Protocol for Increasing the Sensitivity of MS-Based Protein Detection in Human Chorionic Villi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Clinical Specimen Collection and Preparation
2.3. Protein Extraction
2.4. 1DE-Gel Concentration and In-Gel Digestion
2.5. In-Solution Tryptic Digestion
2.6. LC–MS/MS Analysis
2.7. Data Processing
3. Results and Discussion
3.1. 1DE-Gel Concentration for Analyzing the Chorionic Villi Proteome
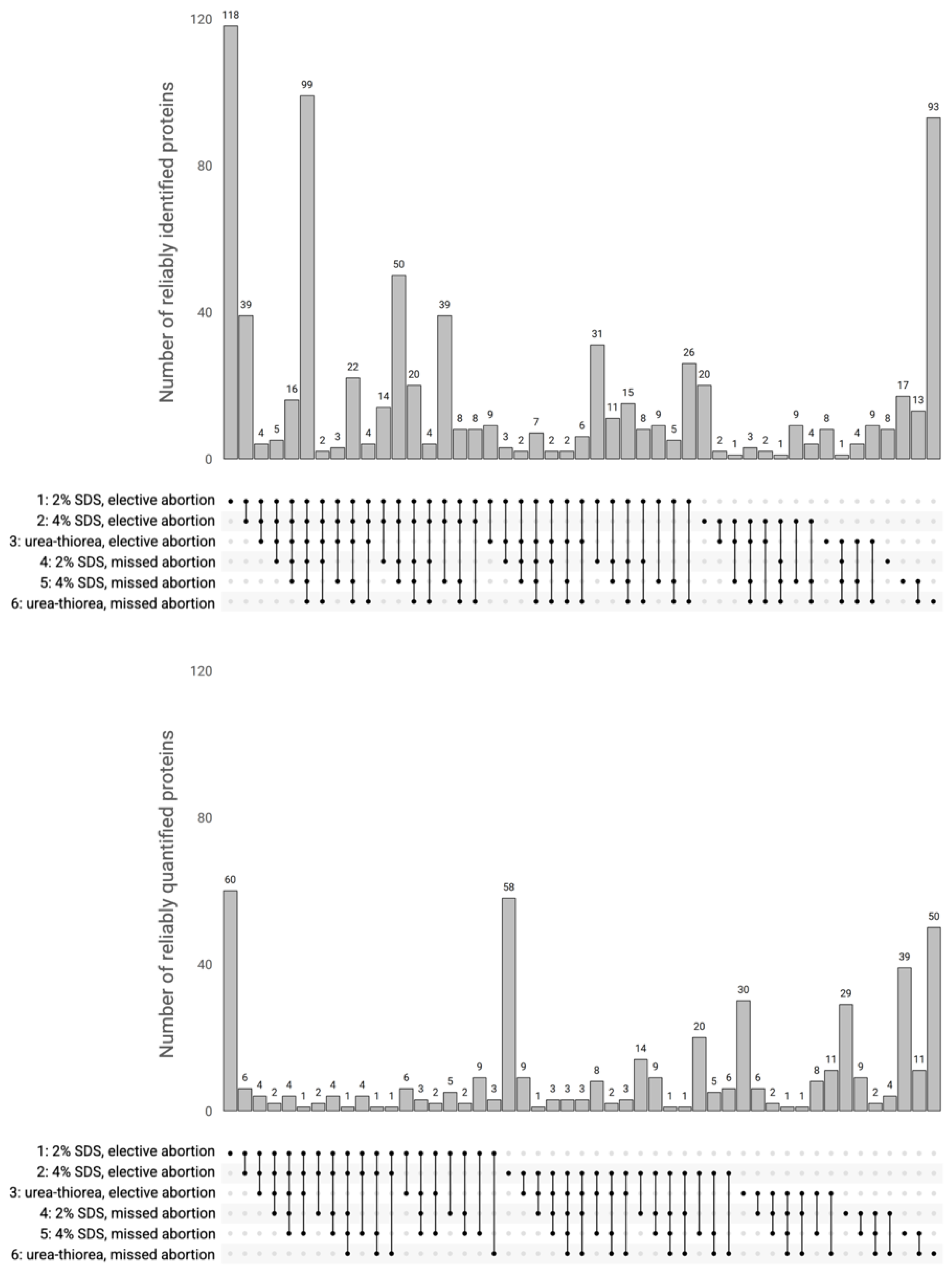
3.2. Assessment of the Variability of Protein Identification and Quantification
3.3. The 1DE-Gel Concentration for Detecting Low-Abundance Proteins in the Placenta
3.4. The Partly Lost Pregnancy-Specific Glycoproteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bell, L.; Saxena, R.; Mattar, S.; You, J.; Wang, M.; Chalasani, N. Utility of formalin-fixed, paraffin-embedded liver biopsy specimens for global proteomic analysis in nonalcoholic steatohepatitis. Proteom. Clin. Appl. 2011, 5, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Wong, L.; Goh, W. Understanding missing proteins: A functional perspective. Drug Discov. Today 2018, 23, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.; Ray, S.; Srivastava, S. The quest of the human proteome and the missing proteins: Digging deeper. Omics J. Integr. Biol. 2015, 19, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Elguoshy, A.; Magdeldin, S.; Xu, B.; Hirao, Y.; Zhang, Y.; Kinoshita, N.; Takisawa, Y.; Nameta, M.; Yamamoto, K.; El-Refy, A.; et al. Why are they missing?: Bioinformatics characterization of missing human proteins. J. Proteom. 2016, 149, 7–14. [Google Scholar] [CrossRef]
- Tarrade, A.; Lai Kuen, R.; Malassiné, A.; Tricottet, V.; Blain, P.; Vidaud, M.; Evain-Brion, D. Characterization of human villous and extravillous trophoblasts isolated from first trimester placenta. Lab. Investig. 2001, 81, 1199–1211. [Google Scholar] [CrossRef] [Green Version]
- Harper, C.; Henderson, J.; Darney, P. Abortion in the United States. Annu. Rev. Public Health 2005, 26, 501–512. [Google Scholar] [CrossRef]
- The Human Protein Atlas: Tissue Atlas, Placenta-Specific Proteome. Available online: https://www.proteinatlas.org/humanproteome/tissue/placenta (accessed on 5 May 2022).
- Mori, M.; Ishikawa, G.; Luo, S.; Mishima, T.; Goto, T.; Robinson, J.; Matsubara, S.; Takeshita, T.; Kataoka, H.; Takizawa, T. The cytotrophoblast layer of human chorionic villi becomes thinner but maintains its structural integrity during gestation. Biol. Reprod. 2007, 76, 164–172. [Google Scholar] [CrossRef]
- Huppertz, B.; Ghosh, D.; Sengupta, J. An integrative view on the physiology of human early placental villi. Prog. Biophys. Mol. Biol. 2014, 114, 33–48. [Google Scholar] [CrossRef]
- Peach, M.; Marsh, N.; Miskiewicz, E.; MacPhee, D. Solubilization of proteins: The importance of lysis buffer choice. Methods Mol. Biol. 2015, 1312, 49–60. [Google Scholar]
- Rabilloud, T.; Luche, S.; Santoni, V.; Chevallet, M. Detergents and chaotropes for protein solubilization before two-dimensional electrophoresis. Methods Mol. Biol. 2007, 355, 111–119. [Google Scholar]
- Wang, F.; Wang, L.; Xu, Z.; Liang, G. Identification and analysis of multi-protein complexes in placenta. PLoS ONE 2013, 8, e62988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, H.; Ding, H.; Fang, M.; Yu, B.; Cheng, Y.; Tan, Y.; Fu, Q.; Lu, B.; Cai, H.; Jin, X.; et al. Proteomics and bioinformatics analysis of altered protein expression in the placental villous tissue from early recurrent miscarriage patients. Placenta 2018, 61, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Luyten, L.; Dieu, M.; Demazy, C.; Fransolet, M.; Nawrot, T.; Renard, P.; Debacq-Chainiaux, F. Optimization of label-free nano LC-MS/MS analysis of the placental proteome. Placenta 2020, 101, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Khorami Sarvestani, S.; Shojaeian, S.; Vanaki, N.; Ghresi-Fard, B.; Amini, M.; Gilany, K.; Soltanghoraee, H.; Arefi, S.; Jeddi-Tehrani, M.; Zarnani, A. Proteome profiling of human placenta reveals developmental stage-dependent alterations in protein signature. Clin. Proteom. 2021, 18, 18. [Google Scholar] [CrossRef]
- Vaisar, T. Thematic review series: Proteomics. Proteomic analysis of lipid-protein complexes. J. Lipid Res. 2009, 50, 781–786. [Google Scholar] [CrossRef] [Green Version]
- Ilavenil, S.; Al-Dhabi, N.; Srigopalram, S.; Kim, Y.; Agastian, P.; Baaru, R.; Choi, K.; Arasu, M.; Park, C.; Park, K. Removal of SDS from biological protein digests for proteomic analysis by mass spectrometry. Proteome Sci. 2016, 14, 11. [Google Scholar] [CrossRef] [Green Version]
- Patole, C.; Bindschedler, L. Chapter 4—Plant proteomics: A guide to improve the proteome coverage. In Advances in Biological Science Research; Meena, S., Naik, M., Eds.; Academic Press: New York, NY, USA, 2019; pp. 45–67. [Google Scholar]
- Brownridge, P.; Beynon, R. The importance of the digest: Proteolysis and absolute quantification in proteomics. Methods 2011, 54, 351–360. [Google Scholar] [CrossRef]
- Miao, Z.; Chen, M.; Wu, H.; Ding, H.; Shi, Z. Comparative proteomic profile of the human placenta in normal and fetal growth restriction subjects. Cell. Physiol. Biochem. 2014, 34, 1701–1710. [Google Scholar] [CrossRef] [Green Version]
- Tubaon, R.; Haddad, P.; Quirino, J. Sample Clean-up Strategies for ESI Mass Spectrometry Applications in Bottom-up Proteomics: Trends from 2012 to 2016. Proteomics 2017, 17, 1700011. [Google Scholar] [CrossRef]
- Vowinckel, J.; Capuano, F.; Campbell, K.; Deery, M.; Lilley, K.; Ralser, M. The beauty of being (label)-free: Sample preparation methods for SWATH-MS and next-generation targeted proteomics. F1000Research 2013, 2, 272. [Google Scholar] [CrossRef] [Green Version]
- Kachuk, C.; Faulkner, M.; Liu, F.; Doucette, A. Automated SDS Depletion for Mass Spectrometry of Intact Membrane Proteins though Transmembrane Electrophoresis. J. Proteome Res. 2016, 15, 2634–2642. [Google Scholar] [CrossRef] [PubMed]
- Smith, B. SDS polyacrylamide gel electrophoresis for N-terminal protein sequencing. Methods Mol. Biol. 1997, 64, 17–24. [Google Scholar]
- Goldman, A.; Beer, L.; Tang, H.; Hembach, P.; Zayas-Bazan, D.; Speicher, D. Proteome Analysis Using Gel-LC-MS/MS. Curr. Protoc. Protein Sci. 2019, 96, e93. [Google Scholar] [CrossRef]
- Gomis-Cebolla, J.; Scaramal Ricietto, A.; Ferré, J. A Genomic and Proteomic Approach to Identify and Quantify the Expressed Bacillus thuringiensis Proteins in the Supernatant and Parasporal Crystal. Toxins 2018, 10, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondratova, V.; Serd’uk, O.; Shelepov, V.; Lichtenstein, A. Concentration and isolation of DNA from biological fluids by agarose gel isotachophoresis. BioTechniques 2005, 39, 695–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luebker, S.; Koepsell, S. Optimization of Urea Based Protein Extraction from Formalin-Fixed Paraffin-Embedded Tissue for Shotgun Proteomics. Int. J. Proteom. 2016, 2016, 4324987. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Kong, T.; Kim, H.; Kim, H. The Proteomic Analysis of Human Placenta with Pre-eclampsia and Normal Pregnancy. J. Korean Med. Sci. 2015, 30, 770–778. [Google Scholar] [CrossRef] [Green Version]
- Centlow, M.; Hansson, S.; Welinder, C. Differential proteome analysis of the preeclamptic placenta using optimized protein extraction. J. Biotechnol. Biomed. 2010, 2010, 458748. [Google Scholar] [CrossRef] [Green Version]
- Ni, X.; Li, X.; Guo, Y.; Zhou, T.; Guo, X.; Zhao, C.; Lin, M.; Zhou, Z.; Shen, R.; Guo, X.; et al. Quantitative proteomics analysis of altered protein expression in the placental villous tissue of early pregnancy loss using isobaric tandem mass tags. Biomed. Res. Int. 2014, 2014, 647143. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.; Krohn, R.; Hermanson, G.; Mallia, A.; Gartner, F.; Provenzano, M.; Fujimoto, E.; Goeke, N.; Olson, B.; Klenk, D. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Rodchenkova, M.; Novikova, S. Optimization of liquid chromatography with mass spectrometric detection method for the qualitative and semi-quantitative proteomic analysis. Analitika 2013, 3, 40–47. [Google Scholar]
- ProteoWizard Home Page. Available online: https://proteowizard.sourceforge.io (accessed on 5 May 2022).
- Vaudel, M.; Barsnes, H.; Berven, F.; Sickmann, A.; Martens, L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 2011, 11, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Burkhart, J.; Zahedi, R.; Oveland, E.; Berven, F.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef]
- The Contaminant Repository for Affinity Purification (CRAPome) Database. Available online: https://reprint-apms.org/?q=chooseworkflow (accessed on 5 May 2022).
- The R Project for Statistical Computing. Available online: https://www.r-project.org (accessed on 5 May 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Ggupset: Combination Matrix Axis for ‘ggplot2’ to Create ‘UpSet’ Plots. Available online: https://CRAN.R-project.org/package=ggupset (accessed on 5 May 2022).
- Galili, T. dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [Green Version]
- Sobhani, N.; Cassidy, A.; Qi, Z.; Yu, J.; Chetty, S.; Norton, M. Gross morphologic appearance of chorionic villi and the risk of chromosomal abnormalities. Reprod. Sci. 2021, 28, 570–574. [Google Scholar] [CrossRef]
- Sadler, T. Langman’s Medical Embryology, 12th ed.; Lippincott Williams and Wilkins: Baltimore, MD, USA, 2012; p. 103. [Google Scholar]
- Lisitsa, A.; Petushkova, N.; Thiele, H.; Moshkovskii, S.; Zgoda, V.; Karuzina, I.; Chernobrovkin, A.; Skipenko, O.; Archakov, A. Application of slicing of one-dimensional gels with subsequent slice-by-slice mass spectrometry for the proteomic profiling of human liver cytochromes P450. J. Proteome Res. 2010, 9, 95–103. [Google Scholar] [CrossRef]
- Vergauwen, G.; Dhondt, B.; Van Deun, J.; De Smedt, E.; Berx, G.; Timmerman, E.; Gevaert, K.; Miinalainen, I.; Cocquyt, V.; Braems, G.; et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci. Rep. 2017, 7, 2704. [Google Scholar] [CrossRef] [Green Version]
- Spinning Around: Tips and Tricks for Using Centrifugal Filters. Available online: https://bitesizebio.com/44738/spinning-around-tips-and-tricks-for-using-centrifugal-filters/ (accessed on 5 May 2022).
- Neilson, K.; Keighley, T.; Pascovici, D.; Cooke, B.; Haynes, P. Label-free quantitative shotgun proteomics using normalized spectral abundance factors. Methods Mol. Biol. 2013, 1002, 205–222. [Google Scholar] [PubMed]
- Bubis, J.; Levitsky, L.; Ivanov, M.; Tarasova, I.; Gorshkov, M. Comparative evaluation of label-free quantification methods for shotgun proteomics. Rapid Commun. Mass Spectrom. 2017, 31, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Barkovits, K.; Pacharra, S.; Pfeiffer, K.; Steinbach, S.; Eisenacher, M.; Marcus, K.; Uszkoreit, J. Reproducibility, Specificity and Accuracy of Relative Quantification Using Spectral Library-based Data-independent Acquisition. Mol. Cell Proteom. 2020, 19, 181–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Female Pregnancy GO Annotations (GO:0007565). Available online: https://www.ebi.ac.uk/QuickGO/annotations?agoUsage=descendants&goUsageRelationships=is_a,part_of,occurs_in&goId=GO:0007565 (accessed on 5 May 2022).
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Baron, M. A note on the historical development of logic diagrams: Leibniz, Euler and Venn. Math. Gaz. 1969, 53, 113–125. [Google Scholar] [CrossRef]
- Jia, A.; Xu, L.; Wang, Y. Venn diagrams in bioinformatics. Brief. Bioinform. 2021, 22, bbab108. [Google Scholar] [CrossRef]
- Finocchi, I.; Petreschi, R. Hierarchical Clustering of Trees: Algorithms and Experiments. In Proceedings of the Third Intranational Workshop Conference: Algorithm Engineering and Experimentation, Washington, DC, USA, 5–6 January 2001. [Google Scholar]
- De Vienne, D. Tanglegrams Are Misleading for Visual Evaluation of Tree Congruence. Mol. Biol. Evol. 2019, 36, 174–176. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual comparisons of grouped data by ranking methods. J. Econ. Entomol. 1946, 39, 269–270. [Google Scholar] [CrossRef]
- Lane, L.; Bairoch, A.; Beavis, R.; Deutsch, E.; Gaudet, P.; Lundberg, E.; Omenn, G. Metrics for the Human Proteome Project 2013–2014 and strategies for finding missing proteins. J. Proteome Res. 2014, 13, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Jamioł, M.; Wawrzykowski, J.; Mojsym, W.; Hoedemaker, M.; Kankofer, M. Activity of selected glycosidases and availability of their substrates in bovine placenta during pregnancy and parturition with and without retained foetal membranes. Reprod. Domest. Anim. 2020, 55, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Sui, L.; An, L.; Tan, K.; Wang, Z.; Wang, S.; Miao, K.; Ren, L.; Tao, L.; He, S.; Yu, Y.; et al. Dynamic proteomic profiles of in vivo- and in vitro-produced mouse postimplantation extraembryonic tissues and placentas. Biol. Reprod. 2014, 91, 155. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Kato, T.; Fujita, A.; Ishizaki, T.; Narumiyam, S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1999, 1, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Wirrig, E.; Snarr, B.; Chintalapudi, M.; O’neal, J.; Phelps, A.; Barth, J.; Fresco, V.; Kern, C.; Mjaatvedt, C.; Toole, B.; et al. Cartilage link protein 1 (Crtl1), an extracellular matrix component playing an important role in heart development. Dev. Biol. 2007, 310, 291–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An Online Catalog of Human Genes and Genetic Disorders: Pregnancy-Specific Beta-1-Glycoprotein 11; PSG11. Available online: https://omim.org/entry/176401 (accessed on 5 May 2022).
- Hammarström, S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999, 9, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Gordon, Y.; Grudzinskas, J.; Jeffrey, D.; Chard, T. Concentrations of pregnancy-specific beta 1-glycoprotein in maternal blood in normal pregnancy and in intrauterine growth retardation. Lancet 1977, 1, 331–333. [Google Scholar] [CrossRef]
- Würz, H.; Geiger, W.; Künzig, H.; Jabs-Lehmann, A.; Bohn, H.; Lüben, G. Radioimmunoassay of SP1 (pregnancy-specific beta1-glycoprotein) in maternal blood and in amniotic fluid normal and pathologic pregnancies. J. Perinat. Med. 1981, 9, 67–78. [Google Scholar] [CrossRef] [Green Version]
- The Carcinoembryonic Antigen Homepage: Human CEA Family. Available online: http://carcinoembryonic-antigen.de/human (accessed on 5 May 2022).
- Rattila, S.; Dunk, C.; Im, M.; Grichenko, O.; Zhou, Y.; Yanez-Mo, M.; Blois, S.; Yamada, K.; Erez, O.; Gomez-Lopez, N.; et al. Interaction of Pregnancy-Specific Glycoprotein 1 With Integrin A5β1 Is a Modulator of Extravillous Trophoblast Functions. Cells 2019, 8, 1369. [Google Scholar] [CrossRef] [Green Version]
- neXtProt: A Knowledge Platform for Human Proteins, Putative Pregnancy-Specific Beta-1-Glycoprotein 7 (PSBG-7). Available online: https://www.nextprot.org/entry/NX_Q13046/ (accessed on 5 May 2022).
- Audain, E.; Uszkoreit, J.; Sachsenberg, T.; Pfeuffer, J.; Liang, X.; Hermjakob, H.; Sanchez, A.; Eisenacher, M.; Reinert, K.; Tabb, D.; et al. In-depth analysis of protein inference algorithms using multiple search engines and well-defined metrics. J. Proteom. 2017, 150, 170–182. [Google Scholar] [CrossRef] [Green Version]
- Levitsky, L.; Ivanov, M.; Lobas, A.; Bubis, J.; Tarasova, I.; Solovyeva, E.; Pridatchenko, M.; Gorshkov, M. IdentiPy: An Extensible Search Engine for Protein Identification in Shotgun Proteomics. J. Proteome Res. 2018, 17, 2249–2255. [Google Scholar] [CrossRef]
- Lisitsa, A.; Petushkova, N.; Levitsky, L.; Zgoda, V.; Larina, O.; Kisrieva, Y.; Frankevich, V.; Gamidov, S. Comparative Analysis of the Performance of Mascot and IdentiPy Algorithms on a Benchmark Dataset Obtained by Tandem Mass Spectrometry Analysis of Testicular Biopsies. Mol. Biol. 2019, 53, 147–155. [Google Scholar] [CrossRef]
- Cho, J.; Lee, H.; Jeong, S.; Kim, K.; Kwon, K.; Yoo, J.; Omenn, G.; Baker, M.; Hancock, W.; Paik, Y. Combination of Multiple Spectral Libraries Improves the Current Search Methods Used to Identify Missing Proteins in the Chromosome-Centric Human Proteome Project. J. Proteome Res. 2015, 14, 4959–4966. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Zhang, J.; Yang, W.; Liu, Y.; Liu, Y.; Shan, Z.; Wang, W. Identification of the Prognosis-Related lncRNAs and Genes in Gastric Cancer. Front. Genet. 2020, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Khan, W.; Hammarström, S. Identification of a new carcinoembryonic antigen (CEA) family member in human fetal liver—cloning and sequence determination of pregnancy-specific glycoprotein 7. Biochem. Biophys. Res. Commun. 1990, 168, 214–225. [Google Scholar] [CrossRef]

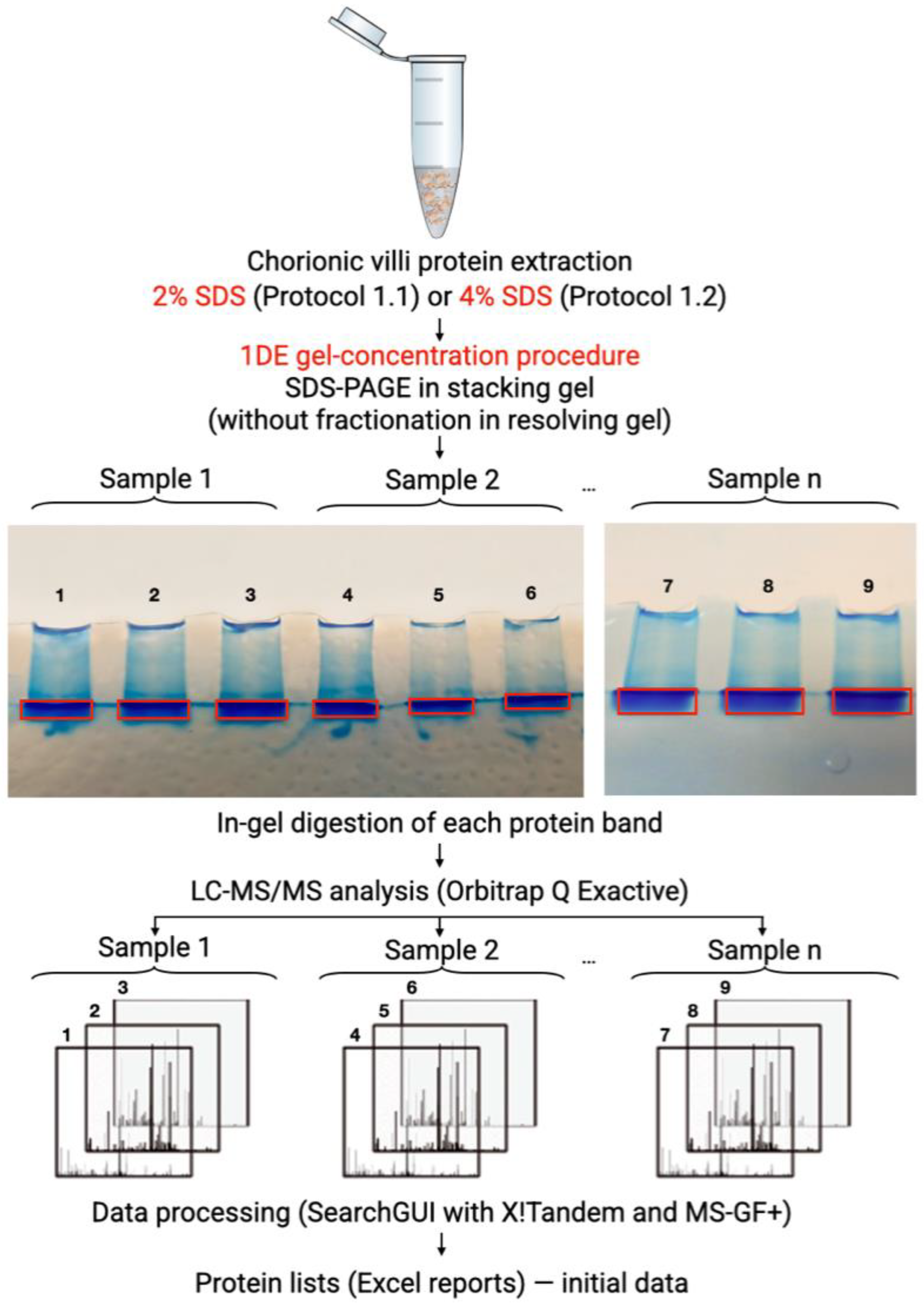

| Dataset | State | Sample Preparation Protocol | Number of Samples | Number of Replicates per Sample |
|---|---|---|---|---|
| 1 | Elective abortion | 2% SDS-based solubilization combined with 1DE-gel concentration and in-gel digestion (Protocol 1.1) | 3 | 2 |
| 2 | Elective abortion | 4% SDS-based solubilization combined with 1DE-gel concentration and in-gel digestion (Protocol 1.2) | 1 | 3 |
| 3 | Elective abortion | Urea-based extraction combined with in-solution digestion (Protocol 2) | 1 | 3 |
| 4 | Surgically treated missed abortion | 2% SDS-based solubilization combined with 1DE-gel concentration and in-gel digestion (Protocol 1.1) | 1 | 2 |
| 5 | Surgically treated missed abortion | 4% SDS-based solubilization combined with 1DE-gel concentration and in-gel digestion (Protocol 1.2) | 1 | 3 |
| 6 | Surgically treated missed abortion | Urea-based extraction combined with in-solution digestion (Protocol 2) | 1 | 3 |
| Dataset | Ptotal | Pplacenta | Isuccessful | Iproteins | Ireliably | Qreliably |
|---|---|---|---|---|---|---|
| 1 | 1377 | 1232 | 884 | 738 | 591 | 120 |
| 2 | 756 | 690 | 460 | 379 | 379 | 176 |
| 3 | 594 | 547 | 298 | 216 | 216 | 113 |
| 4 | 871 | 782 | 546 | 299 | 299 | 112 |
| 5 | 764 | 662 | 449 | 361 | 361 | 154 |
| 6 | 824 | 723 | 447 | 368 | 368 | 110 |
| ## | Accession | Protein Name | Gene | Trophoblastic Cells RNA Expression HPA ** | Cellular Component | Dataset |
|---|---|---|---|---|---|---|
| 1 | P01023 | Alpha-2-macroglobulin | A2M | 744.1 pTPM | secreted | 1, 6 |
| 2 | Q1KMD3 | Heterogeneous nuclear ribonucleo protein U like 2 | HNRNPUL2 | 44.6 pTPM | nucleus | 1 |
| 3 | P07900 | Heat shock protein HSP 90-alpha | HSP90AA1 | 682.3 pTPM | cell membrane, cytoplasm, nucleus | 1, 2, 4, and 5 |
| 4 | Q9Y4L1 | Hypoxia up-regulated 1 | HYOU1 | 35.7 pTPM | endoplasmic reticulum | 2, 3 |
| 5 | Q03252 | Lamin B2 | LMNB2 | 25.5 pTPM | membrane, nucleus | 2, 3 |
| 6 | Q16891 | MICOS complex subunit MIC60 | IMMT | 63.5 pTPM | membrane, mitochondrion | 2 |
| 7 | P00505 | Aspartate aminotransferase, mitochondrial | GOT2 | 42.4 pTPM | membrane, mitochondrion | 2 |
| 8 | Q9BSJ8 | Extended synaptotagmin 1 | ESYT1 | 65.4 pTPM | endoplasmic reticulum | 2 |
| 9 | P22307 | Non-specific lipid-transfer protein | SCP2 | 139.1 pTPM | cytoplasm, mitochondrion | 3, 6 |
| 10 | Q13813 | Spectrin alpha, non-erythrocytic 1 | SPTAN1 | 100.2 pTPM | cytoplasm | 3 |
| 11 | P02751 | Fibronectin 1 | FN1 | 5227.7 pTPM *** | secreted | 3, 4 |
| 12 | P50395 | Rab GDP dissociation inhibitor beta | GDI2 | 224.3 pTPM | cytoplasm, membrane | 4 |
| 13 | P02452 | Collagen alpha-1(I) chain | COL1A1 | 744.6 pTPM | secreted | 5 |
| 14 | Q8IZ83 | Aldehyde dehydrogenase family 16 member A1 | ALDH16A1 | 8.8 pTPM | membrane | 6 |
| 15 | P09110 | 3-ketoacyl-CoA thiolase, peroxisomal | ACAA1 | 28.6 pTPM | peroxisome | 6 |
| ## | Entry | Gene Name | Protein Name | Length, aa | Mass, Da | Sample Preparation Protocol | |
|---|---|---|---|---|---|---|---|
| Elective Abortion | Missed Abortion | ||||||
| 1 | Q00887 | PSG9 | Pregnancy-specific beta-1-glycoprotein 9 | 426 | 48,272 | Protocol 1.1; Protocol 1.2; Protocol 2 | Protocol 1.1; Protocol 1.2 |
| 2 | Q9UQ74 | PSG8 | Pregnancy-specific beta-1-glycoprotein 8 | 426 | 47,772 | Protocol 1.1 | nd |
| 3 | Q13046 | PSG7 | Putative pregnancy-specific beta-1-glycoprotein 7 | 419 | 47,027 | Protocol 1.1 | nd |
| 4 | Q00889 | PSG6 | Pregnancy-specific beta-1-glycoprotein 6 | 435 | 48,814 | Protocol 1.1 | nd |
| 5 | Q15238 | PSG5 | Pregnancy-specific beta-1-glycoprotein 5 | 335 | 37,713 | Protocol 1.1; Protocol 1.2 | Protocol 1.1 |
| 6 | Q00888 | PSG4 | Pregnancy-specific beta-1-glycoprotein 4 | 419 | 47,113 | Protocol 1.1; Protocol 1.2; Protocol 2 | nd |
| 7 | Q16557 | PSG3 | Pregnancy-specific beta-1-glycoprotein 3 | 428 | 47,945 | Protocol 1.1; Protocol 2 | Protocol 1.1; Protocol 1.2 |
| 8 | P11465 | PSG2 | Pregnancy-specific beta-1-glycoprotein 2 | 335 | 37,216 | Protocol 1.1; Protocol 2 | Protocol 1.1 |
| 9 | Q9UQ72 | PSG11 | Pregnancy-specific beta-1-glycoprotein 11 | 335 | 37,146 | Protocol 1.1 | nd |
| 10 | P11464 | PSG1 | Pregnancy-specific beta-1-glycoprotein 1 | 419 | 47,223 | Protocol 1.1 | Protocol 1.1; Protocol 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shkrigunov, T.; Pogodin, P.; Zgoda, V.; Larina, O.; Kisrieva, Y.; Klimenko, M.; Latyshkevich, O.; Klimenko, P.; Lisitsa, A.; Petushkova, N. Protocol for Increasing the Sensitivity of MS-Based Protein Detection in Human Chorionic Villi. Curr. Issues Mol. Biol. 2022, 44, 2069-2088. https://doi.org/10.3390/cimb44050140
Shkrigunov T, Pogodin P, Zgoda V, Larina O, Kisrieva Y, Klimenko M, Latyshkevich O, Klimenko P, Lisitsa A, Petushkova N. Protocol for Increasing the Sensitivity of MS-Based Protein Detection in Human Chorionic Villi. Current Issues in Molecular Biology. 2022; 44(5):2069-2088. https://doi.org/10.3390/cimb44050140
Chicago/Turabian StyleShkrigunov, Timur, Pavel Pogodin, Victor Zgoda, Olesya Larina, Yulia Kisrieva, Maria Klimenko, Oleg Latyshkevich, Peter Klimenko, Andrey Lisitsa, and Natalia Petushkova. 2022. "Protocol for Increasing the Sensitivity of MS-Based Protein Detection in Human Chorionic Villi" Current Issues in Molecular Biology 44, no. 5: 2069-2088. https://doi.org/10.3390/cimb44050140
APA StyleShkrigunov, T., Pogodin, P., Zgoda, V., Larina, O., Kisrieva, Y., Klimenko, M., Latyshkevich, O., Klimenko, P., Lisitsa, A., & Petushkova, N. (2022). Protocol for Increasing the Sensitivity of MS-Based Protein Detection in Human Chorionic Villi. Current Issues in Molecular Biology, 44(5), 2069-2088. https://doi.org/10.3390/cimb44050140






