Amorphigenin from Amorpha fruticosa L. Root Extract Induces Autophagy-Mediated Melanosome Degradation in mTOR-Independent- and AMPK-Dependent Manner
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Chemicals
2.2. Cell Viability Assay
2.3. Melanin Assay
2.4. L-DOPA Oxidation
2.5. Preparation of Root Bark Extract and Isolation of Amorphigenin
2.6. RNA Extraction and Polymerase Chain Reaction (PCR)
2.7. Western Blotting
2.8. Confocal Microscopy
2.9. Gene Silencing
3. Results
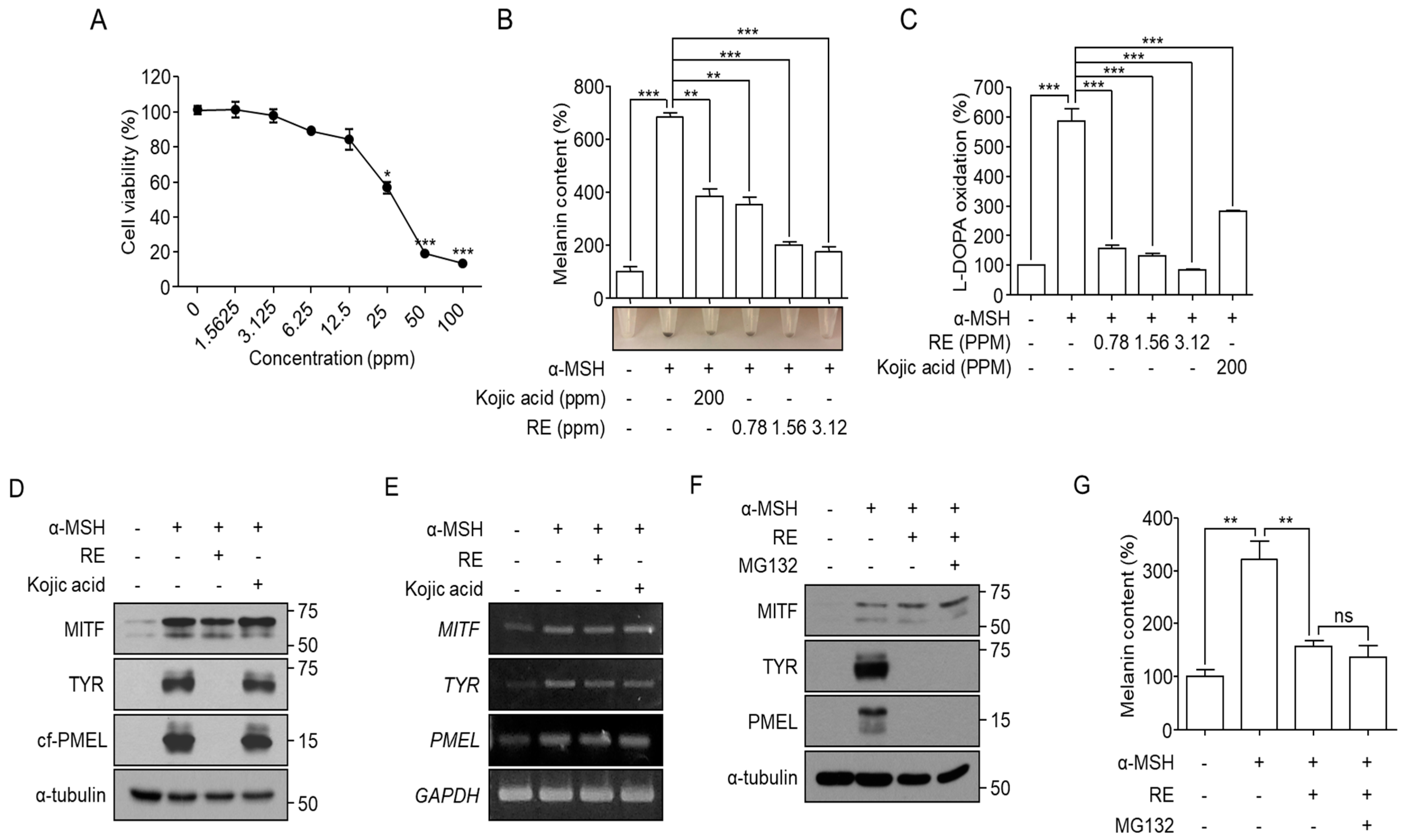
3.1. A. fruticosa RE Induces Depigmentation in α-MSH-Stimulated B16F10 Cells
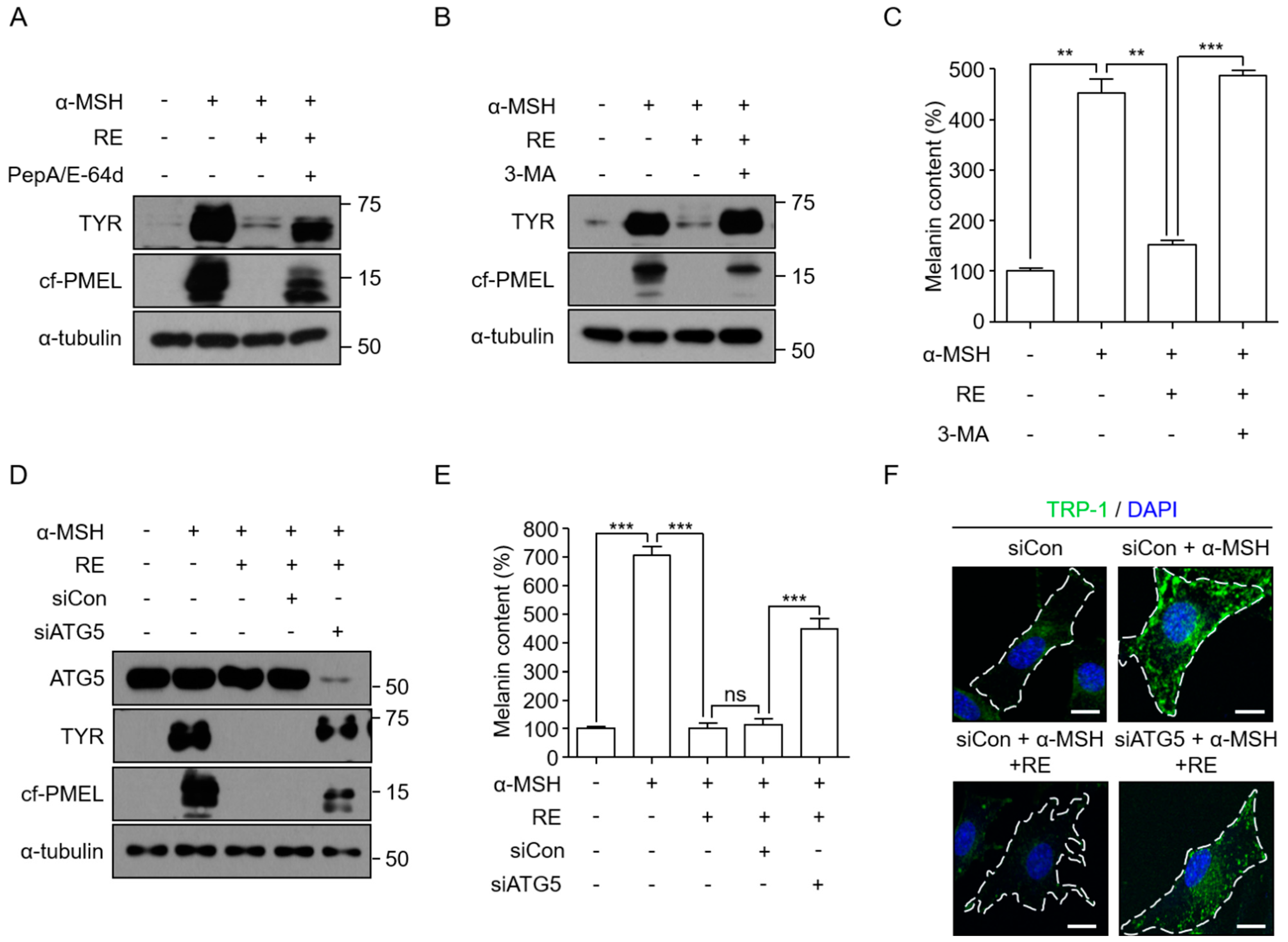
3.2. A. fruticosa RE-Mediated Depigmentation Depends on Autophagy
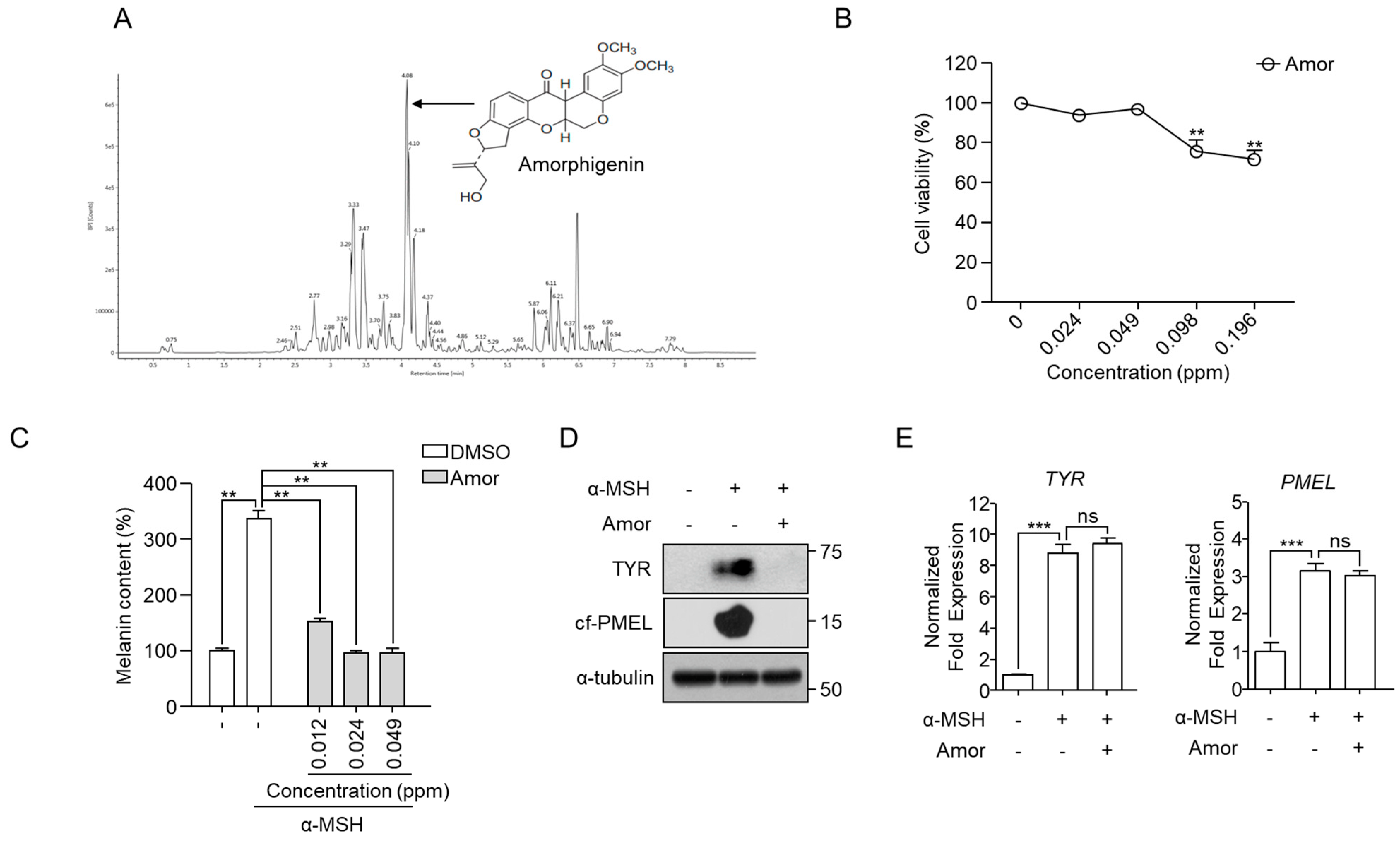
3.3. Identification of Amorphigenin as a Functional Metabolite in A. fruticosa RE
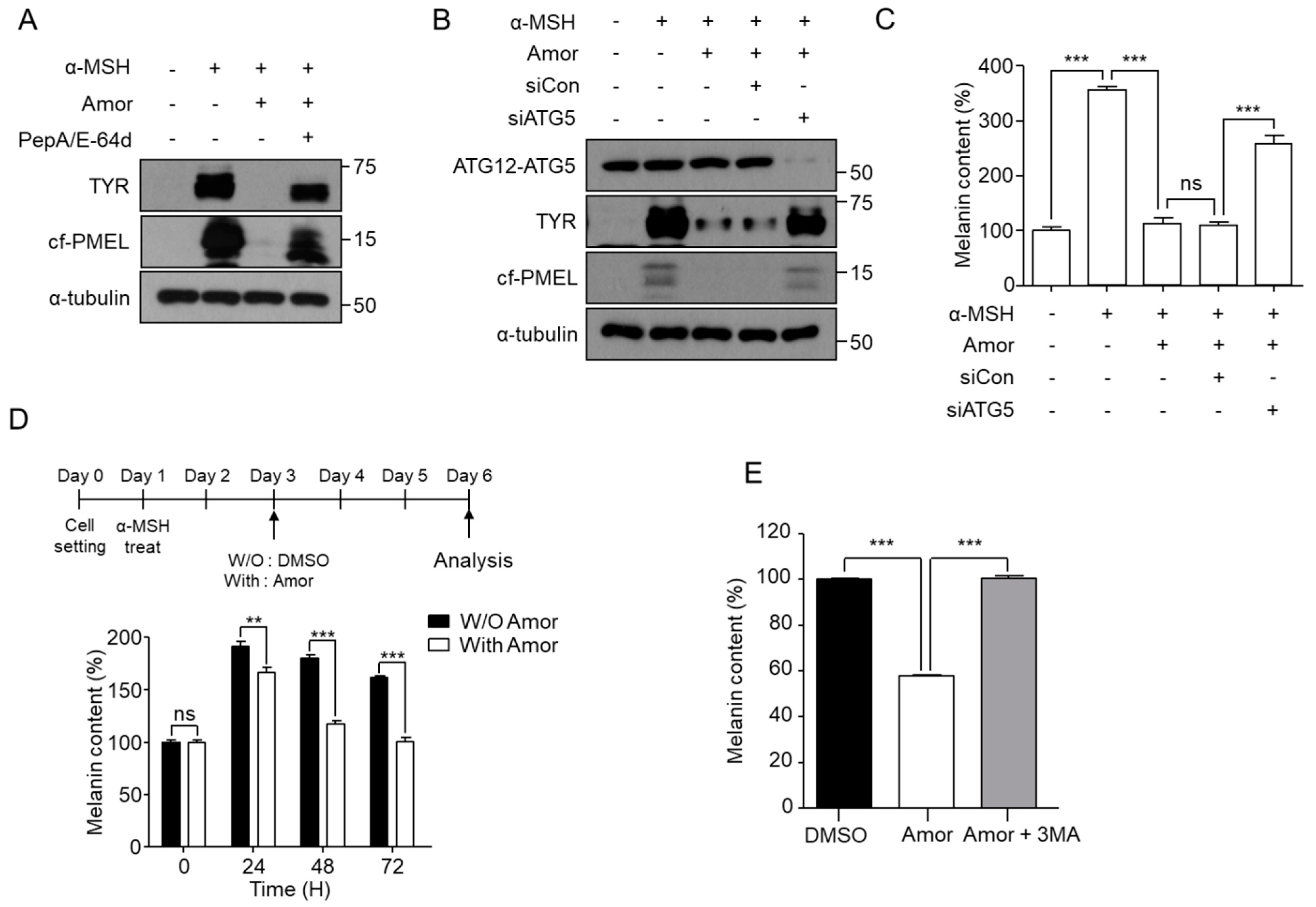
3.4. Amorphigenin-Mediated Depigmentation Depends on Autophagy
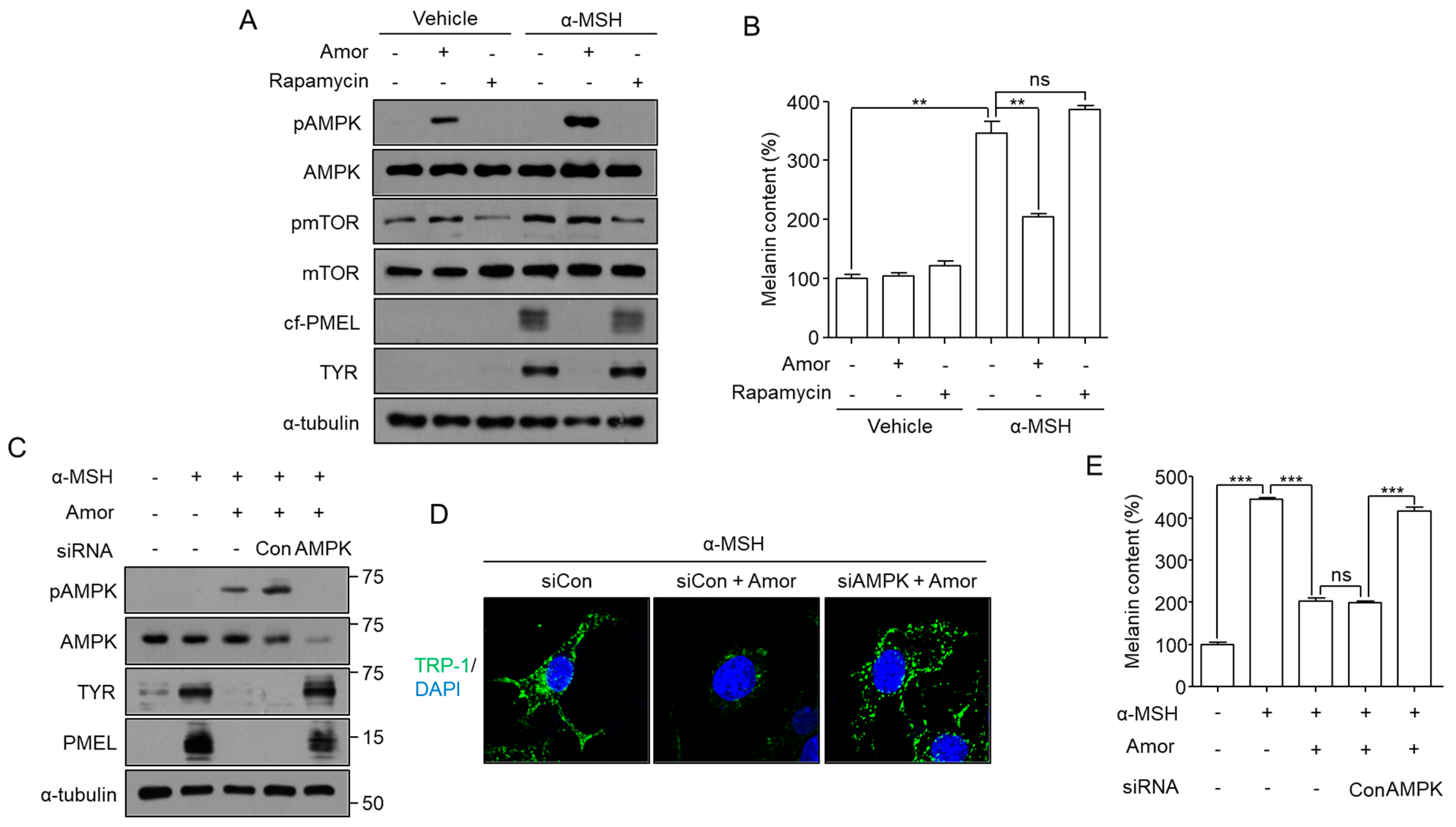
3.5. Amorphigenin Induces Degradation of Melanosomes in an mTOR-Independent and AMPK-Dependent Manner
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, J.Y.; Fisher, D.E. Melanocyte biology and skin pigmentation. Nature 2007, 445, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Bertolotto, C.; Abbe, P.; Hemesath, T.J.; Bille, K.; Fisher, D.E.; Ortonne, J.P.; Ballotti, R. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J. Cell Biol. 1998, 142, 827–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasumoto, K.; Yokoyama, K.; Takahashi, K.; Tomita, Y.; Shibahara, S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J. Biol. Chem. 1997, 272, 503–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Miller, A.J.; Widlund, H.R.; Horstmann, M.A.; Ramaswamy, S.; Fisher, D.E. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am. J. Pathol. 2003, 163, 333–343. [Google Scholar] [CrossRef]
- Di Pietro, S.M.; Falcon-Perez, J.M.; Tenza, D.; Setty, S.R.; Marks, M.S.; Raposo, G.; Dell’Angelica, E.C. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell 2006, 17, 4027–4038. [Google Scholar] [CrossRef] [Green Version]
- Sitaram, A.; Dennis, M.K.; Chaudhuri, R.; De Jesus-Rojas, W.; Tenza, D.; Setty, S.R.; Wood, C.S.; Sviderskaya, E.V.; Bennett, D.C.; Raposo, G.; et al. Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol. Biol. Cell 2012, 23, 3178–3192. [Google Scholar] [CrossRef]
- Huizing, M.; Sarangarajan, R.; Strovel, E.; Zhao, Y.; Gahl, W.A.; Boissy, R.E. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol. Biol. Cell 2001, 12, 2075–2085. [Google Scholar] [CrossRef] [Green Version]
- Theos, A.C.; Tenza, D.; Martina, J.A.; Hurbain, I.; Peden, A.A.; Sviderskaya, E.V.; Stewart, A.; Robinson, M.S.; Bennett, D.C.; Cutler, D.F.; et al. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell 2005, 16, 5356–5372. [Google Scholar] [CrossRef]
- Raposo, G.; Tenza, D.; Murphy, D.M.; Berson, J.F.; Marks, M.S. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 2001, 152, 809–824. [Google Scholar] [CrossRef] [Green Version]
- Brenner, M.; Hearing, V.J. The protective role of melanin against UV damage in human skin. Photochem. Photobiol. 2008, 84, 539–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natale, C.A.; Duperret, E.K.; Zhang, J.; Sadeghi, R.; Dahal, A.; O’Brien, K.T.; Cookson, R.; Winkler, J.D.; Ridky, T.W. Sex steroids regulate skin pigmentation through nonclassical membrane-bound receptors. eLife 2016, 5, e15104. [Google Scholar] [CrossRef] [PubMed]
- Filoni, A.; Mariano, M.; Cameli, N. Melasma: How hormones can modulate skin pigmentation. J. Cosmet. Derm. 2019, 18, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.C.; Miot, L.D.; Miot, H.A. Melasma: A clinical and epidemiological review. An. Bras. Derm. 2014, 89, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Murase, D.; Hachiya, A.; Takano, K.; Hicks, R.; Visscher, M.O.; Kitahara, T.; Hase, T.; Takema, Y.; Yoshimori, T. Autophagy has a significant role in determining skin color by regulating melanosome degradation in keratinocytes. J. Investig. Dermatol. 2013, 133, 2416–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazova, R.; Pawelek, J.M. Why do melanomas get so dark? Exp. Dermatol. 2009, 18, 934–938. [Google Scholar] [CrossRef]
- Lazova, R.; Klump, V.; Pawelek, J. Autophagy in cutaneous malignant melanoma. J. Cutan. Pathol. 2010, 37, 256–268. [Google Scholar] [CrossRef]
- Bomirski, A.; Wrzolkowa, T.; Arendarczyk, M.; Bomirska, M.; Kuklinska, E.; Slominski, A.; Moellmann, G. Pathology and ultrastructural characteristics of a hypomelanotic variant of transplantable hamster melanoma with elevated tyrosinase activity. J. Investig. Dermatol. 1987, 89, 469–473. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Kim, J.; Ahn, Y.; Lee, E.J.; Hwang, S.; Almurayshid, A.; Park, K.; Chung, H.J.; Kim, H.J.; Lee, S.H.; et al. Autophagy induction can regulate skin pigmentation by causing melanosome degradation in keratinocytes and melanocytes. Pigment. Cell Melanoma Res. 2020, 33, 403–415. [Google Scholar] [CrossRef]
- Kim, E.S.; Shin, J.H.; Seok, S.H.; Kim, J.B.; Chang, H.; Park, S.J.; Jo, Y.K.; Choi, E.S.; Park, J.S.; Yeom, M.H.; et al. Autophagy mediates anti-melanogenic activity of 3′-ODI in B16F1 melanoma cells. Biochem. Biophys. Res. Commun. 2013, 442, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.S.; Shin, J.H.; Jo, D.S.; Shin, D.W.; Choi, D.H.; Kim, W.J.; Park, K.; Kim, J.K.; Joo, C.G.; Lee, J.S.; et al. Anti-melanogenic activity of schaftoside in Rhizoma Arisaematis by increasing autophagy in B16F1 cells. Biochem. Biophys. Res. Commun. 2018, 503, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Jo, Y.K.; Park, S.J.; Chang, H.; Shin, J.H.; Choi, E.S.; Kim, J.B.; Seok, S.H.; Kim, J.S.; Oh, J.S.; et al. ARP101 inhibits alpha-MSH-stimulated melanogenesis by regulation of autophagy in melanocytes. FEBS Lett. 2013, 587, 3955–3960. [Google Scholar] [CrossRef]
- Lee, K.W.; Ryu, H.W.; Oh, S.S.; Park, S.; Madhi, H.; Yoo, J.; Park, K.H.; Kim, K.D. Depigmentation of alpha-melanocyte-stimulating hormone-treated melanoma cells by beta-mangostin is mediated by selective autophagy. Exp. Dermatol. 2017, 26, 585–591. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ryu, Y.B.; Curtis-Long, M.J.; Yuk, H.J.; Cho, J.K.; Kim, J.Y.; Kim, K.D.; Lee, W.S.; Park, K.H. Flavanones and rotenoids from the roots of Amorpha fruticosa L. that inhibit bacterial neuraminidase. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2011, 49, 1849–1856. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Daido, S.; Yamamoto, A.; Kobayashi, R.; Yokoyama, T.; Aoki, H.; Iwado, E.; Shinojima, N.; Kondo, Y.; Kondo, S. Pivotal role of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 in apoptosis and autophagy. J. Biol. Chem. 2008, 283, 388–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanov, J.; Walczak, M.; Ibiricu, I.; Schuchner, S.; Ogris, E.; Kraft, C.; Martens, S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012, 31, 4304–4317. [Google Scholar] [CrossRef]
- Cui, X.; Guo, J.; Lai, C.S.; Pan, M.H.; Ma, Z.; Guo, S.; Liu, Q.; Zhang, L.; Ho, C.T.; Bai, N. Analysis of bioactive constituents from the leaves of Amorpha fruticosa L. J. Food Drug Anal. 2017, 25, 992–999. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Lee, J.H. The combination of tephrosin with 2-deoxy-D-glucose enhances the cytotoxicity via accelerating ATP depletion and blunting autophagy in human cancer cells. Cancer Biol. 2011, 12, 989–996. [Google Scholar] [CrossRef] [Green Version]
- Fuhr, L.; Rousseau, M.; Plauth, A.; Schroeder, F.C.; Sauer, S. Amorfrutins Are Natural PPARgamma Agonists with Potent Anti-inflammatory Properties. J. Nat. Prod. 2015, 78, 1160–1164. [Google Scholar] [CrossRef]
- Sauer, S. Amorfrutins: A promising class of natural products that are beneficial to health. Chembiochem 2014, 15, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Marks, M.S. Melanosomes--dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007, 8, 786–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Hammer, J.A. Melanosome transfer: It is best to give and receive. Curr. Opin. Cell Biol. 2014, 29, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seiberg, M.; Paine, C.; Sharlow, E.; Andrade-Gordon, P.; Costanzo, M.; Eisinger, M.; Shapiro, S.S. Inhibition of melanosome transfer results in skin lightening. J. Investig. Dermatol. 2000, 115, 162–167. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.S.; Nam, G.; Bae, I.H.; Park, J. Whitening efficacy of ginsenoside F1 through inhibition of melanin transfer in cocultured human melanocytes-keratinocytes and three-dimensional human skin equivalent. J. Ginseng Res. 2019, 43, 300–304. [Google Scholar] [CrossRef]
- Pomerantz, S.H. Separation, purification, and properties of two tyrosinases from hamster melanoma. J. Biol. Chem. 1963, 238, 2351–2357. [Google Scholar] [CrossRef]
- Cabanes, J.; Chazarra, S.; Garcia-Carmona, F. Kojic acid, a cosmetic skin whitening agent, is a slow-binding inhibitor of catecholase activity of tyrosinase. J. Pharm. Pharmacol. 1994, 46, 982–985. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, Y.; Tao, L.; Tao, X.; Su, X.; Wei, D. Tyrosinase inhibitory effects and inhibition mechanisms of nobiletin and hesperidin from citrus peel crude extracts. J. Enzym. Inhib. Med. Chem. 2007, 22, 91–98. [Google Scholar] [CrossRef]
- Itoh, K.; Hirata, N.; Masuda, M.; Naruto, S.; Murata, K.; Wakabayashi, K.; Matsuda, H. Inhibitory effects of Citrus hassaku extract and its flavanone glycosides on melanogenesis. Biol. Pharm. Bull. 2009, 32, 410–415. [Google Scholar] [CrossRef] [Green Version]
- Maresca, V.; Flori, E.; Cardinali, G.; Briganti, S.; Lombardi, D.; Mileo, A.M.; Paggi, M.G.; Picardo, M. Ferritin light chain down-modulation generates depigmentation in human metastatic melanoma cells by influencing tyrosinase maturation. J. Cell Physiol. 2006, 206, 843–848. [Google Scholar] [CrossRef]
- Ryu, H.W.; Jeong, S.H.; Curtis-Long, M.J.; Jung, S.; Lee, J.W.; Woo, H.S.; Cho, J.K.; Park, K.H. Inhibition effects of mangosenone F from Garcinia mangostana on melanin formation in B16F10 cells. J. Agric. Food Chem. 2012, 60, 8372–8378. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Kim, J.A.; Eom, S.Y.; Lee, S.H.; Min, K.R.; Kim, Y. Inhibitory effect of piperlonguminine on melanin production in melanoma B16 cell line by downregulation of tyrosinase expression. Pigment Cell Res. 2006, 19, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Kim, D.S.; Kim, W.G.; Ryoo, I.J.; Lee, D.H.; Huh, C.H.; Youn, S.W.; Yoo, I.D.; Park, K.C. Terrein: A new melanogenesis inhibitor and its mechanism. Cell Mol. Life Sci. 2004, 61, 2878–2885. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Lee, D.H.; Goldberg, A.L. Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 1998, 8, 397–403. [Google Scholar] [CrossRef]
- Jackson, M.P.; Hewitt, E.W. Cellular proteostasis: Degradation of misfolded proteins by lysosomes. Essays Biochem. 2016, 60, 173–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [Green Version]
- Alers, S.; Loffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012, 32, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Tian, W.; Hu, Z.; Chen, G.; Huang, L.; Li, W.; Zhang, X.; Xue, P.; Zhou, C.; Liu, L.; et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014, 15, 566–575. [Google Scholar] [CrossRef] [Green Version]
- Drake, J.C.; Wilson, R.J.; Laker, R.C.; Guan, Y.; Spaulding, H.R.; Nichenko, A.S.; Shen, W.; Shang, H.; Dorn, M.V.; Huang, K.; et al. Mitochondria-localized AMPK responds to local energetics and contributes to exercise and energetic stress-induced mitophagy. Proc. Natl. Acad. Sci. USA 2021, 118, e2025932118. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.W.; Nguyen, D.T.; Kim, M.; Lee, S.H.; Lim, S.; Kim, J.; Park, K.H.; Kim, J.Y.; Yoo, J.; Hwangbo, C.; et al. Amorphigenin from Amorpha fruticosa L. Root Extract Induces Autophagy-Mediated Melanosome Degradation in mTOR-Independent- and AMPK-Dependent Manner. Curr. Issues Mol. Biol. 2022, 44, 2856-2867. https://doi.org/10.3390/cimb44070196
Lee KW, Nguyen DT, Kim M, Lee SH, Lim S, Kim J, Park KH, Kim JY, Yoo J, Hwangbo C, et al. Amorphigenin from Amorpha fruticosa L. Root Extract Induces Autophagy-Mediated Melanosome Degradation in mTOR-Independent- and AMPK-Dependent Manner. Current Issues in Molecular Biology. 2022; 44(7):2856-2867. https://doi.org/10.3390/cimb44070196
Chicago/Turabian StyleLee, Ki Won, Dang Thi Nguyen, Minju Kim, Si Hyeon Lee, Seyeon Lim, Jisu Kim, Ki Hun Park, Jeong Yoon Kim, Jiyun Yoo, Cheol Hwangbo, and et al. 2022. "Amorphigenin from Amorpha fruticosa L. Root Extract Induces Autophagy-Mediated Melanosome Degradation in mTOR-Independent- and AMPK-Dependent Manner" Current Issues in Molecular Biology 44, no. 7: 2856-2867. https://doi.org/10.3390/cimb44070196
APA StyleLee, K. W., Nguyen, D. T., Kim, M., Lee, S. H., Lim, S., Kim, J., Park, K. H., Kim, J. Y., Yoo, J., Hwangbo, C., & Kim, K. D. (2022). Amorphigenin from Amorpha fruticosa L. Root Extract Induces Autophagy-Mediated Melanosome Degradation in mTOR-Independent- and AMPK-Dependent Manner. Current Issues in Molecular Biology, 44(7), 2856-2867. https://doi.org/10.3390/cimb44070196







