Pharmacophore-Model-Based Virtual-Screening Approaches Identified Novel Natural Molecular Candidates for Treating Human Neuroblastoma
Abstract
1. Introduction
2. Results
2.1. Development of Pharmacophore Model
2.2. Validation of Pharmacophore Model
2.3. Dataset Identification for Pharmacophore Screening
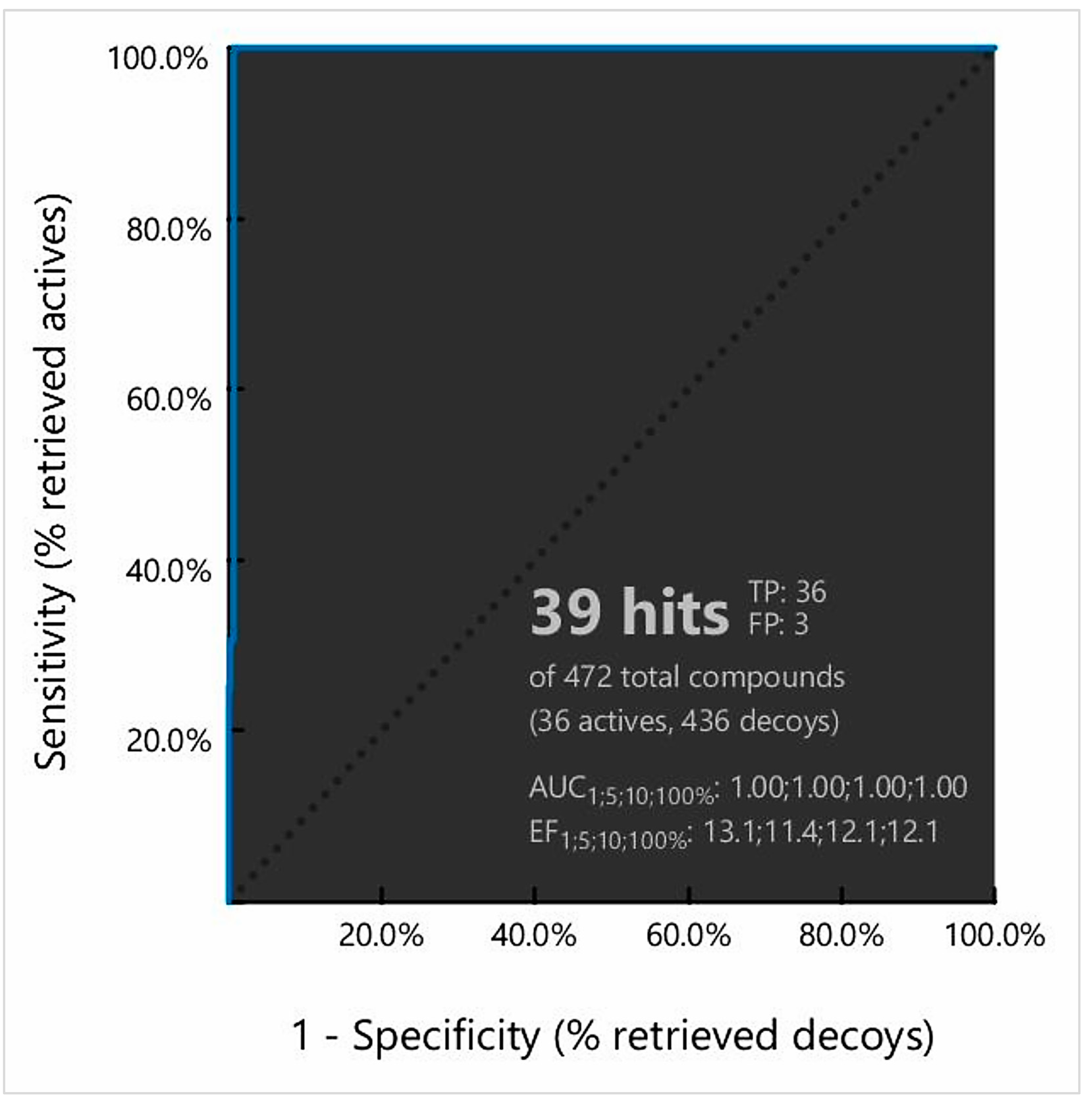
2.4. Pharmacophore Model-Based Virtual Screening
2.5. Docking-Based Virtual Screening
2.5.1. Binding-Site Detection and Ligand-Protein Interaction Prediction
2.5.2. Molecular Docking
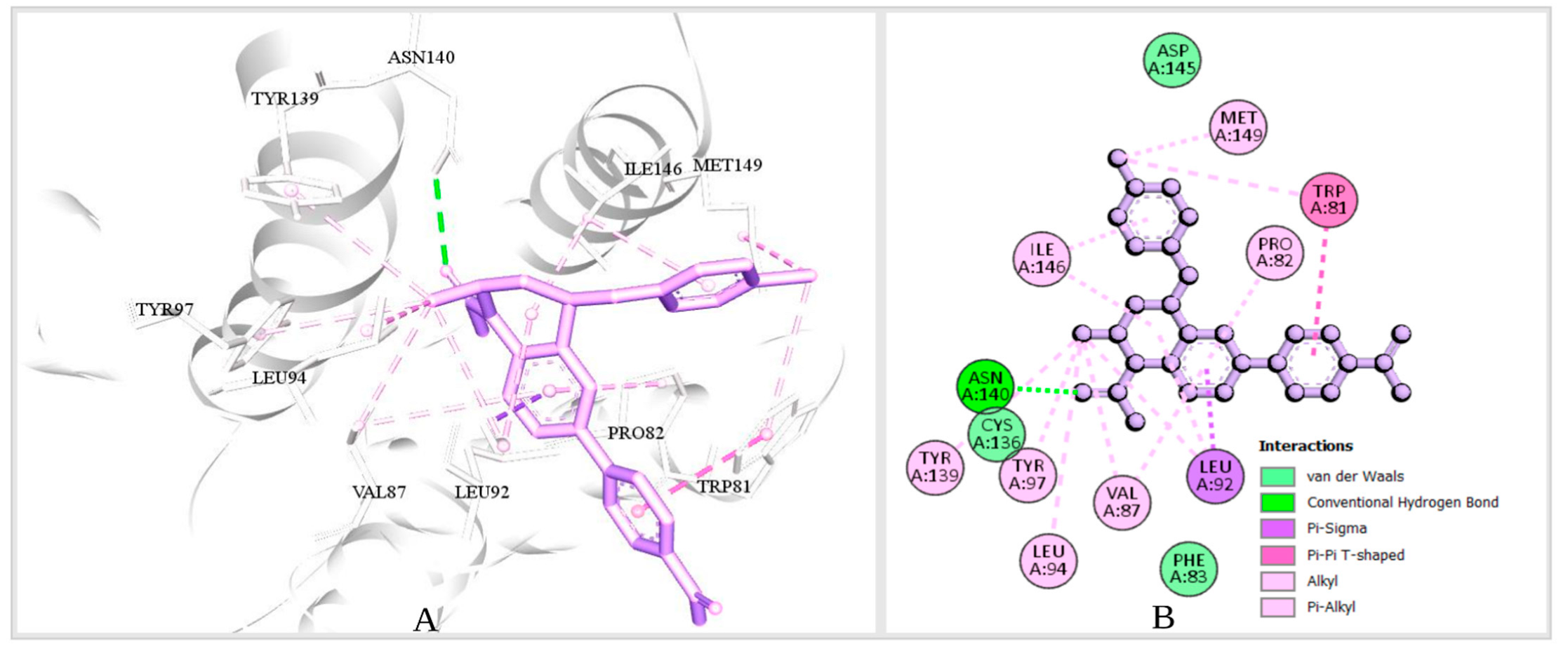
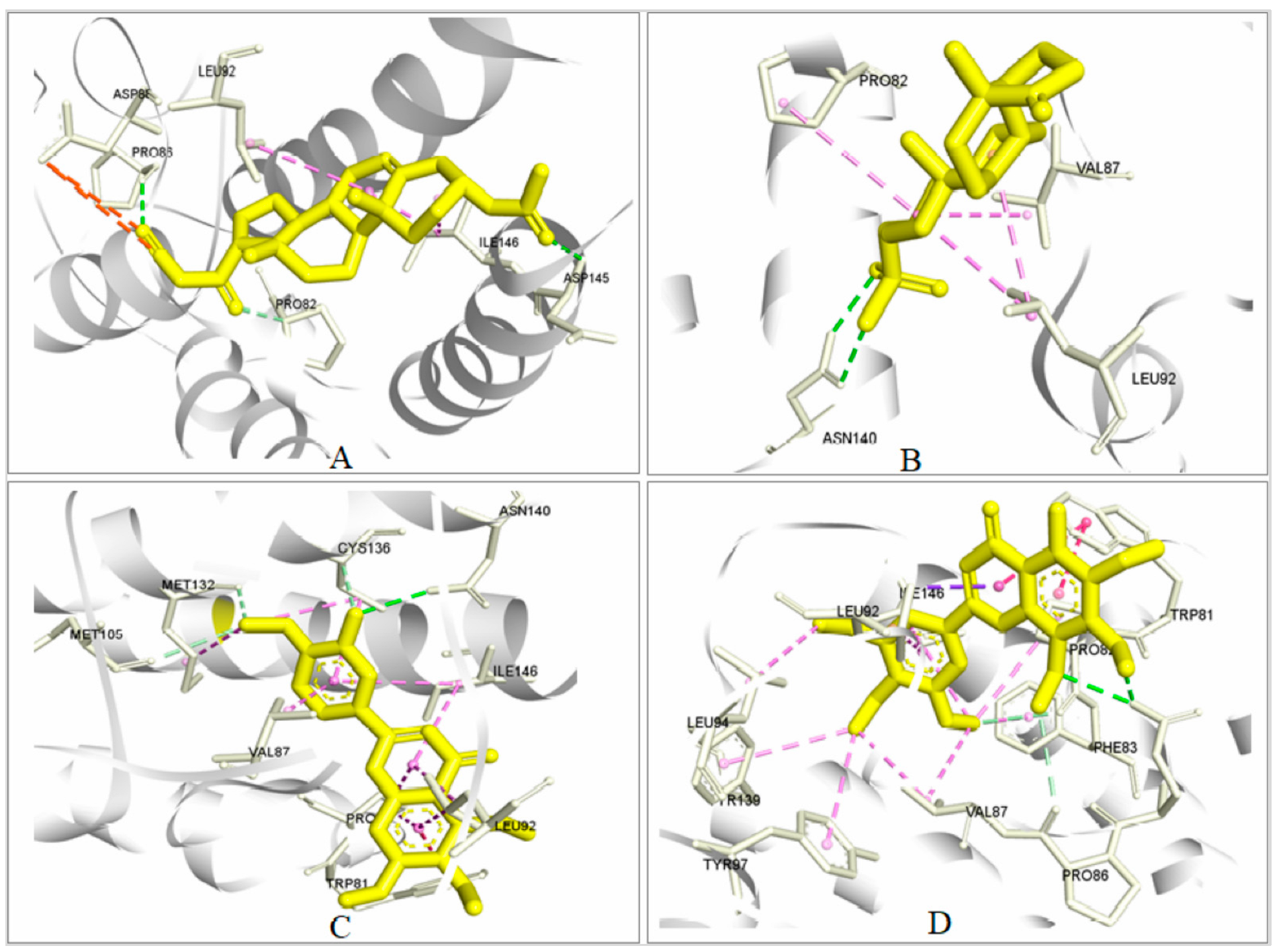
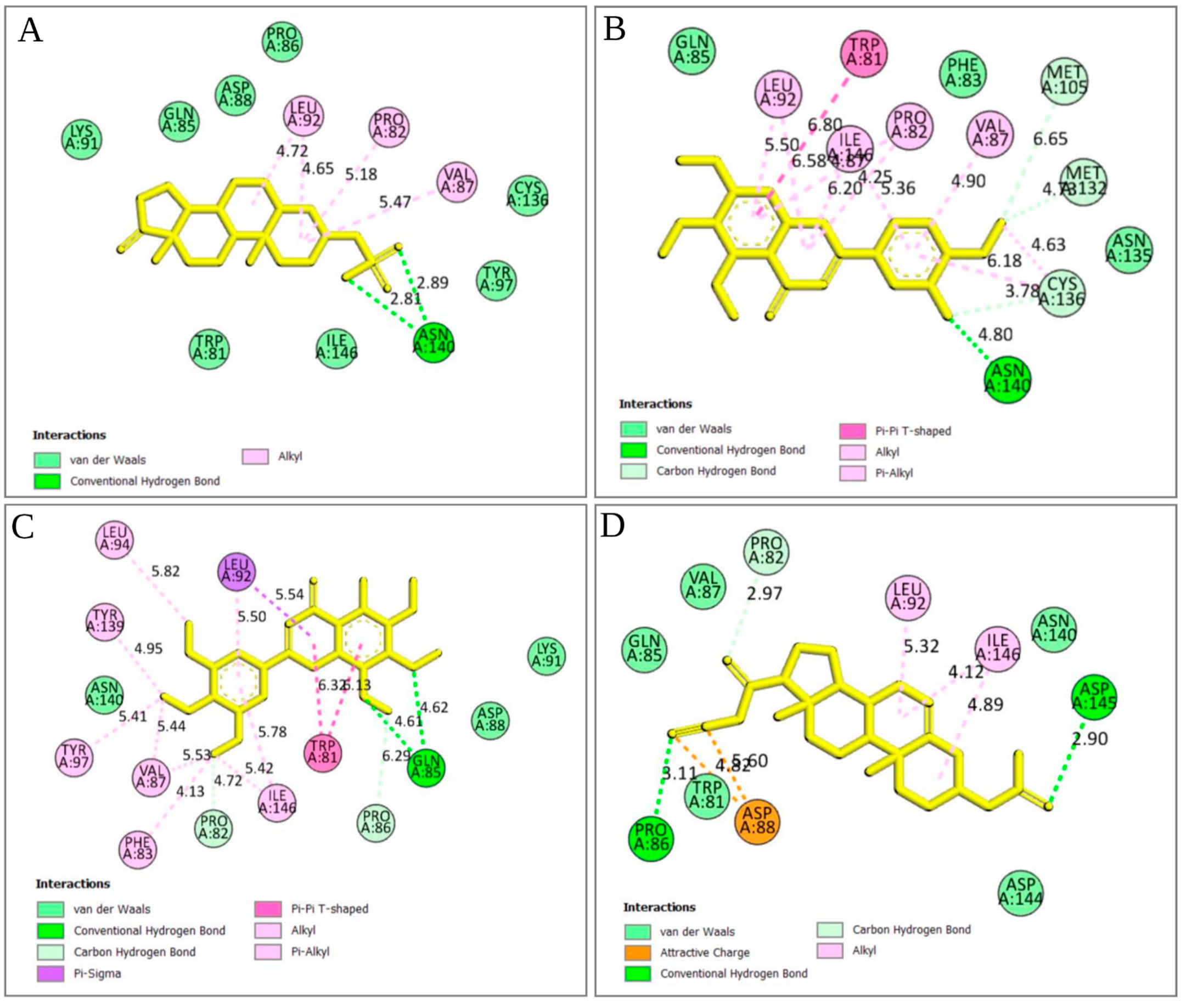
2.5.3. Analysis of Protein–Ligand Interaction
2.5.4. Pharmacophore Features Analysis
2.6. MM/GBSA Analysis
2.7. Pharmacokinetics and Toxicity Properties Analysis
2.7.1. Pharmacokinetic (ADME) Evaluation
2.7.2. Toxicity Determination
2.8. Protein–Ligand Interaction Analysis through Dynamic Simulation
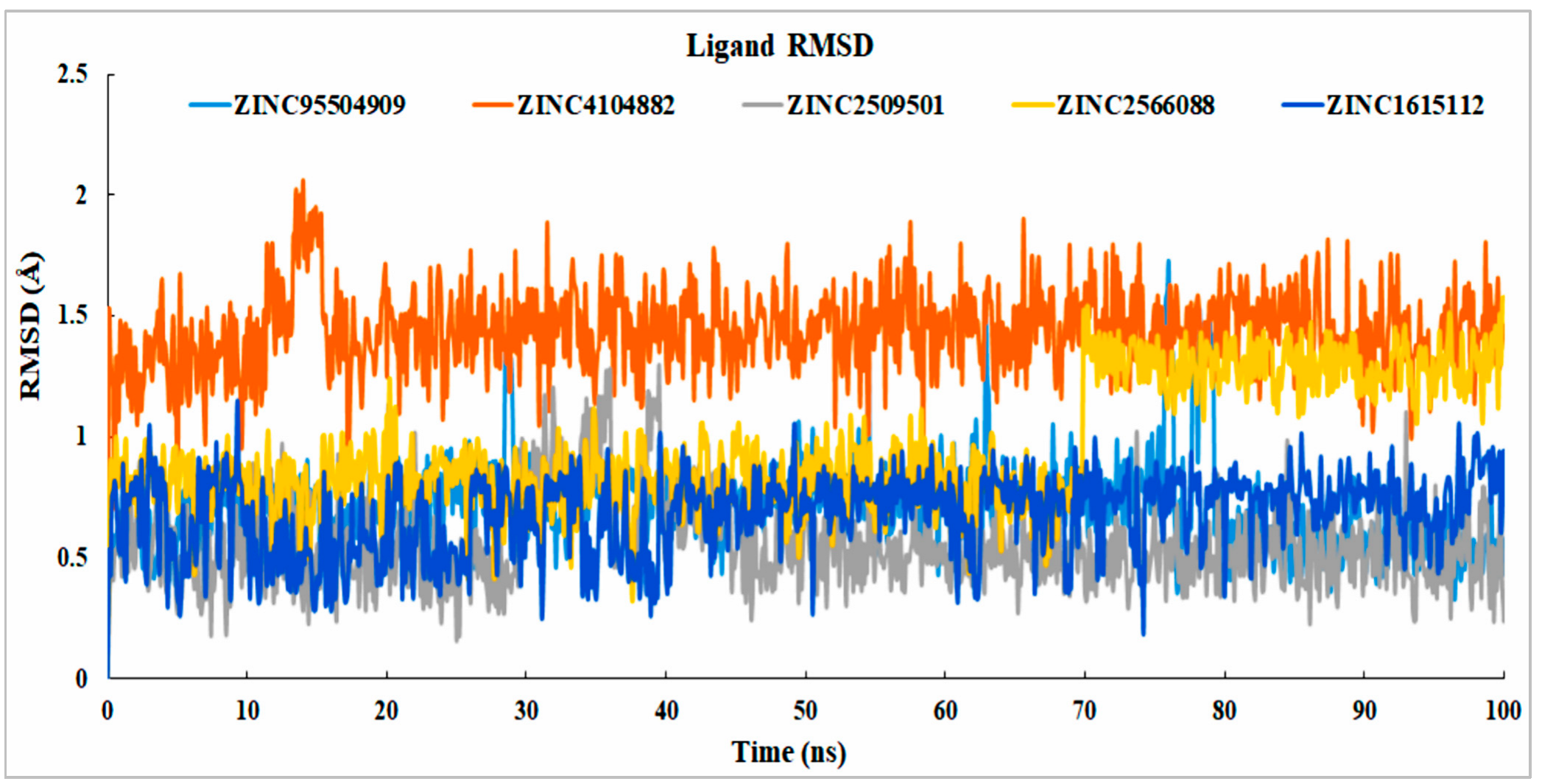
2.8.1. RMSD Value Analysis
Analysis of Protein RMSD
Analysis of Ligand RMSD
2.8.2. RMSF Analysis
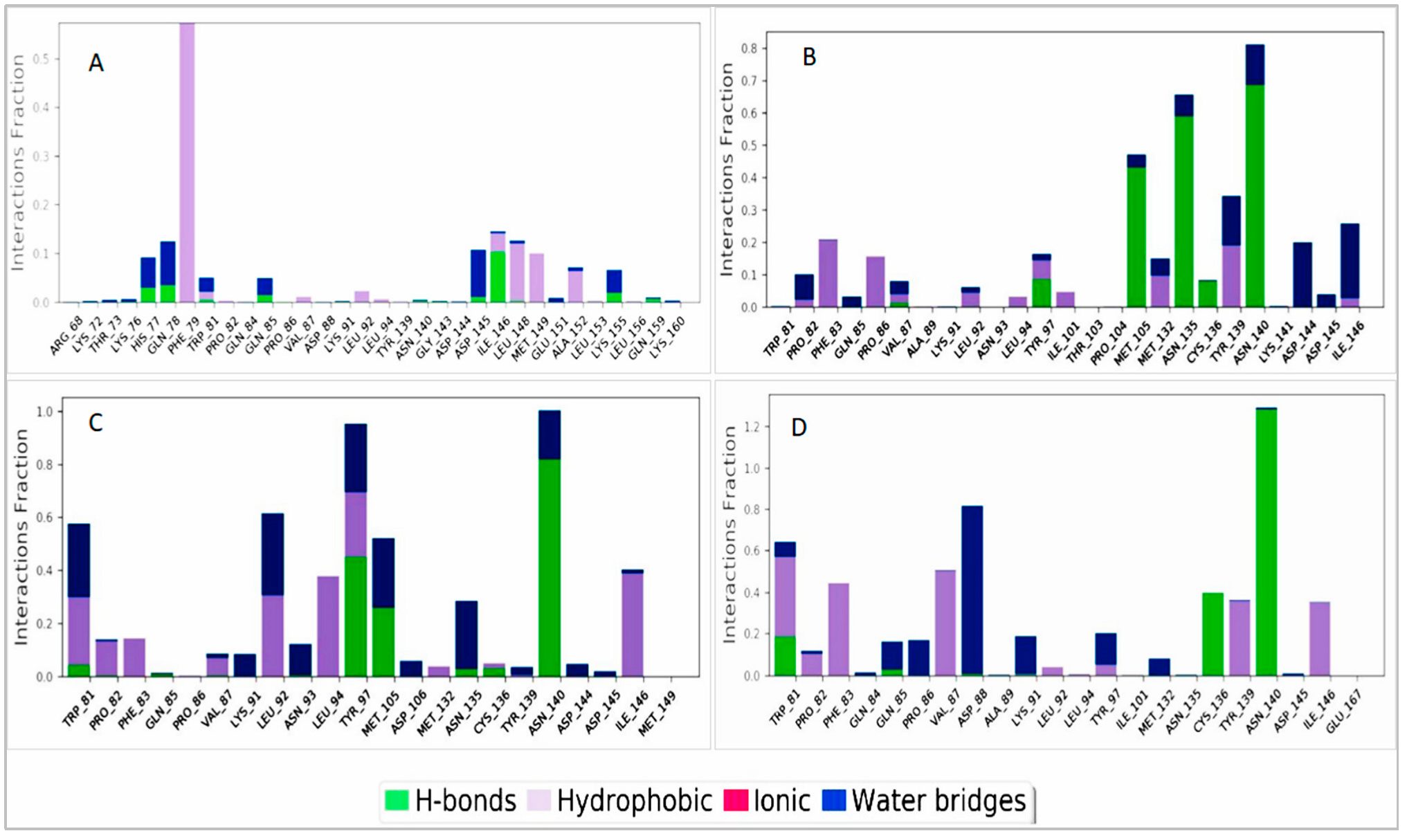
2.8.3. Protein–Ligand Interaction Analysis
3. Discussion
4. Materials and Methods
4.1. Generation of the Pharmacophore Model
4.1.1. Structure-Based Pharmacophore Modeling
4.1.2. Model Validation
4.1.3. Dataset Preparation and Virtual Screening
4.2. Molecular-Docking-Based Virtual Screening
4.2.1. Protein and Ligand Preparation
4.2.2. Grid Preparation and Active-Site Identification
4.2.3. Binding Affinity Determination by Docking
4.3. MM/GBSA Data Calculation
4.4. Toxicity Class Prediction and Evaluation for Drug Candidate
4.4.1. Pharmacokinetic and Pharmacodynamic Evaluation
4.4.2. Toxicity Analysis
4.5. Molecular Dynamic Simulation
4.5.1. Preparation of the Protein and Ligand for Simulation
4.5.2. Trajectory File Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stathis, A.; Bertoni, F. BET Proteins as Targets for Anticancer Treatment. Cancer Discov. 2018, 8, 24–36. [Google Scholar] [CrossRef]
- Morgado-Pascual, J.L.; Rayego-Mateos, S.; Tejedor, L.; Suarez-Alvarez, B.; Ruiz-Ortega, M. Bromodomain and Extraterminal Proteins as Novel Epigenetic Targets for Renal Diseases. Front. Pharmacol. 2019, 10, 1315. [Google Scholar] [CrossRef]
- Kim, S.K.; Liu, X.; Park, J.; Um, D.; Kilaru, G.; Chiang, C.M.; Kang, M.; Huber, K.M.; Kang, K.; Kim, T.K. Functional Coordination of BET Family Proteins Underlies Altered Transcription Associated with Memory Impairment in Fragile X Syndrome. Sci. Adv. 2021, 7, eabf7346. [Google Scholar] [CrossRef]
- Wyce, A.; Ganji, G.; Smitheman, K.N.; Chung, C.W.; Korenchuk, S.; Bai, Y.; Barbash, O.; Le, B.C.; Craggs, P.D.; McCabe, M.T.; et al. BET Inhibition Silences Expression of MYCN and BCL2 and Induces Cytotoxicity in Neuroblastoma Tumor Models. PLoS ONE 2013, 8, e72967. [Google Scholar] [CrossRef] [PubMed]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target C-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Penn, L.Z. Reflecting on 25 Years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Huang, M.; Weiss, W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef] [PubMed]
- Wartiovaara, K.; Barnabe-Heider, F.; Miller, F.D.; Kaplan, D.R. N-Myc Promotes Survival and Induces S-Phase Entry of Postmitotic Sympathetic Neurons. J. Neurosci. 2002, 22, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.S.; Clarke, S.; Veschi, V.; Thiele, C.J. Targeting MYCN in Pediatric and Adult Cancers. Front. Oncol. 2021, 10, 3252. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, W.W.; Zejnullahu, K.; Bradner, J.E.; Varmus, H. Sensitivity of Human Lung Adenocarcinoma Cell Lines to Targeted Inhibition of BET Epigenetic Signaling Proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 19408–19413. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Gong, Y.; Ma, Y.; Lu, K.; Lu, X.; Pierce, L.A.; Thompson, R.C.; Muller, S.; Knapp, S.; Wang, J. Inhibition of BET Bromodomain Targets Genetically Diverse Glioblastoma. Clin. Cancer Res. 2013, 19, 1748–1759. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.I.; Robson, S.C.; Chung, C.W.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET Recruitment to Chromatin as an Effective Treatment for MLL-Fusion Leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Cotterill, S.J.; Parker, L.; Malcolm, A.J.; Reid, M.; More, L.; Craft, A.W. Incidence and Survival for Cancer in Children and Young Adults in the North of England, 1968-1995: A Report from the Northern Region Young Persons’ Malignant Disease Registry. Br. J. Cancer 2000, 83, 397–403. [Google Scholar] [CrossRef]
- Schwab, M.; Varmus, H.E.; Bishop, J.M.; Grzeschik, K.H.; Naylor, S.L.; Sakaguchi, A.Y.; Brodeur, G.; Trent, J. Chromosome Localization in Normal Human Cells and Neuroblastomas of a Gene Related to C-Myc. Nature 1984, 308, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M. Neuroblastoma: Biological Insights into a Clinical Enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Maris, J.M. The Biologic Basis for Neuroblastoma Heterogeneity and Risk Stratification. Curr. Opin. Pediatr. 2005, 17, 7–13. [Google Scholar] [CrossRef]
- Thiele, C.J.; Reynolds, C.P.; Israel, M.A. Decreased Expression of N-Myc Precedes Retinoic Acid-Induced Morphological Differentiation of Human Neuroblastoma. Nature 1985, 313, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Vakoc, C.R. The Mechanisms behind the Therapeutic Activity of BET Bromodomain Inhibition. Mol. Cell 2014, 54, 728. [Google Scholar] [CrossRef] [PubMed]
- Zuber, J.; Shi, J.; Wang, E.; Rappaport, A.R.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; et al. RNAi Screen Identifies Brd4 as a Therapeutic Target in Acute Myeloid Leukaemia. Nature 2011, 478, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Speleman, F.; Park, J.R.; Henderson, T.O. Neuroblastoma: A Tough Nut to Crack. Am. Soc. Clin. Oncol. Educ. B. 2016, 36, e548–e557. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, S.; Bouback, T.A.; Samad, A.; Nur, S.M.; Alam, R.; Abdullah-Al-Mamun, M.; Nain, Z.; Imon, R.R.; Talukder, M.E.K.; Tareq, M.M.I.; et al. Spike Protein Recognizer Receptor ACE2 Targeted Identification of Potential Natural Antiviral Drug Candidates against SARS-CoV-2. Int. J. Biol. Macromol. 2021, 191, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Opo, F.A.D.M.; Rahman, M.M.; Ahammad, F.; Ahmed, I.; Bhuiyan, M.A.; Asiri, A.M. Structure Based Pharmacophore Modeling, Virtual Screening, Molecular Docking and ADMET Approaches for Identification of Natural Anti-Cancer Agents Targeting XIAP Protein. Sci. Rep. 2021, 11, 4049. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Maudsley, S.; Gesty-Palmer, D. Translating in Vitro Ligand Bias into in Vivo Efficacy. Cell. Signal. 2018, 41, 46. [Google Scholar] [CrossRef]
- Dain Md Opo, F.A.; Alsaiari, A.A.; Rahman Molla, M.H.; Ahmed Sumon, M.A.; Yaghmour, K.A.; Ahammad, F.; Mohammad, F.; Simal-Gandara, J. Identification of Novel Natural Drug Candidates against BRAF Mutated Carcinoma; An Integrative in-Silico Structure-Based Pharmacophore Modeling and Virtual Screening Process. Front. Chem. 2022, 10, 1209. [Google Scholar] [CrossRef]
- Gallo, J.M. Pharmacokinetic/Pharmacodynamic-Driven Drug Development. Mt. Sinai J. Med. 2010, 77, 381. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014, 66, 334. [Google Scholar] [CrossRef] [PubMed]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Torres, P.H.M.; Sodero, A.C.R.; Jofily, P.; Silva, F.P., Jr. Key Topics in Molecular Docking for Drug Design. Int. J. Mol. Sci. 2019, 20, 4574. [Google Scholar] [CrossRef]
- Bouback, T.A.; Pokhrel, S.; Albeshri, A.; Aljohani, A.M.; Samad, A.; Alam, R.; Hossen, M.S.; Al-Ghamdi, K.; Talukder, M.E.K.; Ahammad, F.; et al. Pharmacophore-Based Virtual Screening, Quantum Mechanics Calculations, and Molecular Dynamics Simulation Approaches Identified Potential Natural Antiviral Drug Candidates against MERS-CoV S1-NTD. Molecules 2021, 26, 4961. [Google Scholar] [CrossRef]
- Rahman, M.M.; Opo, F.A.D.M.; Asiri, A.M.; Rahman, M.M.; Opo, F.A.D.M.; Asiri, A.M. Comprehensive Studies of Different Cancer Diseases among Less-Developed Countries. Healthcare 2022, 10, 424. [Google Scholar] [CrossRef]
- Neophytou, C.M.; Trougakos, I.P.; Erin, N.; Papageorgis, P. Apoptosis Deregulation and the Development of Cancer Multi-Drug Resistance. Cancers 2021, 13, 4363. [Google Scholar] [CrossRef]
- Sager, R. Expression Genetics in Cancer: Shifting the Focus from DNA to RNA. Proc. Natl. Acad. Sci. USA 1997, 94, 952. [Google Scholar] [CrossRef]
- Barr, E.K.; Applebaum, M.A. Genetic Predisposition to Neuroblastoma. Children 2018, 5, 119. [Google Scholar] [CrossRef]
- Osuna-Marco, M.P.; López-Barahona, M.; López-Ibor, B.; Tejera, Á.M. Ten Reasons Why People With Down Syndrome Are Protected From the Development of Most Solid Tumors—A Review. Front. Genet. 2021, 12, 2220. [Google Scholar] [CrossRef]
- Kruidenier, L.; Chung, C.W.; Cheng, Z.; Liddle, J.; Che, K.; Joberty, G.; Bantscheff, M.; Bountra, C.; Bridges, A.; Diallo, H.; et al. A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response. Nature 2012, 488, 404–408. [Google Scholar] [CrossRef]
- Picaud, S.; Wells, C.; Felletar, I.; Brotherton, D.; Martin, S.; Savitsky, P.; Diez-Dacal, B.; Philpott, M.; Bountra, C.; Lingard, H.; et al. RVX-208, an Inhibitor of BET Transcriptional Regulators with Selectivity for the Second Bromodomain. Proc. Natl. Acad. Sci. USA 2013, 110, 19754–19759. [Google Scholar] [CrossRef]
- Healy, J.R.; Hart, L.S.; Shazad, A.L.; Gagliardi, M.E.; Tsang, M.; Elias, J.; Ruden, J.; Farrel, A.; Rokita, J.L.; Li, Y.; et al. Limited Anti-Tumor Activity of Combined BET and MEK Inhibition in Neuroblastoma. Pediatr. Blood Cancer 2020, 67, e28267. [Google Scholar] [CrossRef]
- Yu, Z.; Ku, A.F.; Anglin, J.L.; Sharma, R.; Ucisik, M.N.; Faver, J.C.; Li, F.; Nyshadham, P.; Simmons, N.; Sharma, K.L.; et al. Discovery and Characterization of Bromodomain 2-Specific Inhibitors of BRDT. Proc. Natl. Acad. Sci. USA 2021, 118, e2021102118. [Google Scholar] [CrossRef]
- Dzieran, J.; Garcia, A.R.; Westermark, U.K.; Henley, A.B.; Sánchez, E.E.; Träger, C.; Johansson, H.J.; Lehtiö, J.; Arsenian-Henriksson, M. MYCN-Amplified Neuroblastoma Maintains an Aggressive and Undifferentiated Phenotype by Deregulation of Estrogen and NGF Signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E1229–E1238. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Knapp, S. The Bromodomain Interaction Module. FEBS Lett. 2012, 586, 2692–2704. [Google Scholar] [CrossRef]
- Xu, Y.; Vakoc, C.R. Targeting Cancer Cells with BET Bromodomain Inhibitors. Cold Spring Harb. Perspect. Med. 2017, 7, a026674. [Google Scholar] [CrossRef]
- Samad, A.; Ahammad, F.; Nain, Z.; Alam, R.; Imon, R.R.; Hasan, M.; Rahman, M.S. Designing a Multi-Epitope Vaccine against SARS-CoV-2: An Immunoinformatics Approach. J. Biomol. Struct. Dyn. 2020, 40, 14–30. [Google Scholar] [CrossRef]
- Aljahdali, M.O.; Molla, M.H.R.; Ahammad, F. Compounds Identified from Marine Mangrove Plant (Avicennia Alba) as Potential Antiviral Drug Candidates Against WDSV, an In-Silico Approach. Mar. Drugs 2021, 19, 253. [Google Scholar] [CrossRef] [PubMed]
- Valasani, K.R.; Vangavaragu, J.R.; Day, V.W.; Yan, S.S. Structure Based Design, Synthesis, Pharmacophore Modeling, Virtual Screening, and Molecular Docking Studies for Identification of Novel Cyclophilin D Inhibitors. J. Chem. Inf. Model. 2014, 54, 902–912. [Google Scholar] [CrossRef]
- He, G.; Gong, B.; Li, J.; Song, Y.; Li, S.; Lu, X. An Improved Receptor-Based Pharmacophore Generation Algorithm Guided by Atomic Chemical Characteristics and Hybridization Types. Front. Pharmacol. 2018, 9, 1463. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449. [Google Scholar] [CrossRef]
- Ahmed, T.A. Pharmacokinetics of Drugs Following IV Bolus, IV Infusion, and Oral Administration. Basic Pharmacokinet. Concepts Some Clin. Appl. 2015. [Google Scholar] [CrossRef]
- Benedetti, M.S.; Whomsley, R.; Poggesi, I.; Cawello, W.; Mathy, F.X.; Delporte, M.L.; Papeleu, P.; Watelet, J.B. Drug Metabolism and Pharmacokinetics. Drug Metab. Rev. 2009, 41, 344–390. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.D.; Nicholson, J.K. Gut Microbiome Interactions with Drug Metabolism, Efficacy and Toxicity. Transl. Res. 2017, 179, 204. [Google Scholar] [CrossRef] [PubMed]
- Stillhart, C.; Vučićević, K.; Augustijns, P.; Basit, A.W.; Batchelor, H.; Flanagan, T.R.; Gesquiere, I.; Greupink, R.; Keszthelyi, D.; Koskinen, M.; et al. Impact of Gastrointestinal Physiology on Drug Absorption in Special Populations––An UNGAP Review. Eur. J. Pharm. Sci. 2020, 147, 105280. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Lepailleur, A.; Mignani, S.M.; Dallemagne, P.; Rochais, C. HERG Toxicity Assessment: Useful Guidelines for Drug Design. Eur. J. Med. Chem. 2020, 195, 112290. [Google Scholar] [CrossRef]
 ), red arrows (
), red arrows ( ) indicating H-bond acceptors, and one red star (
) indicating H-bond acceptors, and one red star ( ) depicting negative ionizable groups.
) depicting negative ionizable groups.
 ), red arrows (
), red arrows ( ) indicating H-bond acceptors, and one red star (
) indicating H-bond acceptors, and one red star ( ) depicting negative ionizable groups.
) depicting negative ionizable groups.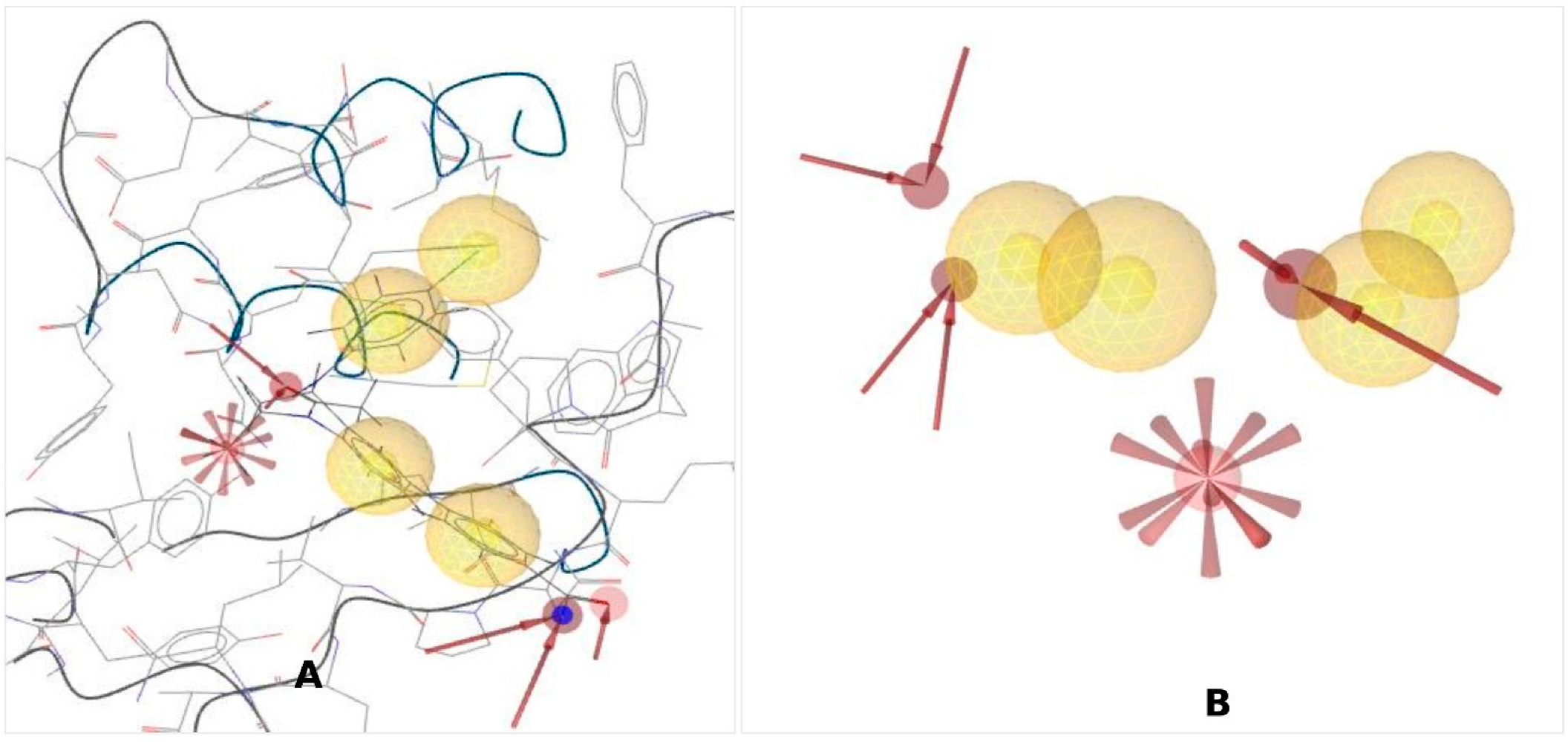




| ZINC ID | Compound Name | Molecular Formula | Binding Affinity (kcal/mol) |
|---|---|---|---|
| ZINC4104882 | [17-(2-diazoacetyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-yl] acetate | C23H32N2O3 | −8.0 |
| ZINC2509501 | [(3S,8R,9S,10S,13S,14S)-10,13-Dimethyl-17-oxo-1,2,3,4,5,6,7,8,9,11,12,14,15,16-tetradecahydrocyclopenta[a]phenanthren-3-yl] hydrogen sulfate | C19H30O5S | −7.9 |
| ZINC2566088 | 3′-Hydroxy-5,6,7,4′-tetramethoxyflavone | C19H18O7 | −7.6 |
| ZINC1615112 | Gardenin A | C21H22O9 | −7.2 |
| Complex Name | ∆GB | ∆GBC | ∆GBCo | ∆GBH | ∆GBL | ∆GBP | ∆GBS | ∆GBV |
|---|---|---|---|---|---|---|---|---|
| ZINC4104882 | −15.34 ± 1.04 | −14.85 ± 2.22 | 2.102 ± 0.96 | −1.87 ± 0.76 | −7.56 ± 2.72 | −0.331 ± 0.11 | 12.028 ± 2.09 | −22.755 ± 3.77 |
| ZINC2509501 | −19.91 ± 2.92 | −15.95 ± 1.36 | 2.260 ± 0.55 | −2.89 ± 0.52 | −7.48 ± 2.39 | −0.315 ± 0.20 | 14.85 ± 2.69 | −23.67 ± 2.11 |
| ZINC2566088 | −16.69 ± 1.53 | −14.75 ± 2.36 | 2.80 ± 0.104 | −1.70 ± 0.81 | −5.48 ± 2.52 | −0.34 ± 0.12 | 15.82 ± 1.66 | −26.33 ± 2.45 |
| ZINC1615112 | −17.98 ± 2.19 | −15.33 ± 1.017 | 2.58 ± 0.407 | −1.08 ± 0.64 | −8.70 ± 1.68 | −0.21 ± 0.11 | 13.93 ± 1.07 | −18.26 ± 1.78 |
| ZINC95504909 | −14.45 ± 2.04 | −12.51 ± 1.33 | 2.22 ± 0.98 | −2.22 ± 0.98 | −4.56 ± 2.12 | −0.451 ± 0.13 | 11.028 ± 1.08 | −20.566 ± 2.88 |
| Properties | Parameter | ZINC4104882 | ZINC2509501 | ZINC2566088 | ZINC1615112 |
|---|---|---|---|---|---|
| Physico-chemical properties | MW (g/mol) | 384.51 | 370.50 | 358.34 | 418.39 |
| Heavy atoms | 28 | 25 | 26 | 30 | |
| Arom. heavy atoms | 0 | 0 | 16 | 16 | |
| Rotatable bonds | 4 | 2 | 5 | 7 | |
| H-bond acceptors | 5 | 5 | 7 | 9 | |
| H-bond donors | 0 | 1 | 1 | 1 | |
| Molar refractivity | 108.36 | 96.29 | 95.91 | 108.89 | |
| Lipophilicity | Log Po/w | 3.48 | 2.48 | 3.31 | 3.79 |
| Water solubility | Log S (ESOL) | Moderate | Soluble | Moderate | Moderate |
| Pharmacokinetics | GI absorption | High | High | High | High |
| CYP3A4 inhibitor | No | No | Yes | Yes | |
| BBB permeant | No | No | No | No | |
| Drug likeness | Lipinski violation | Yes | Yes | Yes | Yes |
| Bioavailability score | 0.55 | 0.56 | 0.55 | 0.55 | |
| Medi. chemistry | Synth. accessibility | 5.23 | 4.82 | 3.57 | 3.91 |
| Endpoint | Target | ZINC4104882 | ZINC2509501 | ZINC2566088 | ZINC1615112 |
|---|---|---|---|---|---|
| Organ toxicity | Hepatotoxicity | Inactive | Inactive | Inactive | Inactive |
| Toxicity endpoints | Carcinogenicity | Active | Inactive | Inactive | Inactive |
| Immunotoxicity | Active | Inactive | Active | Active | |
| Mutagenicity | Inactive | Inactive | Inactive | Inactive | |
| Cytotoxicity | Active | Inactive | Inactive | Inactive | |
| LD50 (mg/kg) | 5000 | 6000 | 4000 | 5000 | |
| Toxicity class | 5 | 6 | 5 | 5 | |
| Tox21-nuclear-receptor signaling pathways | Androgen receptor (AR) | Active | Inactive | Inactive | Inactive |
| Aryl hydrocarbon receptor (AhR) | Inactive | Inactive | Active | Active | |
| Tox21-stress-response pathway | Heat shock factor response element | Inactive | Inactive | Inactive | Inactive |
| Mitochondrial membrane potential (MMP) | Inactive | Inactive | Active | Active | |
| Phosphoprotein (tumor suppressor) p53 | Inactive | Inactive | Inactive | Inactive | |
| Fathead minnow LC50 (96 h) | mg/L | N/A | 4.99 | 0.53 | 0.55 |
| 48-h Daphnia magna LC50 | mg/L | N/A | N/A | 8.48 | 15.42 |
| Developmental toxicity | value | 0.96 | 1.19 | 0.84 | 0.76 |
| Oral rat LD50 | mg/kg | N/A | N/A | 142.54 | 972.5 |
| Bioaccumulation factor | Log10 | N/A | N/A | 1.76 | 1.94 |
| Water solubility (25 °C) | mg/L | N/A | 32.56 | 57.67 | 27.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Opo, F.A.D.M.; Alkarim, S.; Alrefaei, G.I.; Molla, M.H.R.; Alsubhi, N.H.; Alzahrani, F.; Ahammad, F. Pharmacophore-Model-Based Virtual-Screening Approaches Identified Novel Natural Molecular Candidates for Treating Human Neuroblastoma. Curr. Issues Mol. Biol. 2022, 44, 4838-4858. https://doi.org/10.3390/cimb44100329
Opo FADM, Alkarim S, Alrefaei GI, Molla MHR, Alsubhi NH, Alzahrani F, Ahammad F. Pharmacophore-Model-Based Virtual-Screening Approaches Identified Novel Natural Molecular Candidates for Treating Human Neuroblastoma. Current Issues in Molecular Biology. 2022; 44(10):4838-4858. https://doi.org/10.3390/cimb44100329
Chicago/Turabian StyleOpo, F A Dain Md, Saleh Alkarim, Ghadeer I. Alrefaei, Mohammad Habibur Rahman Molla, Nouf H. Alsubhi, Faisal Alzahrani, and Foysal Ahammad. 2022. "Pharmacophore-Model-Based Virtual-Screening Approaches Identified Novel Natural Molecular Candidates for Treating Human Neuroblastoma" Current Issues in Molecular Biology 44, no. 10: 4838-4858. https://doi.org/10.3390/cimb44100329
APA StyleOpo, F. A. D. M., Alkarim, S., Alrefaei, G. I., Molla, M. H. R., Alsubhi, N. H., Alzahrani, F., & Ahammad, F. (2022). Pharmacophore-Model-Based Virtual-Screening Approaches Identified Novel Natural Molecular Candidates for Treating Human Neuroblastoma. Current Issues in Molecular Biology, 44(10), 4838-4858. https://doi.org/10.3390/cimb44100329


_Kim.png)








