation, clearance, and spreading. Dev. Cell 2015, 32, 491–501. [Google Scholar] [CrossRef]
- Wang, Y.C.; Lauwers, E.; Verstreken, P. Presynaptic protein homeostasis and neuronal function. Curr. Opin. Genet. Dev. 2017, 44, 38–46. [Google Scholar] [CrossRef] [PubMed]
- De Wilde, M.C.; Overk, C.R.; Sijben, J.W.; Masliah, E. Meta-analysis of synaptic pathology in Alzheimer’s disease reveals selective molecular vesicular machinery vulnerability. Alzheimers Dement. 2016, 12, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Spires-Jones, T.L.; Hyman, B.T. The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron 2014, 82, 756–771. [Google Scholar] [CrossRef]
- Herrera-Landero, A.; Amaya-Sanchez, L.E.; Solorzano-Santos, F.; Gordillo-Perez, M.G. Borrelia burgdorferi as a risk factor for Alzheimer’s dementia and mild cognitive impairment. Eur. Geriatr. Med. 2019, 10, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Senejani, A.G.; Maghsoudlou, J.; El-Zohiry, D.; Gaur, G.; Wawrzeniak, K.; Caravaglia, C.; Khatri, V.A.; MacDonald, A.; Sapi, E. Borrelia burgdorferi Co-Localizing with Amyloid Markers in Alzheimer’s Disease Brain Tissues. J. Alzheimers Dis. 2022, 85, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Haahr, R.; Tetens, M.M.; Dessau, R.B.; Krogfelt, K.A.; Bodilsen, J.; Andersen, N.S.; Moller, J.K.; Roed, C.; Christiansen, C.B.; Ellermann-Eriksen, S.; et al. Risk of Neurological Disorders in Patients With European Lyme Neuroborreliosis: A Nationwide, Population-Based Cohort Study. Clin. Infect. Dis. 2020, 71, 1511–1516. [Google Scholar] [CrossRef]
- Ramesh, G.; Borda, J.T.; Dufour, J.; Kaushal, D.; Ramamoorthy, R.; Lackner, A.A.; Philipp, M.T. Interaction of the Lyme disease spirochete Borrelia burgdorferi with brain parenchyma elicits inflammatory mediators from glial cells as well as glial and neuronal apoptosis. Am. J. Pathol. 2008, 173, 1415–1427. [Google Scholar] [CrossRef] [PubMed]
- Poole, S.; Singhrao, S.K.; Kesavalu, L.; Curtis, M.A.; Crean, S. Determining the presence of periodontopathic virulence factors in short-term postmortem Alzheimer’s disease brain tissue. J. Alzheimers Dis. 2013, 36, 665–677. [Google Scholar] [CrossRef]
- Díaz-Zúñiga, J.; Muñoz, Y.; Melgar-Rodríguez, S.; More, J.; Bruna, B.; Lobos, P.; Monasterio, G.; Vernal, R.; Paula-Lima, A. Serotype b of Aggregatibacter actinomycetemcomitans triggers pro-inflammatory responses and amyloid beta secretion in hippocampal cells: A novel link between periodontitis and Alzheimer’s disease? J. Oral Microbiol. 2019, 11, 1586423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellen, R.P.; Galimanas, V.B. Spirochetes at the forefront of periodontal infections. Periodontology 2000 2005, 38, 13–32. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Crincoli, V.; Laino, L.; Alovisi, M.; Sovereto, D.; Mastrangelo, F.; Lo Russo, L.; Lo Muzio, L. The role of periodontitis and periodontal bacteria in the onset and progression of Alzheimer’s disease: A systematic review. J. Clin. Med. 2020, 9, 495. [Google Scholar] [CrossRef] [PubMed]
- Miklossy, J. Historic evidence to support a causal relationship between spirochetal infections and Alzheimer’s disease. Front. Aging Neurosci. 2015, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Foschi, F.; Izard, J.; Sasaki, H.; Sambri, V.; Prati, C.; Müller, R.; Stashenko, P. Treponema denticola in disseminating endodontic infections. J. Dent. Res. 2006, 85, 761–765. [Google Scholar] [CrossRef]
- Riviere, G.R.; Riviere, K.H.; Smith, K.S. Molecular and immunological evidence of oral Treponema in the human brain and their association with Alzheimer’s disease. Oral. Microbiol. Immunol. 2002, 17, 113–118. [Google Scholar] [CrossRef]
- Zhuang, Z.Q.; Shen, L.L.; Li, W.W.; Fu, X.; Zeng, F.; Gui, L.; Lu, Y.; Cai, M.; Zhu, C.; Tan, Y.L.; et al. Gut Microbiota is Altered in Patients with Alzheimer’s Disease. J. Alzheimers Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef]
- Henderson, G.; Jaber, T.; Carpenter, D.; Wechsler, S.L.; Jones, C. Identification of herpes simplex virus type 1 proteins encoded within the first 1.5 kb of the latency-associated transcript. J. Neurovirol. 2009, 15, 439–448. [Google Scholar] [CrossRef]
- Porcellini, E.; Carbone, I.; Ianni, M.; Licastro, F. Alzheimer’s disease gene signature says: Beware of brain viral infections. Immun. Ageing 2010, 7, 16. [Google Scholar] [CrossRef]
- Wozniak, M.; Bell, T.; Denes, A.; Falshaw, R.; Itzhaki, R. Anti-HSV1 activity of brown algal polysaccharides and possible relevance to the treatment of Alzheimer’s disease. Int. J. Biol. Macromol. 2015, 74, 530–540. [Google Scholar] [CrossRef]
- Westman, G.; Blomberg, J.; Yun, Z.; Lannfelt, L.; Ingelsson, M.; Eriksson, B.M. Decreased HHV-6 IgG in Alzheimer’s Disease. Front. Neurol. 2017, 8, 40. [Google Scholar] [CrossRef] [Green Version]
- Itzhaki, R.F. Herpes simplex virus type 1 and Alzheimer’s disease: Possible mechanisms and signposts. FASEB J. 2017, 31, 3216–3226. [Google Scholar] [CrossRef] [PubMed]
- De Chiara, G.; Marcocci, M.E.; Sgarbanti, R.; Civitelli, L.; Ripoli, C.; Piacentini, R.; Garaci, E.; Grassi, C.; Palamara, A.T. Infectious agents and neurodegeneration. Mol. Neurobiol. 2012, 46, 614–638. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.M.; Clement, C.; Pogue, A.I.; Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. Pathogenic microbes, the microbiome, and Alzheimer’s disease (AD). Front. Aging Neurosci. 2014, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Cairns, D.M.; Rouleau, N.; Parker, R.N.; Walsh, K.G.; Gehrke, L.; Kaplan, D.L. A 3D human brain-like tissue model of herpes-induced Alzheimer’s disease. Sci. Adv. 2020, 6, eaay8828. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, M.J.; Venkatesan, A. Herpes simplex virus-1 encephalitis in adults: Pathophysiology, diagnosis, and management. Neurotherapeutics 2016, 13, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Herpes Simplex Virus. World Health Organization (WHO). Available online: https://www.who.int/news-room/fact-sheets/detail/herpes-simplex-virus (accessed on 3 July 2022).
- Protto, V.; Marcocci, M.E.; Miteva, M.T.; Piacentini, R.; Li Puma, D.D.; Grassi, C.; Palamara, A.T.; De Chiara, G. Role of HSV-1 in Alzheimer’s disease pathogenesis: A challenge for novel preventive/therapeutic strategies. Curr. Opin. Pharmacol. 2022, 63, 102200. [Google Scholar] [CrossRef]
- Harris, S.A.; Harris, E.A. Herpes simplex virus type 1 and other pathogens are key causative factors in sporadic Alzheimer’s disease. J. Alzheimers Dis. 2015, 48, 319–353. [Google Scholar] [CrossRef]
- Itzhaki, R.F.; Lin, W.R.; Shang, D.; Wilcock, G.K.; Faragher, B.; Jamieson, G.A. Herpes simplex virus type 1 in brain and risk of Alzheimer’s disease. Lancet 1997, 349, 241–244. [Google Scholar] [CrossRef]
- Itzhaki, R.F. Herpes simplex virus type 1 and Alzheimer’s disease: Increasing evidence for a major role of the virus. Front. Aging Neurosci. 2014, 6, 202. [Google Scholar] [CrossRef]
- Mawanda, F.; Wallace, R. Can infections cause Alzheimer’s disease? Epidemiol. Rev. 2013, 35, 161–180. [Google Scholar] [CrossRef] [Green Version]
- Carter, C. Interactions between the products of the Herpes simplex genome and Alzheimer’s disease susceptibility genes: Relevance to pathological-signalling cascades. Neurochem. Int. 2008, 52, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.A.; Harris, E.A. Molecular Mechanisms for Herpes Simplex Virus Type 1 Pathogenesis in Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Campadelli-Fiume, G.; Mirandola, P.; Menotti, L. Human herpesvirus 6: An emerging pathogen. Emerg. Infect. Dis. 1999, 5, 353–366. [Google Scholar] [CrossRef]
- Readhead, B.; Haure-Mirande, J.V.; Funk, C.C.; Richards, M.A.; Shannon, P.; Haroutunian, V.; Sano, M.; Liang, W.S.; Beckmann, N.D.; Price, N.D.; et al. Multiscale Analysis of Independent Alzheimer’s Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus. Neuron 2018, 99, 64–82.e67. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Pisa, D.; Fernandez-Fernandez, A.M.; Carrasco, L. Infection of Fungi and Bacteria in Brain Tissue From Elderly Persons and Patients With Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Pisa, D.; Marina, A.I.; Morato, E.; Rabano, A.; Carrasco, L. Fungal infection in patients with Alzheimer’s disease. J. Alzheimers Dis. 2014, 41, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Pisa, D.; Rábano, A.; Carrasco, L. Alzheimer’s disease and disseminated mycoses. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1125–1132. [Google Scholar] [CrossRef]
- Perez, J.C. The interplay between gut bacteria and the yeast Candida albicans. Gut Microbes 2021, 13, 1979877. [Google Scholar] [CrossRef]
- Garcia, R.I.; Henshaw, M.M.; Krall, E.A. Relationship between periodontal disease and systemic health. Periodontology 2000 2001, 25, 21–36. [Google Scholar] [CrossRef]
- Abbayya, K.; Puthanakar, N.Y.; Naduwinmani, S.; Chidambar, Y. Association between periodontitis and Alzheimer’s disease. N. Am. J. Med. Sci. 2015, 7, 241. [Google Scholar] [CrossRef] [Green Version]
- Tonsekar, P.P.; Jiang, S.S.; Yue, G. Periodontal disease, tooth loss and dementia: Is there a link? A systematic review. Gerodontology 2017, 34, 151–163. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef] [PubMed]
- Kamer, A.R.; Fortea, J.O.; Videla, S.; Mayoral, A.; Janal, M.; Carmona-Iragui, M.; Benejam, B.; Craig, R.G.; Saxena, D.; Corby, P.; et al. Periodontal disease’s contribution to Alzheimer’s disease progression in Down syndrome. Alzheimers Dement. 2016, 2, 49–57. [Google Scholar] [CrossRef]
- Franceschi, F.; Ojetti, V.; Candelli, M.; Covino, M.; Cardone, S.; Potenza, A.; Simeoni, B.; Gabrielli, M.; Sabia, L.; Gasbarrini, G.; et al. Microbes and Alzheimer’ disease: Lessons from H. pylori and GUT microbiota. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Varesi, A.; Pierella, E.; Romeo, M.; Piccini, G.B.; Alfano, C.; Bjorklund, G.; Oppong, A.; Ricevuti, G.; Esposito, C.; Chirumbolo, S.; et al. The Potential Role of Gut Microbiota in Alzheimer’s Disease: From Diagnosis to Treatment. Nutrients 2022, 14, 668. [Google Scholar] [CrossRef] [PubMed]
- Kinney, J.W.; Bemiller, S.M.; Murtishaw, A.S.; Leisgang, A.M.; Salazar, A.M.; Lamb, B.T. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. 2018, 4, 575–590. [Google Scholar] [CrossRef]
- Sankowski, R.; Mader, S.; Valdes-Ferrer, S.I. Systemic inflammation and the brain: Novel roles of genetic, molecular, and environmental cues as drivers of neurodegeneration. Front. Cell Neurosci. 2015, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bi, W.; Xiao, S.; Lan, X.; Cheng, X.; Zhang, J.; Lu, D.; Wei, W.; Wang, Y.; Li, H.; et al. Neuroinflammation induced by lipopolysaccharide causes cognitive impairment in mice. Sci. Rep. 2019, 9, 5790. [Google Scholar] [CrossRef] [PubMed]
- Gutsmann, T.; Muller, M.; Carroll, S.F.; MacKenzie, R.C.; Wiese, A.; Seydel, U. Dual role of lipopolysaccharide (LPS)-binding protein in neutralization of LPS and enhancement of LPS-induced activation of mononuclear cells. Infect. Immun. 2001, 69, 6942–6950. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, H.; Takeuchi, S.; Kubota, K.; Kobayashi, Y.; Kozakai, S.; Ukai, I.; Shichiku, A.; Okubo, M.; Numasaki, M.; Kanemitsu, Y.; et al. Lipopolysaccharide (LPS)-binding protein stimulates CD14-dependent Toll-like receptor 4 internalization and LPS-induced TBK1-IKK-IRF3 axis activation. J. Biol. Chem. 2018, 293, 10186–10201. [Google Scholar] [CrossRef] [Green Version]
- Balin, B.J.; Gérard, H.C.; Arking, E.J.; Appelt, D.M.; Branigan, P.J.; Abrams, J.T.; Whittum-Hudson, J.A.; Hudson, A.P. Identification and localization of Chlamydia pneumoniae in the Alzheimer’s brain. Med. Microbiol. Immunol. 1998, 187, 23–42. [Google Scholar] [CrossRef]
- Burns, A.; Jacoby, R.; Luthert, P.; Levy, R. Cause of death in Alzheimer’s disease. Age Ageing 1990, 19, 341–344. [Google Scholar] [CrossRef]
- Liu, G.; Lanham, C.; Buchan, J.R.; Kaplan, M.E. High-throughput transformation of Saccharomyces cerevisiae using liquid handling robots. PLoS ONE 2017, 12, e0174128. [Google Scholar] [CrossRef]
- Dhakal, S.; Macreadie, I. Protein homeostasis networks and the use of yeast to guide interventions in Alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 8014. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, P.; Martins, R.; Macreadie, I. Yeast as a model for studying Alzheimer’s disease. FEMS Yeast Res. 2010, 10, 961–969. [Google Scholar] [CrossRef]
- Rencus-Lazar, S.; DeRowe, Y.; Adsi, H.; Gazit, E.; Laor, D. Yeast Models for the Study of Amyloid-Associated Disorders and Development of Future Therapy. Front. Mol. Biosci. 2019, 6, 15. [Google Scholar] [CrossRef]
- Angelucci, F.; Cechova, K.; Amlerova, J.; Hort, J. Antibiotics, gut microbiota, and Alzheimer’s disease. J. Neuroinflamm. 2019, 16, 1–10. [Google Scholar] [CrossRef]
- Seo, D.O.; Holtzman, D.M. Gut Microbiota: From the Forgotten Organ to a Potential Key Player in the Pathology of Alzheimer’s Disease. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Piacentini, E.; Sanchez, B.; Arauzo, V.; Calbo, E.; Cuchi, E.; Nava, J.M. Procalcitonin levels are lower in intensive care unit patients with H1N1 influenza A virus pneumonia than in those with community-acquired bacterial pneumonia. A pilot study. J. Crit. Care 2011, 26, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Han, Y.; Zheng, Z.; Peng, G.; Liu, P.; Yue, S.; Zhu, S.; Chen, J.; Lv, H.; Shao, L.; et al. Altered Gut Microbial Metabolites in Amnestic Mild Cognitive Impairment and Alzheimer’s Disease: Signals in Host-Microbe Interplay. Nutrients 2021, 13, 228. [Google Scholar] [CrossRef]
- Galland, L. The gut microbiome and the brain. J. Med. Food 2014, 17, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Bibi, F.; Yasir, M.; Sohrab, S.S.; Azhar, E.I.; Al-Qahtani, M.H.; Abuzenadah, A.M.; Kamal, M.A.; Naseer, M.I. Link between chronic bacterial inflammation and Alzheimer disease. CNS Neurol. Disord. Drug Targets Former. Curr. Drug Targets-CNS Neurol. Disord. 2014, 13, 1140–1147. [Google Scholar] [CrossRef]
- Zhao, Y.; Jaber, V.; Lukiw, W.J. Secretory Products of the Human GI Tract Microbiome and Their Potential Impact on Alzheimer’s Disease (AD): Detection of Lipopolysaccharide (LPS) in AD Hippocampus. Front. Cell Infect. Microbiol. 2017, 7, 318. [Google Scholar] [CrossRef]
- Johnson, K.V.; Foster, K.R. Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 2018, 16, 647–655. [Google Scholar] [CrossRef]
- Bostanciklioğlu, M. Intestinal bacterial flora and Alzheimer’s disease. Neurophysiology 2018, 50, 140–148. [Google Scholar] [CrossRef]
- Li, B.; He, Y.; Ma, J.; Huang, P.; Du, J.; Cao, L.; Wang, Y.; Xiao, Q.; Tang, H.; Chen, S. Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement. 2019, 15, 1357–1366. [Google Scholar] [CrossRef]
- He, Y.; Li, B.; Sun, D.; Chen, S. Gut Microbiota: Implications in Alzheimer’s Disease. J. Clin. Med. 2020, 9, 2042. [Google Scholar] [CrossRef]
- Wang, Y.K.; Kuo, F.C.; Liu, C.J.; Wu, M.C.; Shih, H.Y.; Wang, S.S.; Wu, J.Y.; Kuo, C.H.; Huang, Y.K.; Wu, D.C. Diagnosis of Helicobacter pylori infection: Current options and developments. World J. Gastroenterol. 2015, 21, 11221–11235. [Google Scholar] [CrossRef] [PubMed]
- Bu, X.L.; Yao, X.Q.; Jiao, S.S.; Zeng, F.; Liu, Y.H.; Xiang, Y.; Liang, C.R.; Wang, Q.H.; Wang, X.; Cao, H.Y. A study on the association between infectious burden and A lzheimer’s disease. Eur. J. Neurol. 2015, 22, 1519–1525. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Cattane, N.; Galluzzi, S.; Provasi, S.; Lopizzo, N.; Festari, C.; Ferrari, C.; Guerra, U.P.; Paghera, B.; Muscio, C. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 2017, 49, 60–68. [Google Scholar] [CrossRef]
- Hudec, J.; Kobida, L.; Canigova, M.; Lacko-Bartosova, M.; Lozek, O.; Chlebo, P.; Mrazova, J.; Ducsay, L.; Bystricka, J. Production of gamma-aminobutyric acid by microorganisms from different food sources. J. Sci. Food Agric. 2015, 95, 1190–1198. [Google Scholar] [CrossRef]
- Mitew, S.; Kirkcaldie, M.T.; Dickson, T.C.; Vickers, J.C. Altered synapses and gliotransmission in Alzheimer’s disease and AD model mice. Neurobiol. Aging 2013, 34, 2341–2351. [Google Scholar] [CrossRef]
- Brown, G.C. The endotoxin hypothesis of neurodegeneration. J. Neuroinflamm. 2019, 16, 1–10. [Google Scholar] [CrossRef]
- Zhan, X.; Stamova, B.; Sharp, F.R. Lipopolysaccharide Associates with Amyloid Plaques, Neurons and Oligodendrocytes in Alzheimer’s Disease Brain: A Review. Front. Aging Neurosci. 2018, 10, 42. [Google Scholar] [CrossRef]
- Hoogland, I.C.; Houbolt, C.; van Westerloo, D.J.; van Gool, W.A.; van de Beek, D. Systemic inflammation and microglial activation: Systematic review of animal experiments. J. Neuroinflamm. 2015, 12, 114. [Google Scholar] [CrossRef]
- Miklossy, J. Emerging roles of pathogens in Alzheimer disease. Expert Rev. Mol. Med. 2011, 13, e30. [Google Scholar] [CrossRef]
- Cheon, M.S.; Bajo, M.; Gulesserian, T.; Cairns, N.; Lubec, G. Evidence for the relation of herpes simplex virus type 1 to Down syndrome and Alzheimer’s disease. Electrophoresis 2001, 22, 445–448. [Google Scholar] [CrossRef]
- Lin, W.R.; Wozniak, M.A.; Cooper, R.J.; Wilcock, G.K.; Itzhaki, R.F. Herpesviruses in brain and Alzheimer’s disease. J. Pathol. 2002, 197, 395–402. [Google Scholar] [CrossRef]
- Strandberg, T.E.; Pitkala, K.; Eerola, J.; Tilvis, R.; Tienari, P.J. Interaction of herpesviridae, APOE gene, and education in cognitive impairment. Neurobiol. Aging 2005, 26, 1001–1004. [Google Scholar] [CrossRef]
- Katan, M.; Moon, Y.P.; Paik, M.C.; Sacco, R.L.; Wright, C.B.; Elkind, M.S. Infectious burden and cognitive function: The Northern Manhattan Study. Neurology 2013, 80, 1209–1215. [Google Scholar] [CrossRef]
- Wright, C.B.; Gardener, H.; Dong, C.; Yoshita, M.; DeCarli, C.; Sacco, R.L.; Stern, Y.; Elkind, M.S. Infectious Burden and Cognitive Decline in the Northern Manhattan Study. J. Am. Geriatr. Soc. 2015, 63, 1540–1545. [Google Scholar] [CrossRef]
- Emery, D.C.; Shoemark, D.K.; Batstone, T.E.; Waterfall, C.M.; Coghill, J.A.; Cerajewska, T.L.; Davies, M.; West, N.X.; Allen, S.J. 16S rRNA next generation sequencing analysis shows bacteria in Alzheimer’s post-mortem brain. Front. Aging Neurosci. 2017, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Alkasir, R.; Li, J.; Li, X.; Jin, M.; Zhu, B. Human gut microbiota: The links with dementia development. Protein Cell 2017, 8, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Kamer, A.R.; Craig, R.G.; Pirraglia, E.; Dasanayake, A.P.; Norman, R.G.; Boylan, R.J.; Nehorayoff, A.; Glodzik, L.; Brys, M.; de Leon, M.J. TNF-alpha and antibodies to periodontal bacteria discriminate between Alzheimer’s disease patients and normal subjects. J. Neuroimmunol. 2009, 216, 92–97. [Google Scholar] [CrossRef]
- Sparks Stein, P.; Steffen, M.J.; Smith, C.; Jicha, G.; Ebersole, J.L.; Abner, E.; Dawson, D., 3rd. Serum antibodies to periodontal pathogens are a risk factor for Alzheimer’s disease. Alzheimers Dement. 2012, 8, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, H.; Eribe, E.R.; Singhrao, S.K.; Olsen, I. High throughput sequencing detect gingivitis and periodontal oral bacteria in Alzheimer’s disease autopsy brains. J. Neurosci. Res. 2019, 1, 1–10. [Google Scholar] [CrossRef]
- Beydoun, M.A.; Beydoun, H.A.; Weiss, J.; Hossain, S.; El-Hajj, Z.W.; Zonderman, A.B. Helicobacter pylori, periodontal pathogens, and their interactive association with incident all-cause and Alzheimer’s disease dementia in a large national survey. Mol. Psychiatry 2021, 26, 6038–6053. [Google Scholar] [CrossRef]
- Kanagasingam, S.; Chukkapalli, S.S.; Welbury, R.; Singhrao, S.K. Porphyromonas gingivalis is a Strong Risk Factor for Alzheimer’s Disease. J. Alzheimers Dis. Rep. 2020, 4, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Gale, S.D.; Brown, B.L.; Erickson, L.; Berrett, A.; Hedges, D.W. Association between latent toxoplasmosis and cognition in adults: A cross-sectional study. Parasitology 2015, 142, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Lovheim, H.; Olsson, J.; Weidung, B.; Johansson, A.; Eriksson, S.; Hallmans, G.; Elgh, F. Interaction between Cytomegalovirus and Herpes Simplex Virus Type 1 Associated with the Risk of Alzheimer’s Disease Development. J. Alzheimers Dis. 2018, 61, 939–945. [Google Scholar] [CrossRef]
- Hemling, N.; Roytta, M.; Rinne, J.; Pollanen, P.; Broberg, E.; Tapio, V.; Vahlberg, T.; Hukkanen, V. Herpesviruses in brains in Alzheimer’s and Parkinson’s diseases. Ann. Neurol. 2003, 54, 267–271. [Google Scholar] [CrossRef]
- Carbone, I.; Lazzarotto, T.; Ianni, M.; Porcellini, E.; Forti, P.; Masliah, E.; Gabrielli, L.; Licastro, F. Herpes virus in Alzheimer’s disease: Relation to progression of the disease. Neurobiol. Aging 2014, 35, 122–129. [Google Scholar] [CrossRef]
- Alonso, R.; Pisa, D.; Aguado, B.; Carrasco, L. Identification of fungal species in brain tissue from Alzheimer’s disease by next-generation sequencing. J. Alzheimers Dis. 2017, 58, 55–67. [Google Scholar] [CrossRef]
- Alonso, R.; Pisa, D.; Fernández-Fernández, A.M.; Rábano, A.; Carrasco, L. Fungal infection in neural tissue of patients with amyotrophic lateral sclerosis. Neurobiol. Dis. 2017, 108, 249–260. [Google Scholar] [CrossRef]
- Brandscheid, C.; Schuck, F.; Reinhardt, S.; Schafer, K.H.; Pietrzik, C.U.; Grimm, M.; Hartmann, T.; Schwiertz, A.; Endres, K. Altered Gut Microbiome Composition and Tryptic Activity of the 5xFAD Alzheimer’s Mouse Model. J. Alzheimers Dis. 2017, 56, 775–788. [Google Scholar] [CrossRef]
- Bonfili, L.; Cecarini, V.; Berardi, S.; Scarpona, S.; Suchodolski, J.S.; Nasuti, C.; Fiorini, D.; Boarelli, M.C.; Rossi, G.; Eleuteri, A.M. Microbiota modulation counteracts Alzheimer’s disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci. Rep. 2017, 7, 2426. [Google Scholar] [CrossRef] [PubMed]
- Singhrao, S.K.; Harding, A.; Poole, S.; Kesavalu, L.; Crean, S. Porphyromonas gingivalis Periodontal Infection and Its Putative Links with Alzheimer’s Disease. Mediat. Inflamm. 2015, 2015, 137357. [Google Scholar] [CrossRef]
- Minter, M.R.; Zhang, C.; Leone, V.; Ringus, D.L.; Zhang, X.; Oyler-Castrillo, P.; Musch, M.W.; Liao, F.; Ward, J.F.; Holtzman, D.M.; et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci. Rep. 2016, 6, 30028. [Google Scholar] [CrossRef] [PubMed]
- Minter, M.R.; Hinterleitner, R.; Meisel, M.; Zhang, C.; Leone, V.; Zhang, X.; Oyler-Castrillo, P.; Zhang, X.; Musch, M.W.; Shen, X.; et al. Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1DeltaE9 murine model of Alzheimer’s disease. Sci. Rep. 2017, 7, 10411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harach, T.; Marungruang, N.; Duthilleul, N.; Cheatham, V.; Mc Coy, K.D.; Frisoni, G.; Neher, J.J.; Fak, F.; Jucker, M.; Lasser, T.; et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci. Rep. 2017, 7, 41802. [Google Scholar] [CrossRef]
- Bauerl, C.; Collado, M.C.; Diaz Cuevas, A.; Vina, J.; Perez Martinez, G. Shifts in gut microbiota composition in an APP/PSS1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett. Appl. Microbiol. 2018, 66, 464–471. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Sugahara, H.; Shimada, K.; Mitsuyama, E.; Kuhara, T.; Yasuoka, A.; Kondo, T.; Abe, K.; Xiao, J.Z. Therapeutic potential of Bifidobacterium breve strain A1 for preventing cognitive impairment in Alzheimer’s disease. Sci. Rep. 2017, 7, 13510. [Google Scholar] [CrossRef]
- Nimgampalle, M.; Kuna, Y. Anti-Alzheimer Properties of Probiotic, Lactobacillus plantarum MTCC 1325 in Alzheimer’s Disease induced Albino Rats. J. Clin. Diagn. Res. 2017, 11, KC01–KC05. [Google Scholar] [CrossRef]
- Azm, S.A.N.; Djazayeri, A.; Safa, M.; Azami, K.; Ahmadvand, B.; Sabbaghziarani, F.; Sharifzadeh, M.; Vafa, M. Lactobacilli and bifidobacteria ameliorate memory and learning deficits and oxidative stress in beta-amyloid (1-42) injected rats. Appl. Physiol. Nutr. Metab. 2018, 43, 718–726. [Google Scholar] [CrossRef]
- Wu, S.C.; Cao, Z.S.; Chang, K.M.; Juang, J.L. Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat. Commun. 2017, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, M.D.S.; Nguyen, V.T.T.; Reinhardt, C.; Endres, K. Impact of Gut Microbiome Manipulation in 5xFAD Mice on Alzheimer’s Disease-Like Pathology. Microorganisms 2021, 9, 815. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Vogel, H. Drosophila models of neurodegenerative diseases. Annu. Rev. Pathol. 2009, 4, 315–342. [Google Scholar] [CrossRef]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef]
- Hazan, S. Rapid improvement in Alzheimer’s disease symptoms following fecal microbiota transplantation: A case report. J. Int. Med. Res. 2020, 48, 300060520925930. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, C.; Santangelo, R. Alzheimer’s disease and gut microbiota modifications: The long way between preclinical studies and clinical evidence. Pharmacol. Res. 2018, 129, 329–336. [Google Scholar] [CrossRef]
- Leblhuber, F.; Egger, M.; Schuetz, B.; Fuchs, D. Commentary: Effect of Probiotic Supplementation on Cognitive Function and Metabolic Status in Alzheimer’s Disease: A Randomized, Double-Blind and Controlled Trial. Front. Aging Neurosci. 2018, 10, 54. [Google Scholar] [CrossRef]
- Jiang, C.; Li, G.; Huang, P.; Liu, Z.; Zhao, B. The Gut Microbiota and Alzheimer’s Disease. J. Alzheimers Dis. 2017, 58, 1–15. [Google Scholar] [CrossRef]
- Zhan, G.; Yang, N.; Li, S.; Huang, N.; Fang, X.; Zhang, J.; Zhu, B.; Yang, L.; Yang, C.; Luo, A. Abnormal gut microbiota composition contributes to cognitive dysfunction in SAMP8 mice. Aging 2018, 10, 1257–1267. [Google Scholar] [CrossRef]
- Vassar, R.; Kandalepas, P.C. The beta-secretase enzyme BACE1 as a therapeutic target for Alzheimer’s disease. Alzheimers Res. Ther. 2011, 3, 20. [Google Scholar] [CrossRef]
- Hampel, H.; Vassar, R.; De Strooper, B.; Hardy, J.; Willem, M.; Singh, N.; Zhou, J.; Yan, R.; Vanmechelen, E.; De Vos, A. The β-secretase BACE1 in Alzheimer’s disease. Biol. Psychiatry 2021, 89, 745–756. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, R.; Guo, X.; Yan, C.; Lei, J.; Shi, Y. Structural basis of gamma-secretase inhibition and modulation by small molecule drugs. Cell 2021, 184, 521–533.e514. [Google Scholar] [CrossRef]
- Congdon, E.E.; Sigurdsson, E.M. Tau-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 399–415. [Google Scholar] [CrossRef]
- Zhang, X.; Song, W. The role of APP and BACE1 trafficking in APP processing and amyloid-beta generation. Alzheimers Res. Ther. 2013, 5, 46. [Google Scholar] [CrossRef]
- Cole, S.L.; Vassar, R. The Alzheimer’s disease β-secretase enzyme, BACE1. Mol. Neurodegener. 2007, 2, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Venugopal, C.; Demos, C.M.; Rao, K.S.; Pappolla, M.A.; Sambamurti, K. Beta-secretase: Structure, function, and evolution. CNS Neurol. Disord. Drug Targets 2008, 7, 278–294. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Osswald, H.L. BACE1 (β-secretase) inhibitors for the treatment of Alzheimer’s disease. Chem. Soc. Rev. 2014, 43, 6765–6813. [Google Scholar] [CrossRef]
- Chu, J.; Li, J.G.; Hoffman, N.E.; Madesh, M.; Praticò, D. Degradation of gamma secretase activating protein by the ubiquitin–proteasome pathway. J. Neurochem. 2015, 133, 432–439. [Google Scholar] [CrossRef]
- Wolfe, M.S. gamma-Secretase as a target for Alzheimer’s disease. Adv. Pharmacol. 2012, 64, 127–153. [Google Scholar] [CrossRef]
- Gertsik, N.; Chiu, D.; LI, Y. Complex regulation of gamma-secretase: From obligatory to modulatory subunits. Front. Aging Neurosci. 2015, 6, 342. [Google Scholar] [CrossRef]
- Haugabook, S.; Sambamurti, K. Gamma Secretase. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–10. [Google Scholar]
- Reddy, P.H.; Williams, J.; Smith, F.; Bhatti, J.S.; Kumar, S.; Vijayan, M.; Kandimalla, R.; Kuruva, C.S.; Wang, R.; Manczak, M.; et al. MicroRNAs, Aging, Cellular Senescence, and Alzheimer’s Disease. Prog. Mol. Biol. Transl. Sci. 2017, 146, 127–171. [Google Scholar] [CrossRef]
- Haass, C.; Kaether, C.; Thinakaran, G.; Sisodia, S. Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2012, 2, a006270. [Google Scholar] [CrossRef]
- Talwar, P.; Gupta, R.; Kushwaha, S.; Agarwal, R.; Saso, L.; Kukreti, S.; Kukreti, R. Viral Induced Oxidative and Inflammatory Response in Alzheimer’s Disease Pathogenesis with Identification of Potential Drug Candidates: A Systematic Review using Systems Biology Approach. Curr. Neuropharmacol. 2019, 17, 352–365. [Google Scholar] [CrossRef]
- Medeiros, R.; Baglietto-Vargas, D.; LaFerla, F.M. The role of tau in Alzheimer’s disease and related disorders. CNS Neurosci. Ther. 2011, 17, 514–524. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, R.J.; Gustafson, D.R.; Hardy, J. The genetic architecture of Alzheimer’s disease: Beyond APP, PSENs and APOE. Neurobiol. Aging 2012, 33, 437–456. [Google Scholar] [CrossRef]
- Cuccaro, M.L.; Carney, R.M.; Zhang, Y.; Bohm, C.; Kunkle, B.W.; Vardarajan, B.N.; Whitehead, P.L.; Cukier, H.N.; Mayeux, R.; George-Hyslop, P.S. SORL1 mutations in early-and late-onset Alzheimer disease. Neurol. Genet. 2016, 2, e116. [Google Scholar] [CrossRef]
- D’Argenio, V.; Sarnataro, D. New insights into the molecular bases of familial Alzheimer’s disease. J. Pers. Med. 2020, 10, 26. [Google Scholar] [CrossRef]
- Higgins, L.S.; Catalano, R.; Quon, D.; Cordell, B. Transgenic mice expressing human beta-APP751, but not mice expressing beta-APP695, display early Alzheimer’s disease-like histopathology. Ann. N. Y. Acad. Sci. 1993, 695, 224–227. [Google Scholar] [CrossRef]
- Hooli, R.E.T.B. The Genetic Basis of Alzheimer’s Disease: Findings from Genome-Wide Studies. In State, Genomics, Circuits, and Pathways in Clinical Neuropsychiatry; Lehner, B.L.M.T., Matthew, W., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 547–571. [Google Scholar]
- Eratne, D.; Loi, S.M.; Farrand, S.; Kelso, W.; Velakoulis, D.; Looi, J.C. Alzheimer’s disease: Clinical update on epidemiology, pathophysiology and diagnosis. Australas. Psychiatry 2018, 26, 347–357. [Google Scholar] [CrossRef]
- Sevigny, J.; Chiao, P.; Bussiere, T.; Weinreb, P.H.; Williams, L.; Maier, M.; Dunstan, R.; Salloway, S.; Chen, T.; Ling, Y.; et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 2016, 537, 50–56. [Google Scholar] [CrossRef]
- Kastanenka, K.V.; Bussiere, T.; Shakerdge, N.; Qian, F.; Weinreb, P.H.; Rhodes, K.; Bacskai, B.J. Immunotherapy with Aducanumab Restores Calcium Homeostasis in Tg2576 Mice. J. Neurosci. 2016, 36, 12549–12558. [Google Scholar] [CrossRef]
- Padda, I.S.; Parmar, M. Aducanumab; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Arndt, J.W.; Qian, F.; Smith, B.A.; Quan, C.; Kilambi, K.P.; Bush, M.W.; Walz, T.; Pepinsky, R.B.; Bussiere, T.; Hamann, S.; et al. Structural and kinetic basis for the selectivity of aducanumab for aggregated forms of amyloid-beta. Sci. Rep. 2018, 8, 6412. [Google Scholar] [CrossRef]
- Cummings, J.; Aisen, P.; Lemere, C.; Atri, A.; Sabbagh, M.; Salloway, S. Aducanumab produced a clinically meaningful benefit in association with amyloid lowering. Alzheimers Res. Ther. 2021, 13, 98. [Google Scholar] [CrossRef]
- Fujimoto, T.; Matsushita, Y.; Gouda, H.; Yamaotsu, N.; Hirono, S. In silico multi-filter screening approaches for developing novel β-secretase inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 2771–2775. [Google Scholar] [CrossRef]
- AID 332040. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/332040 (accessed on 10 April 2022).
- Huang, D.; Luthi, U.; Kolb, P.; Edler, K.; Cecchini, M.; Audetat, S.; Barberis, A.; Caflisch, A. Discovery of cell-permeable non-peptide inhibitors of beta-secretase by high-throughput docking and continuum electrostatics calculations. J. Med. Chem. 2005, 48, 5108–5111. [Google Scholar] [CrossRef]
- AID 239369. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/239369 (accessed on 10 April 2022).
- AID 240366. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/240366 (accessed on 10 April 2022).
- AID 240368. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/240368 (accessed on 10 April 2022).
- Rajamani, R.; Reynolds, C.H. Modeling the binding affinities of beta-secretase inhibitors: Application to subsite specificity. Bioorg. Med. Chem. Lett. 2004, 14, 4843–4846. [Google Scholar] [CrossRef]
- AID 238396. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/238396 (accessed on 10 April 2022).
- AID 240791. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/240791 (accessed on 10 April 2022).
- Jeon, S.Y.; Bae, K.; Seong, Y.H.; Song, K.S. Green tea catechins as a BACE1 (beta-secretase) inhibitor. Bioorg. Med. Chem. Lett. 2003, 13, 3905–3908. [Google Scholar] [CrossRef]
- AID 44249. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/44249 (accessed on 10 April 2022).
- AID 44250. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/44250 (accessed on 10 April 2022).
- AID 242654. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/242654 (accessed on 10 April 2022).
- Hom, R.K.; Fang, L.Y.; Mamo, S.; Tung, J.S.; Guinn, A.C.; Walker, D.E.; Davis, D.L.; Gailunas, A.F.; Thorsett, E.D.; Sinha, S.; et al. Design and synthesis of statine-based cell-permeable peptidomimetic inhibitors of human beta-secretase. J. Med. Chem. 2003, 46, 1799–1802. [Google Scholar] [CrossRef]
- AID 44238. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/44238 (accessed on 10 April 2022).
- Mekala, S.; Nelson, G.; Li, Y.M. Recent developments of small molecule gamma-secretase modulators for Alzheimer’s disease. RSC Med. Chem 2020, 11, 1003–1022. [Google Scholar] [CrossRef]
- AID 1678948. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678948 (accessed on 10 April 2022).
- AID 1678949. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678949 (accessed on 10 April 2022).
- AID 1678953. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678953 (accessed on 10 April 2022).
- AID 1678954. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678954 (accessed on 10 April 2022).
- AID 1678955. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678955 (accessed on 10 April 2022).
- AID 1678967. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678967 (accessed on 10 April 2022).
- AID 1678968. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678968 (accessed on 10 April 2022).
- AID 1678971. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1678971 (accessed on 10 April 2022).
- Czirr, E.; Leuchtenberger, S.; Dorner-Ciossek, C.; Schneider, A.; Jucker, M.; Koo, E.H.; Pietrzik, C.U.; Baumann, K.; Weggen, S. Insensitivity to Aβ42-lowering Nonsteroidal Anti-inflammatory Drugs and γ-Secretase Inhibitors Is Common among Aggressive Presenilin-1 Mutations. J. Biol. Chem. 2007, 282, 24504–24513. [Google Scholar] [CrossRef]
- AID 359781. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/359781 (accessed on 10 April 2022).
- Bakshi, P.; Jin, C.; Broutin, P.; Berhane, B.; Reed, J.; Mullan, M. Structural optimization of a CXCR2-directed antagonist that indirectly inhibits γ-secretase and reduces Aβ. Bioorg. Med. Chem. 2009, 17, 8102–8112. [Google Scholar] [CrossRef]
- AID 452857. PubChem Bioassay Record. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/452857 (accessed on 10 April 2022).
- Liu, X.Y.; Zhang, N.; Zhang, S.X.; Xu, P. Potential new therapeutic target for Alzheimer’s disease: Glucagon-like peptide-1. Eur. J. Neurosci. 2021, 54, 7749–7769. [Google Scholar] [CrossRef]
- Jia, Q.; Deng, Y.; Qing, H. Potential therapeutic strategies for Alzheimer’s disease targeting or beyond beta-amyloid: Insights from clinical trials. BioMed. Res. Int. 2014, 2014, 837157. [Google Scholar] [CrossRef]
- Yu, T.W.; Lane, H.Y.; Lin, C.H. Novel Therapeutic Approaches for Alzheimer’s Disease: An Updated Review. Int. J. Mol. Sci. 2021, 22, 8208. [Google Scholar] [CrossRef]
- Guo, L.; Xu, J.; Du, Y.; Wu, W.; Nie, W.; Zhang, D.; Luo, Y.; Lu, H.; Lei, M.; Xiao, S.; et al. Effects of gut microbiota and probiotics on Alzheimer’s disease. Transl. Neurosci. 2021, 12, 573–580. [Google Scholar] [CrossRef]
- Kruger, J.F.; Hillesheim, E.; Pereira, A.; Camargo, C.Q.; Rabito, E.I. Probiotics for dementia: A systematic review and meta-analysis of randomized controlled trials. Nutr. Rev. 2021, 79, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Walsham, N.E.; Sherwood, R.A. Fecal calprotectin in inflammatory bowel disease. Clin. Exp. Gastroenterol. 2016, 9, 21–29. [Google Scholar] [CrossRef]
- Wang, C.; Klechikov, A.G.; Gharibyan, A.L.; Warmlander, S.K.; Jarvet, J.; Zhao, L.; Jia, X.; Narayana, V.K.; Shankar, S.K.; Olofsson, A.; et al. The role of pro-inflammatory S100A9 in Alzheimer’s disease amyloid-neuroinflammatory cascade. Acta Neuropathol. 2014, 127, 507–522. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Y.; Gilthorpe, J.; van der Maarel, J.R. MRP14 (S100A9) protein interacts with Alzheimer beta-amyloid peptide and induces its fibrillization. PLoS ONE 2012, 7, e32953. [Google Scholar] [CrossRef]
- Leblhuber, F.; Geisler, S.; Steiner, K.; Fuchs, D.; Schutz, B. Elevated fecal calprotectin in patients with Alzheimer’s dementia indicates leaky gut. J. Neural Transm. 2015, 122, 1319–1322. [Google Scholar] [CrossRef] [PubMed]
- Marizzoni, M.; Provasi, S.; Cattaneo, A.; Frisoni, G.B. Microbiota and neurodegenerative diseases. Curr. Opin. Neurol. 2017, 30, 630–638. [Google Scholar] [CrossRef]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015, 39, 567–591. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef] [Green Version]
- Konig, J.; Wells, J.; Cani, P.D.; Garcia-Rodenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.J. Human Intestinal Barrier Function in Health and Disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef]
- Choi, V.M.; Herrou, J.; Hecht, A.L.; Teoh, W.P.; Turner, J.R.; Crosson, S.; Wardenburg, J.B. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat. Med. 2016, 22, 563–567. [Google Scholar] [CrossRef]
- Yu, Y.; Zhao, F. Microbiota-gut-brain axis in autism spectrum disorder. J. Genet. Genom. 2021, 48, 755–762. [Google Scholar] [CrossRef]
- Lanoiselee, H.M.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef]
- Chen, C.J.; Ou, Y.C.; Li, J.R.; Chang, C.Y.; Pan, H.C.; Lai, C.Y.; Liao, S.L.; Raung, S.L.; Chang, C.J. Infection of pericytes in vitro by Japanese encephalitis virus disrupts the integrity of the endothelial barrier. J. Virol. 2014, 88, 1150–1161. [Google Scholar] [CrossRef]
- Kim, K.S. How pathogens penetrate the blood-brain barrier: To reach the central nervous system, meningitis-causing strains of E. coli and other pathogens exploit microbial and host factors. Microbe 2014, 9, 487–492. [Google Scholar] [CrossRef]
- Xi, J.; Ding, D.; Zhu, H.; Wang, R.; Su, F.; Wu, W.; Xiao, Z.; Liang, X.; Zhao, Q.; Hong, Z.; et al. Disturbed microbial ecology in Alzheimer’s disease: Evidence from the gut microbiota and fecal metabolome. BMC Microbiol. 2021, 21, 226. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, N.; Quigley, C.; Paw, C.; Butala, S.; Schneider, E.; Pandiyan, P. Role of Short Chain Fatty Acids in Controlling Tregs and Immunopathology During Mucosal Infection. Front. Microbiol. 2018, 9, 1995. [Google Scholar] [CrossRef]
- Zhang, Y.; Geng, R.; Tu, Q. Gut microbial involvement in Alzheimer’s disease pathogenesis. Aging 2021, 13, 13359–13371. [Google Scholar] [CrossRef]
- Dhillon, S. Aducanumab: First Approval. Drugs 2021, 81, 1437–1443. [Google Scholar] [CrossRef]
- Vaz, M.; Silva, V.; Monteiro, C.; Silvestre, S. Role of Aducanumab in the Treatment of Alzheimer’s Disease: Challenges and Opportunities. Clin. Interv. Aging 2022, 17, 797–810. [Google Scholar] [CrossRef]
- Fleisher, A.S.; Raman, R.; Siemers, E.R.; Becerra, L.; Clark, C.M.; Dean, R.A.; Farlow, M.R.; Galvin, J.E.; Peskind, E.R.; Quinn, J.F.; et al. Phase 2 safety trial targeting amyloid beta production with a gamma-secretase inhibitor in Alzheimer disease. Arch. Neurol. 2008, 65, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Salloway, S.; Sperling, R.; Fox, N.C.; Blennow, K.; Klunk, W.; Raskind, M.; Sabbagh, M.; Honig, L.S.; Porsteinsson, A.P.; Ferris, S.; et al. Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer’s disease. N. Engl. J. Med. 2014, 370, 322–333. [Google Scholar] [CrossRef]
- Wischik, C.; Staff, R. Challenges in the conduct of disease-modifying trials in AD: Practical experience from a phase 2 trial of Tau-aggregation inhibitor therapy. J. Nutr. Health Aging 2009, 13, 367–369. [Google Scholar] [CrossRef]
- Doody, R.S.; Farlow, M.; Aisen, P.S. Phase 3 trials of solanezumab and bapineuzumab for Alzheimer’s disease. N. Engl. J. Med. 2014, 370, 1460. [Google Scholar] [CrossRef]
- Khan, T.K.; Alkon, D.L. Alzheimer’s Disease Cerebrospinal Fluid and Neuroimaging Biomarkers: Diagnostic Accuracy and Relationship to Drug Efficacy. J. Alzheimers Dis. 2015, 46, 817–836. [Google Scholar] [CrossRef]
- Saumier, D.; Aisen, P.S.; Gauthier, S.; Vellas, B.; Ferris, S.H.; Duong, A.; Suhy, J.; Oh, J.; Lau, W.; Garceau, D.; et al. Lessons learned in the use of volumetric MRI in therapeutic trials in Alzheimer’s disease: The ALZHEMED (Tramiprosate) experience. J. Nutr. Health Aging 2009, 13, 370–372. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Mirra, S.S.; Heyman, A.; McKeel, D.; Sumi, S.M.; Crain, B.J.; Brownlee, L.M.; Vogel, F.S.; Hughes, J.P.; van Belle, G.; Berg, L. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 1991, 41, 479–486. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, D.; Zeng, Y.; Huang, T.Y.; Xu, H.; Zhao, Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 1–37. [Google Scholar] [CrossRef]
- Vigasova, D.; Nemergut, M.; Liskova, B.; Damborsky, J. Multi-pathogen infections and Alzheimer’s disease. Microb. Cell Fact. 2021, 20, 25. [Google Scholar] [CrossRef] [PubMed]
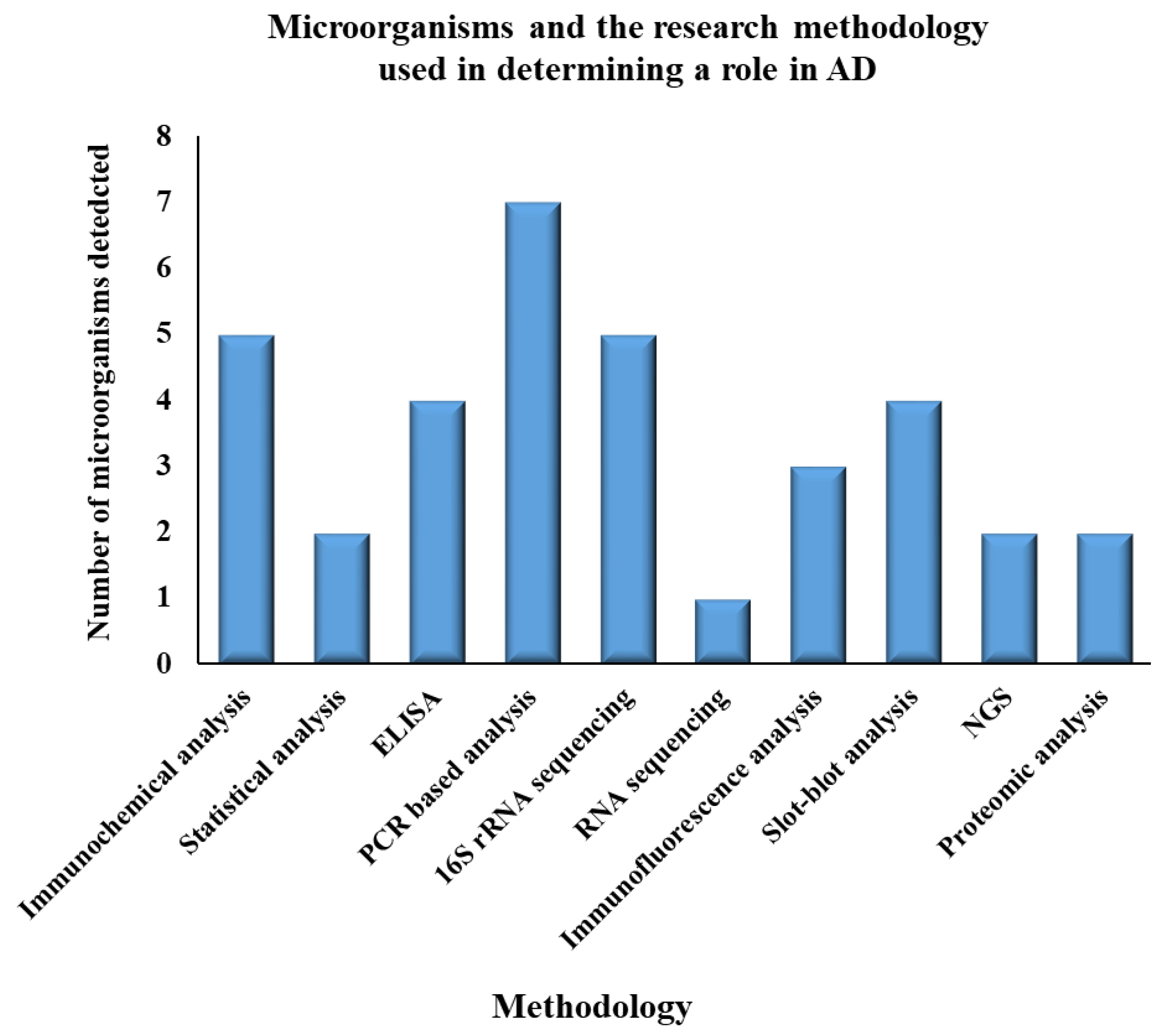
| S.No. | Name of Microorganisms | Role in AD | Methodology | Year | Reference |
|---|---|---|---|---|---|
| 1 | Helicobacter pylori | ▪ Upregulation of Tau protein ▪ Neuroinflammation ▪ Neurotoxicity | ▪ Immunochemical analysis ▪ Statistical analysis ▪ ELISA ▪ PCR based analysis | 2005, 2013, 2015 | [92,102,103,104] |
| 2 | Actinobacteria | Modulation of specific gene expressions | 16S rRNA sequencing | 2017 | [105] |
| 3 | Firmicutes | Cerebral Aβ amyloidosis | 16S rRNA sequencing | 2017 | [105,106] |
| 4 | Proteobacteria | Release of proinflammatory cytokines | 16S rRNA sequencing | 2017 | [105] |
| 5 | Treponemadenticola | Inflammation in brain | ▪ 16S rDNA sequencing ▪ ELISA | 2009, 2012, 2019, 2020 | [107,108,109,110] |
| 6 | Gingivitis bacteria | ▪ Alteration of tau and ubiquitin ▪ Leading to chronic inflammation | 16S rDNA sequencing | 2019 | [109] |
| 7 | Porphyromonas gingivalis | ▪ Cleavage of Tau protein ▪ Phosphorylate neuronal tau | Serological studies (ELISA) | 2009 | [107,111] |
| 8 | Tannerella forsythia | Inflammation | ▪ PCR-based analysis ▪ Immunochemical analysis | 2019 | [31] |
| 9 | Toxoplasma gondii | ▪ Inflammation in CNS ▪ Activation of T cells ▪ Oxidative stress | Immunochemical analysis | 2015 | [112] |
| 10 | Herpes simplex Virus (HSV) | ▪ Neurotrophic ▪ Neuroinvasive ▪ Brain infection ▪ Encephalitis in adults ▪ Meningitis in neonates ▪ Lesions in infected individuals’ brains | ▪ ELISA ▪ PCR-based analysis ▪ RNA sequencing ▪ Statistical analysis | 2015, 2003, 2002, 2014, 2018 | [56,101,104,113,114,115] |
| 11 | Saccharomyces cerevisiae | Oxidative stress | ▪ Immunofluorescence analysis ▪ Slot-blot analysis, ▪ Proteomic analysis ▪ PCR- based analysis | 2014, 2017 | [58,59,116,117] |
| 12 | Malassezia species | Neuroinflammatory responsethrough T-cell activation | ▪ PCR-based analysis ▪ Immunochemical analysis ▪ Immunofluorescence analysis ▪ Slot- blot analysis | 2014 | [58,59] |
| 13 | Candida species | Activation of NF-κB leading to proinflammatory cytokines IL-6, IL-1, and TNF | ▪ Proteomic analysis ▪ PCR-based analysis ▪ Immunochemical analysis ▪ Slot- blot analysis ▪ NGS | 2014, 2017 | [58,59,116,117] |
| 14 | Cladosporium cryptococcus | Neuroinflammation | ▪ PCR-based analysis ▪ Slot- blot analysis ▪ NGS | 2014, 2017 | [58,59,116,117] |
| S.No. | Name of Microorganisms | Treatment to Model | Result of Experiment | Year | Reference |
|---|---|---|---|---|---|
| 1 | APOE−/− mice | Porphyromonas gingivalis active invasion | Activation in APOE−/− mice brain | 2015 | [120] |
| 2 | APP/PS1 mice | Antibiotic treatment to Tg mice | ▪ Changes in composition of intestinal microbiome ▪ Decrease concentration in Aβ deposition and soluble Aβ increased ▪ Reactive gliosis decreased ▪ Expansion of Lachnospiraceae ▪ Aβ deposition in aged Tg mice is reduced ▪ Plaque-localized microglia and astrocytes reduced in antibiotic-exposed mice | 2016, 2017, 2018, | [121,122,123,124] |
| 3 | 5XFAD mice | Composition of fecal microbiota changed along with age | ▪ Enzyme trypsin in human fecal matter decreased ▪ APP expression seen in the gut tissues. | 2017 | [118] |
| 4 | 3XTg-AD mice | Mice treated with probiotics | ▪ Concentration of plasma influenced in the case of inflammatory cytokines and gut hormones ▪ Accumulation of Aβ aggregates and brain damage reduced | 2017 | [119] |
| 5 | AD mouse model (ICV injection of Aβ) | Bifidobacterium breve strain A1given orally | Function of hippocampus enhanced | 2017 | [125] |
| 6 | AD rat model (IP injection of D- galactose) | Mice treated with Lactobacillus plantarum MTCC 1325 | Acetylcholine level is restored, formation of Aβ plaque attenuated | 2017 | [126] |
| 7 | AD rat model (intrahippocampal injection of Aβ) | Mice treated with Lactobacillus and Bifidobacterium | Learning problems and oxidative stress observed | 2018 | [127] |
| 8 | Transgenic flies: Drosophila | Drosophila exposed to Enterobacteria infection | ▪ Neuroinflammation is reduced when hemocytes are genetically depleted ▪ Neurodegeneration reduced | 2017 | [128] |
| 9 | 5XFAD mice | Multi-antibiotic treatment - Gentamicin (0.1251 mg/mL) - Vancomycin (0.0635 mg/mL) - Metronidazole (0.25 mg/mL) - Neomycin (0.0635 mg/mL) - Ampicillin (0.1251 mg/mL) - Kanamycin (0.3753 mg/mL) - Colistin (7,506,000 U/mL) - Cefoperazone (0.1251 mg/mL) | ▪ Manipulation of the gut microbiome ▪ Mice revealed antidiabetic properties ▪ Effect on nest building quality | 2021 | [129] |
| No. | IUPAC Names of Compounds | CID | SID | AID | IC50 (µM) | EC50 (µM) | Ki Value (µM) | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 1-N-[(2S,3S,5R)-3-amino-6-(4-fluoroani-li-no)-5-methyl-6-oxo-1-phenylhexan-2-yl] -3-N,3-N-dipropyl- benzene-1,3-dicarboxamide | 5494423 | 103477859 | 332040 | 0.026 | [165,166] | ||
| 2 | 3-benzoyl-N-[(2S,3R)-4-(cyclopropylam-ino)-3-hydroxy-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]benzamide | 5327063 | 103478410 | 332040 | 0.098 | [165,166] | ||
| 3 | 3-N-[(2S,3R)-4-(cyclopropylamino)-3-hydroxy-1-phenylbutan-2-yl]-5-[me-thyl(methylsulfonyl)amino]-1-N-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | 5287532 | 103492819 | 332040 | 0.015 | [165,166] | ||
| 4 | [3-[2-(5-aminopentylamino)-2-oxoethoxy]-5-[[(1R)-1-(4-fluorophenyl)ethyl]carbamoyl]phenyl] phenylmethanesulfonate | 448772 | 103511935 | 332040 | 1.4 | [165,166] | ||
| 5 | 1-carbazol-9-yl-3-[4-(3-carbazol-9-yl-2-hydroxypropyl)piperazin-1-yl]propan-2-ol | 2729022 | 103564170 | 332040 | 7 | [165,166] | ||
| 6 | 1-[4-(3-carbazol-9-yl-2-hydroxypropyl)piperazin-1-yl]-3-(3,6-dichlorocarbazol-9-yl)propan-2-ol | 2802372 | 103564215 | 332040 | 5 | [165,166] | ||
| 7 | 1-(3-acetylphenyl)-3-(3-ethylsulfanyl-1,2,4-thiadiazol-5-yl)urea | 11198061 | 103459736 | 239369 | 28.8 | [167,168] | ||
| 8 | 5-[[3,5-bis(trifluoromethyl)phenyl]car-bam-oylamino]-2-(dimethylamino)-N-(3-morpholin-4-ylpropyl)benzamide | 4243074 | 103459765 | 239369 | 8.1 | [167,168] | ||
| 9 | 5-[[3,5-bis(trifluoromethyl)phenyl]car-bam-oylamino]-2-(dimethylamino)-N-(3-methoxypropyl)benzamide | 5040262 | 103459791 | 239369 | 9.4 | [167,168] | ||
| 10 | 5-[(3-chlorophenyl)carbamoylamino]-2-(dimethylamino)-N-(3-ethoxypropyl)benzamide | 3948694 | 103460050 | 239369 | 9.8 | [167,168] | ||
| 11 | 1-(3-acetylphenyl)-3-(3-ethylsulfanyl-1,2,4-thiadiazol-5-yl)urea | 11198061 | 103459736 | 240366 | 0.0114 | [167,169] | ||
| 12 | 5-[[3,5-bis(trifluoromethyl)phenyl]car-bam-oylamino]-2-(dimethylamino)-N-(3-methoxypropyl)benzamide | 5040262 | 103459791 | 240366 | 27 | [167,169] | ||
| 13 | 5-[(3-chlorophenyl)carbamoylamino]-2-(dimethylamino)-N-(3-ethoxypropyl)benzamide | 3948694 | 103460050 | 240366 | 0.0233 | [167,169] | ||
| 14 | 5-[[3,5-bis(trifluoromethyl)phenyl]car-bam-oylamino]-2-(dimethylamino)-N-(3-morpholin-4-ylpropyl)benzamide | 4243074 | 103459765 | 240368 | 0.0035 | [167,170] | ||
| 15 | tert-butyl N-[1-[[(4S)-8-[[1-(benzyl-amino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-4-methylsulfanyl-1-oxobutan-2-yl]carbamate | 44305543 | 103256754 | 238396 | 5.808 | [171,172] | ||
| 16 | tert-butyl N-[(2S)-1-[[1-[[(4S)-8-[[1-(benzylamino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-4-methylsulfonyl-1-oxobutan-2-yl]amino]-3-methyl-1-oxobutan-2-yl]carbamate | 44305867 | 103257369 | 238396 | 0.008 | [171,172] | ||
| 17 | tert-butyl N-[(2S)-1-[[1-[[(4S)-8-[[1-(benzylamino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-4-methylsulfanyl-1-oxobutan-2-yl]amino]-3-methyl-1-oxobutan-2-yl]carbamate | 44305868 | 103257370 | 238396 | 0.0025 | [171,172] | ||
| 18 | tert-butyl N-[(2S)-1-[[1-[[(4S)-8-[[1-(benzylamino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-3-methylsulfonyl-1-oxopropan-2-yl]amino]-3-methyl-1-oxobutan-2-yl]carbamate | 44305869 | 103257371 | 238396 | 0.0094 | [171,172] | ||
| 19 | tert-butyl N-[(2S)-1-[[4-amino-1-[[(4S)-8-[[1-(benzylamino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-1,4-dioxobutan-2-yl]amino]-3-methyl-1-oxobutan-2-yl]carbamate | 44305870 | 103257372 | 238396 | 0.0059 | [171,172] | ||
| 20 | tert-butyl N-[(2S)-1-[[4-amino-1-[[(4S)-8-[[1-(benzylamino)-1-oxopropan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-1,4-dioxobutan-2-yl]amino]-3-methyl-1-oxobutan-2-yl]carbamate | 44305918 | 103257473 | 238396 | 0.0614 | [171,172] | ||
| 21 | tert-butyl N-[(2S)-1-[[1-[[(4S)-8-[[1-(benzylamino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-3-methylsulfanyl-1-oxopropan-2-yl]amino]-3-methyl-1-oxobutan-2-yl]carbamate | 44305946 | 103257506 | 238396 | 0.0501 | [171,172] | ||
| 22 | (4S)-4-amino-5-[[(2S)-1-[[(2S)-4-amino-1-[[(4S,5S,7R)-8-[[(2S)-1-[[(2S)-4-carboxy-1-[[(1S)-1-carboxy-2-phenylethyl]amino]-1-oxobutan-2-yl]amino]-1-oxopropan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-1,4-dioxobutan-2-yl]amino]-3-methyl-1-oxobutan-2-yl]amino]-5-oxopentanoic acid | 445649 | 103282992 | 238396 | 0.0016 | [171,172] | ||
| 23 | tert-butyl N-[(3S)-1-amino-5-[[(4S)-8-[[1-(benzylamino)-1-oxopropan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-1,4-dioxopentan-3-yl]carbamate | 44394717 | 103451640 | 238396 | 22.423 | [171,172] | ||
| 24 | tert-butyl N-[(3S)-1-amino-5-[[(4S)-8-[[1-(benzylamino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-1,4-dioxopentan-3-yl]carbamate | 44394718 | 103451641 | 238396 | 3.134 | [171,172] | ||
| 25 | tert-butyl N-[(2R)-4-[[(4S)-8-[[1-(benzylamino)-3-methyl-1-oxobutan-2-yl]amino]-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]amino]-1-methylsulfonyl-3-oxobutan-2-yl]carbamate | 44394719 | 103451642 | 238396 | 1.129 | [171,172] | ||
| 26 | (2R,5S)-5-[[(2S)-2-[[(2R,4S,5S)-5-[[(2S)-2-[[(2S)-2-[[(2S)-2-amino-4-carboxybutanoyl]amino]-4-methylpentanoyl]amino]-3-carboxypropanoyl]amino]-4-hydroxy-2,7-dimethyloctanoyl]amino]-3-methylbutanoyl]amino]-2-benzyl-4-oxooctanedioic acid | 44394812 | 103451744 | 238396 | 0.00032 | [171,172] | ||
| 27 | 1-N-[(2S,3S,5R)-5-(benzylcarbamoyl)-1-(3,5-difluorophenyl)-3-hydroxyheptan-2-yl]-3-N,3-N-dipropylbenzene-1,3-dicarboxamide | 10121628 | 103222903 | 240791 | 1.4 | [171,173] | ||
| 28 | 1-N-[(2S,3S,5R)-1-(3,5-difluorophenyl)-5-[[(3S,5R)-3,5-dimethoxycyclohexyl]carbamoyl]-3-hydroxyheptan-2-yl]-3-N,3-N-dipropylbenzene-1,3-dicarboxamide | 11354322 | 103222904 | 240791 | 6.6 | [171,173] | ||
| 29 | 5-[[(2R,4S,5S)-6-(3,5-difluorophenyl)-5-[[3-(dipropylcarbamoyl)benzoyl]amino]-2-ethyl-4-hydroxyhexanoyl]amino]pentanoic acid | 10282555 | 103222935 | 240791 | 0.4 | [171,173] | ||
| 30 | 3-[[(2R,4S,5S)-6-(3,5-difluorophenyl)-5-[[3-(dipropylcarbamoyl)benzoyl]amino]-2-ethyl-4-hydroxyhexanoyl]amino]propanoic acid | 10167359 | 240791 | 2.8 | - | [171,173] | ||
| 31 | 4-[[[(2R,4S,5S)-6-(3,5-difluorophenyl)-5-[[3-(dipropylcarbamoyl)benzoyl]amino]-2-ethyl-4-hydroxyhexanoyl]amino]methyl]cyclohexane-1-carboxylic acid | 9809811 | 103223159 | 240791 | 0.05 | [171,173] | ||
| 32 | [(2S,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 65056 | 103359341 | 44249 | 4.5 | [95,96] | ||
| 33 | (2R,3S)-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol | 65084 | 103359373 | 44249 | 2.5 | [174,175] | ||
| 34 | [(2R,3S)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 5276890 | 103359448 | 44249 | 1.8 | [174,175] | ||
| 35 | (2S,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol | 182232 | 103359466 | 44249 | 28 | [174,175] | ||
| 36 | (2S,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol | 182232 | 103359466 | 44249 | 23 | [174,175] | ||
| 37 | (2S,3S)-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol | 10425234 | 103359508 | 44249 | 2.4 | [174,175] | ||
| 38 | [(2S,3R)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 6419835 | 103359509 | 44249 | 6 | [174,175] | ||
| 39 | [(2S,3S)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 2824823 | 103359580 | 44249 | 1.6 | [174,175] | ||
| 40 | (2S,3R)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol | 73160 | 103474775 | 44249 | 30 | [174,175] | ||
| 41 | (2R,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol | 9064 | 123094711 | 44249 | 35 | [174,175] | ||
| 42 | [(2S,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 65056 | 103359341 | 44250 | 5.3 | [174,176] | ||
| 43 | [(2R,3S)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 5276890 | 103359448 | 44250 | 0.17 | [174,176] | ||
| 44 | [(2S,3S)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl] 3,4,5-trihydroxybenzoate | 2824823 | 103359580 | 44250 | 0.21 | [174,176] | ||
| 45 | 5-[[3,5-bis(trifluoromethyl)phenyl]carbamoylamino]-2-(dimethyl- amino)-N-(3-morpholin-4-ylpropyl)benzamide | 4243074 | 103459765 | 242654 | 0.0578 | [167,177] | ||
| 46 | 5-[(3-chlorophenyl)carbamoylamino]-2-(dimethylamino)-N-(3-ethoxypropyl)benzamide | 3948694 | 103460050 | 242654 | 0.097 | [167,177] | ||
| 47 | (4R)-4-[[(2R)-2-[[(2R)-2-[[(3S)-4-[(4-butoxybenzoyl)amino]-3-hydroxy-6-methylheptanoyl]amino]-3-methylbutanoyl]amino]propanoyl]amino]-5-[[(1R)-1-carboxy-2-phenylethyl]amino]-5-oxopentanoic acid | 44296588 | 103236975 | 44238 | 27 | [178,179] | ||
| 48 | methyl 4-[[(2R)-2-[[(3S)-3-hydroxy-6-methyl-4-[[(2S)-2-[[(2S)-3-methyl-2-[(2-methylpropan-2-yl)oxycarbonylamino]butanoyl]amino]-4-methylsulfanylbutanoyl]amino]heptanoyl]amino]-3-methylbutanoyl]amino]butanoate | 44296483 | 103236754 | 44238 | 47 | [178,179] | ||
| 49 | 3-[[(2R)-2-[[(3S)-3-hydroxy-6-methyl-4-[[(2S)-2-[[(2S)-3-methyl-2-[(2-methylpropan-2-yl)oxycarbonylamino]butanoyl]amino]-4-methylsulfanylbutanoyl]amino]heptanoyl]amino]-3-methylbutanoyl]amino]propanoic acid | 44296454 | 103236696 | 44238 | 30 | [178,179] | ||
| 50 | 4-[[[(2R)-2-[[(3S)-3-hydroxy-6-methyl-4-[[(2S)-2-[[(2S)-3-methyl-2-[(2-methylpropan-2-yl)oxycarbonylamino]butanoyl]amino]-4-methylsulfanylbutanoyl]amino]heptanoyl]amino]-3-methylbutanoyl]amino]methyl]benzoic acid | 44296453 | 103236695 | 44238 | 4 | [178,179] |
| No. | IUPAC Names of Compounds | CID | SID | AID | IC50 (µM) | EC50 (µM) | PMID | Reference |
| 1 | (2R)-2-[4-[(R)-(4-fluorophenyl)-(4-methylpiperidin-1-yl)methyl]-3-[4-(trifluoromethyl)phenyl]phenyl]propanoic acid | 53493001 | 163326581 | 1678948 | 0.15 | 33479693 | [180,181] | |
| 2 | (2R)-2-[4-[(R)-(4-fluorophenyl)-(4-methylpiperidin-1-yl)methyl]-3-[4-(trifluoromethyl)phenyl]phenyl]propanoic acid | 53493001 | 163326581 | 1678949 | 0.17 | 33479693 | [180,182] | |
| 3 | 5-[4-[8-(3,4-dichlorophenyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyridin-3-yl]-2-methoxyphenyl]-2-methyl-1,3-oxazole | 66608062 | 318390383 | 1678953 | 0.24 | 33479693 | [180,183] | |
| 4 | 6-chloro-3′-[3-methoxy-4-(2-methyl-1,3-oxazol-5-yl)phenyl]-1-(2,2,2-trifluoroethyl)spiro[5H-4,1-benzoxazepine-3,8′-6,7-dihydro-5H-[1,2,4]triazolo[4,3-a]pyridine]-2-one | 57521199 | 336903002 | 1678953 | 0.03 | 33479693 | [180,183] | |
| 5 | 3′-[3-methoxy-4-(2-methyl-1,3-oxazol-5-yl)phenyl]-1-(2,2,2-trifluoroethyl)-6-(trifluoromethyl)spiro[5H-4,1-benzoxazepine-3,8′-6,7-dihydro-5H-[1,2,4]triazolo[4,3-a]pyridine]-2-one | 127049947 | 336903439 | 1678953 | 0.031 | 33479693 | [180,183] | |
| 6 | 2-[(8R)-3-[3-methoxy-4-(2-methyl-1,3-oxazol-5-yl)phenyl]-8-(3,4,5-trifluorophenoxy)-6,7-dihydro-5H-[1,2,4]triazolo[4,3-a]pyridin-8-yl]propan-2-ol | 56837196 | 336903440 | 1678953 | 0.026 | 33479693 | [180,183] | |
| 7 | 2-[8-(3-chloro-4-fluorophenoxy)-3-[3-methoxy-4-(2-methyl-1,3-oxazol-5-yl)phenyl]-6,7-dihydro-5H-[1,2,4]triazolo[4,3-a]pyridin-8-yl]propan-2-ol | 66603958 | 336905149 | 1678953 | 0.038 | 33479693 | [180,183] | |
| 8 | 8-[(3,4-difluorophenyl)methyl]-3-[3-methoxy-4-(2-methyl-1,3-oxazol-5-yl)phenyl]-N-(2,2,2-trifluoroethyl)-6,7-dihydro-5H-[1,2,4]triazolo[4,3-a]pyridine-8-carboxamide | 66604236 | 336906847 | 1678953 | 0.075 | 33479693 | [180,183] | |
| 9 | 8-(4-fluoro-2-methylphenyl)-N-(1-methylindazol-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-amine | 142607770 | 404669659 | 1678954 | 1.2 | 33479693 | [180,184] | |
| 10 | 8-(4-fluoro-2-methylphenyl)-N-(1-methyl-4,5,6,7-tetrahydroindazol-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-amine | 142607900 | 404671937 | 1678954 | 0.38 | 33479693 | [180,184] | |
| 11 | N-[3,3-difluoro-1-(3-methyl-1,2,4-thiadiazol-5-yl)piperidin-4-yl]-8-(2,3,4-trifluorophenyl)-[1,2,4]triazolo[1,5-a]pyridin-2-amine | 58397894 | 461518428 | 1678954 | 0.143 | 33479693 | [180,184] | |
| 12 | (3E)-1-[(1S)-1-(4-fluorophenyl)ethyl]-3-[[3-methoxy-4-(4-methylimidazol-1-yl)phenyl]methylidene]piperidin-2-one | 11560787 | 104244932 | 1678955 | 0.093 | 33479693 | [180,185] | |
| 13 | (2R)-2-[4-[(R)-(4-fluorophenyl)-(4-methylpiperidin-1-yl)methyl]-3-[4-(trifluoromethyl)phenyl]phenyl]propanoic acid | 53493001 | 163326581 | 1678967 | 0.146 | 33479693 | [180,186] | |
| 14 | (2R)-2-[4-[(R)-(4-fluorophenyl)-(4-methylpiperidin-1-yl)methyl]-3-[4-(trifluoromethyl)phenyl]phenyl]propanoic acid | 53493001 | 163326581 | 1678968 | 0.064 | 33479693 | [180,187] | |
| 15 | (2R)-2-[3-chloro-4-(2,2,2-trifluoroethoxy)-5-[4-(trifluoromethyl)phenyl]phenyl]-3-cyclobutylpropanoic acid | 71623049 | 318386441 | 1678971 | 0.067 | 33479693 | [180,188] | |
| 16 | (2S)-2-[[(2S)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino]-N-[(7S)-5-methyl-6-oxo-7H-benzo[d][1]benzazepin-7-yl]propanamide | 10435235 | 103537689 | 359781 | 0.000119 | 17573346 | [189,190] | |
| 17 | 1-(2-fluorophenyl)-3-(4-methoxy-2-nitrophenyl)urea | 2992227 | 103719617 | 452857 | 10 | 19853461 | [191,192] | |
| 18 | 1-(3,4-dimethylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226838 | 103719560 | 452857 | 15 | 19853461 | [191,192] | |
| 19 | 1-(2-hydroxy-4-nitrophenyl)-3-(2,3,4-trifluorophenyl)urea | 46226843 | 103719569 | 452857 | 8 | 19853461 | [191,192] | |
| 20 | 1-(3,5-dimethylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226853 | 103719584 | 452857 | 2 | 19853461 | [191,192] | |
| 21 | 1-(2-ethyl-6-methylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226857 | 103719593 | 452857 | 7 | 19853461 | [191,192] | |
| 22 | 1-(2-ethyl-6-propan-2-ylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226858 | 103719594 | 452857 | 3 | 19853461 | [191,192] | |
| 23 | 1-(2-fluorophenyl)-3-(2-methoxy-4-nitrophenyl)urea | 310495 | 103719605 | 452857 | 5 | 19853461 | [191,192] | |
| 24 | 1-(3-fluorophenyl)-3-(2-methoxy-4-nitrophenyl)urea | 4580964 | 103719606 | 452857 | 12 | 19853461 | [191,192] | |
| 25 | 1-(2-hydroxy-4-nitrophenyl)-3-(2,4,5-trifluorophenyl)urea | 46226866 | 103719607 | 452857 | 10 | 19853461 | [191,192] | |
| 26 | 1-(2-hydroxy-4-nitrophenyl)-3-(2-methylphenyl)urea | 22011319 | 103719608 | 452857 | 2 | 19853461 | [191,192] | |
| 27 | 1-(2,5-dimethylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226837 | 103719559 | 452857 | 10 | 19853461 | [191,192] | |
| 28 | 1-(2-ethylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 21184795 | 103719618 | 452857 | 0.09 | 19853461 | [191,192] | |
| 29 | 1-(2-hydroxy-4-nitrophenyl)-3-(2-propylphenyl)urea | 46196416 | 103719619 | 452857 | 0.1 | 19853461 | [191,192] | |
| 30 | 1-(2-hydroxy-4-nitrophenyl)-3-[2-(2-methylpropyl)phenyl]urea | 46226874 | 103719620 | 452857 | 1 | 19853461 | [191,192] | |
| 31 | 1-(3-ethylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226884 | 103719634 | 452857 | 4 | 19853461 | [191,192] | |
| 32 | 1-(4-ethylphenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226885 | 103719635 | 452857 | 3 | 19853461 | [190,191] | |
| 33 | 1-(2-hydroxy-4-nitrophenyl)-3-(2-methyl-6-propan-2-ylphenyl)urea | 46226886 | 103719636 | 452857 | 5 | 19853461 | [190,191] | |
| 34 | 1-(2,4-dibromophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 22011336 | 103719649 | 452857 | 0.8 | 19853461 | [191,192] | |
| 35 | 1-(2,4-dichlorophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 22011310 | 103719650 | 452857 | 0.3 | 19853461 | [191,192] | |
| 36 | 1-(2-hydroxy-4-nitrophenyl)-3-(2,3,4-trichlorophenyl)urea | 46226814 | 103719527 | 452857 | 3 | 19853461 | [191,192] | |
| 37 | 1-(2-bromophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 3854666 | 103547644 | 452857 | 0.5 | 19853461 | [191,192] | |
| 38 | 1-[2,6-di(propan-2-yl)phenyl]-3-(2-hydroxy-4-nitrophenyl)urea | 46226772 | 103719467 | 452857 | 8 | 19853461 | [191,192] | |
| 39 | 1-(2,4-difluorophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226786 | 103719483 | 452857 | 15 | 19853461 | [191,192] | |
| 40 | 1-(2,5-dichlorophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 4592749 | 103719484 | 452857 | 0.5 | 19853461 | [191,192] | |
| 41 | 1-(3,4-dichlorophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226804 | 103719508 | 452857 | 0.7 | 19853461 | [191,192] | |
| 42 | 1-(3,4-difluorophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 46226805 | 103719509 | 452857 | 8 | 19853461 | [191,192] | |
| 43 | 1-(2-hydroxy-4-nitrophenyl)-3-(4-propylphenyl)urea | 21184782 | 103719510 | 452857 | 4 | 19853461 | [191,192] | |
| 44 | 1-(2-fluorophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 21184605 | 103719525 | 452857 | 10 | 19853461 | [191,192] | |
| 45 | 1-(2-chlorophenyl)-3-(2-hydroxy-4-nitrophenyl)urea | 9883044 | 103719526 | 452857 | 0.6 | 19853461 | [191,192] | |
| 46 | 1-(2-hydroxy-4-nitrophenyl)-3-phenylurea | 3618472 | 103195215 | 452857 | 12 | 19853461 | [191,192] | |
| 47 | 1-(2-hydroxy-4-nitrophenyl)-3-(2,4,5-trichlorophenyl)urea | 46226815 | 103719528 | 452857 | 3 | 19853461 | [191,192] | |
| 48 | 1-[2-ethyl-6-(2-methylpropyl)phenyl]-3-(2-hydroxy-4-nitrophenyl)urea | 46226816 | 103719529 | 452857 | 15 | 19853461 | [191,192] | |
| 49 | 1-(2-fluorophenyl)-3-(4-nitrophenyl)urea | 2985799 | 2985799 | 452857 | 30 | 19853461 | [191,192] | |
| 50 | 1-(2-hydroxy-4-nitrophenyl)-3-(2,4,6-trifluorophenyl)urea | 46226828 | 103719547 | 452857 | 15 | 19853461 | [191,192] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, P.; Lee, Y.-H.; Panday, H.; Kant, S.; Bajwa, N.; Parashar, R.; Jha, S.K.; Jha, N.K.; Nand, P.; Lee, S.-S.; et al. Implications of Microorganisms in Alzheimer’s Disease. Curr. Issues Mol. Biol. 2022, 44, 4584-4615. https://doi.org/10.3390/cimb44100314
Yadav P, Lee Y-H, Panday H, Kant S, Bajwa N, Parashar R, Jha SK, Jha NK, Nand P, Lee S-S, et al. Implications of Microorganisms in Alzheimer’s Disease. Current Issues in Molecular Biology. 2022; 44(10):4584-4615. https://doi.org/10.3390/cimb44100314
Chicago/Turabian StyleYadav, Pardeep, Yeon-Hee Lee, Hrithika Panday, Shubham Kant, Neha Bajwa, Ritika Parashar, Saurabh Kumar Jha, Niraj Kumar Jha, Parma Nand, Sang-Soo Lee, and et al. 2022. "Implications of Microorganisms in Alzheimer’s Disease" Current Issues in Molecular Biology 44, no. 10: 4584-4615. https://doi.org/10.3390/cimb44100314
APA StyleYadav, P., Lee, Y. -H., Panday, H., Kant, S., Bajwa, N., Parashar, R., Jha, S. K., Jha, N. K., Nand, P., Lee, S. -S., & Jha, A. K. (2022). Implications of Microorganisms in Alzheimer’s Disease. Current Issues in Molecular Biology, 44(10), 4584-4615. https://doi.org/10.3390/cimb44100314









