Genetic and Epigenetic Regulation of MEFV Gene and Their Impact on Clinical Outcome in Auto-Inflammatory Familial Mediterranean Fever Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
Disease Severity
2.2. Methods
2.2.1. Blood Sample Collection
2.2.2. Mutation Analysis
2.2.3. Methylation Analysis
2.2.4. Pyrin Analysis
2.2.5. Receiver Operating Characteristic Curve (ROC Curve)
2.3. Statistical Analysis
3. Results
3.1. Demographic Data
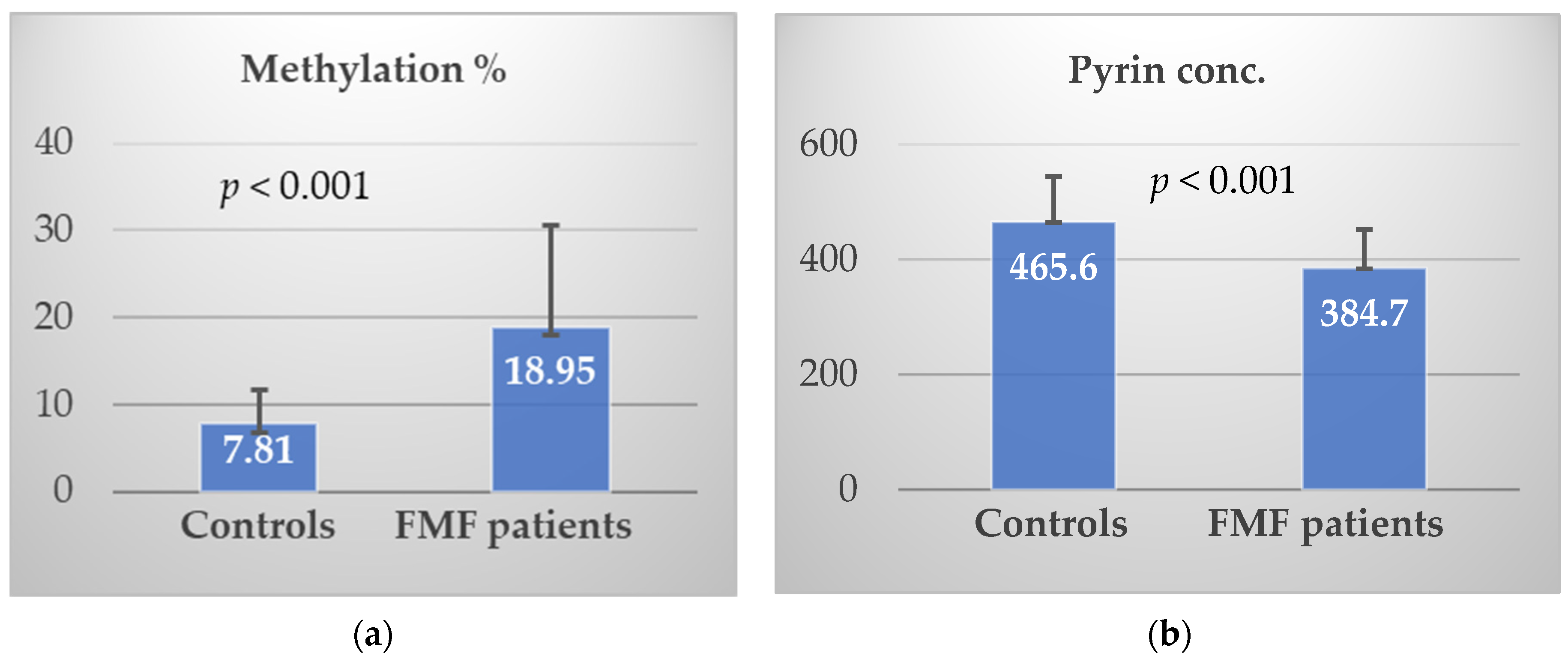
3.2. Methylation% and Pyrin Level
3.3. Genotyping
3.4. Disease Severity and Studied Parameters
3.5. Biomarkers
3.6. Phenotype and Studied Parameters
3.7. Colchicine Response
3.7.1. Colchicine Response among Patients
3.7.2. Colchicine Response, Methylation%, and Pyrin
3.8. Correlations between Studied Biomarkers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, S.; Zhang, J.; Tee, W. Epigenetic Regulation of Inflammatory Signaling and Inflammation-Induced Cancer. Front. Cell Dev. Biol. 2022, 10, 931493. [Google Scholar] [CrossRef] [PubMed]
- Maximilian, H.; Giacomo, C. Molecular Mechanisms of Trans-generational Epigenetic Inheritance. Nat. Rev. Genet. 2022, 23, 325–341. [Google Scholar]
- Alyson, A.; Vincent, C.; Benjamin, P. How does epigenetics influence the course of evolution? Philos. Trans. R. Soc. B 2020, 376, 20200111. [Google Scholar]
- Tuong, Z.; Stewart, B.; Guo, S.; Clatworthy, M. Epigenetics and Tissue Immunity—Translating Environmental Cues into Functional Adaptations. Immunol. Rev. 2022, 305, 111–136. [Google Scholar] [CrossRef]
- Álvarez-Errico, D.; Vento-Tormo, R.; Ballestar, E. Genetic and Epigenetic Determinants in Auto-inflammatory Diseases. Front. Immunol. 2017, 8, 318. [Google Scholar] [CrossRef]
- Saxena, R.; Tiwari, K. Role of Epigenetics in Developing Therapeutic Strategies against COVID-19. J. Clin. Diagn. Res. 2022, 16, GE01–GE08. [Google Scholar] [CrossRef]
- Santaló, J.; Berdasc, M. Ethical Implications of Epigenetic in the Era of Personalized Medicine. Clin. Epigenetics 2022, 14, 44. [Google Scholar] [CrossRef]
- Li, J.; Li, L.; Wang, Y.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. Insights Into the Role of DNA Methylation in Immune Cell Development and Autoimmune Disease. Front. Cell Dev. Biol. 2021, 9, 757318. [Google Scholar] [CrossRef]
- Pasquale, S.; Assunta, S.; Cesar, A.; Giorgio, G.; Giovanni, N.; Francesca, R.; Roberta, T.; Claudio, S. Metabolic Regulation of Epigenetic Modifications and Cell Differentiation in Cancer. Cancers 2020, 12, 3788. [Google Scholar]
- Dogan, E.; Gursoy, S.; Bozkaya, G.; Camlar, S.; Kilicarslan, O.; Soylu, A.; Ulgenalp, A.; Kavukcu, S.; Bozkaya, O.G. The Effects of Epigenetic Regulation on Phenotypic Expressivity in Turkish Patients with Familial Mediterranean fever. Indian J. Rheumatol. 2019, 14, 297–303. [Google Scholar]
- Sebastian, K.; Silvia, D.; Christiane, W.; Michael, B.; Sevi, D.; Lukas, B.; Dirk, S. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet. 2022, 54, 1895–1906. [Google Scholar]
- Yutaka, M.; Izuru, O.; Erik, W.; Daichi, M.; Kenji, S.; Masahiro, S. Structural Insights into Methylated DNA Recognition by the MethylCpG Binding Domain of MBD6 from Arabidopsis thaliana. ACS Omega 2022, 7, 3212–3221. [Google Scholar]
- Sammallahti, S.; Koopman-Verhoef, M.; Binter, A.; Mulder, R.; Cabré-Riera, A.; Kvist, T.; Malmberg, A.; Pesce, G.; Plancoulaine, S.; Heiss, J.; et al. Longitudinal Associations of DNA Methylation and Sleep in Children: A meta-analysis. Clin. Epigenetics 2022, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Zelin, J.; Yun, L. DNA methylation in human diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar]
- Erdem, G.; Erdemir, S.; Abaci, I.; Kirectepe, A.; Everest, E.; Turanli, E. Alternatively spliced MEFV transcript lacking exon 2 and its protein isoform pyrin-2d implies an epigenetic regulation of the gene in inflammatory cell culture models. Genet. Mol. Biol. 2017, 40, 688–697. [Google Scholar]
- Carlos, D.; Octavio, M.; Esteban, B. Understanding the Relevance of DNA Methylation Changes in Immune Differentiation and Disease. Genes 2020, 11, 110. [Google Scholar]
- Krista, S.; Thomas, P.; Robert, J.; Lynn, B. Folate and DNA Methylation: A Review of Molecular Mechanisms and the Evidence for Folate’s Role. Am. Soc. Nutrition. Adv. Nutr. 2012, 3, 21–38. [Google Scholar]
- Funes, S.; Fernández-Fierro, A.; Rebolledo-Zelada, D.; Mackern-Oberti, J.; Kalergis, A. Contribution of Dysregulated DNA Methylation to Autoimmunity. Int. J. Mol. Sci. 2021, 22, 11892. [Google Scholar] [CrossRef] [PubMed]
- Migita, K.; Asano, T.; Sato, S.; Koga, T.; Fujita, Y.; Kawakami, A. Familial Mediterranean fever: Overview of Pathogenesis, Clinical Features and Management. Immunol. Med. 2018, 41, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.; El-Shayeb, A.; ElHabachi, N.; Khodair, M.; Elwazzan, D.; Abdeen, N.; Said, M.; Ebaid, R.; El-Shahawy, N.; Seif, A.; et al. Molecular Patterns of MEFV Gene Mutations in Egyptian Patients with Familial Mediterranean Fever: A Retrospective Cohort Study. Int. J. Inflamm. 2019, 2019, 2578760. [Google Scholar] [CrossRef]
- Mansour, A. Familial Mediterranean fever, review of the literature. Clin. Rheumatol. 2017, 36, 1707–1713. [Google Scholar]
- Abukhalaf, S.; Dandis, B.; Tari, T.; Alzughayyar, A.; Rajabi, Y. Familial Mediterranean fever Complicated by a Triad of Adrenal Crisis: Amyloid Goiter and Cardiac Amyloidosis. Case Rep. Rheumatol. 2020, 2020, 7865291. [Google Scholar] [CrossRef] [PubMed]
- Ozen, S.; Kone-Paut, I.; Gül, A. Colchicine Resistance and Intolerance in Familial Mediterranean fever: Definition, Causes, and Alternative Treatments. Semin. Arthritis Rheum. 2017, 47, 115–120. [Google Scholar] [CrossRef]
- Maggio, M.; Corsello, G. FMF is not always “fever”: From clinical presentation to “treat to target”. Ital. J. Pediatr. 2020, 46, 7. [Google Scholar] [CrossRef]
- Manukyan, G.; Aminov, R. Update on Pyrin Functions and Mechanisms of Familial Mediterranean fever. Front. Microbiol. 2016, 7, 456. [Google Scholar] [CrossRef]
- Yvan, J.; Lucie, L.; Flora, M.; Amandine, M.; Sarah, B.; Omran, A.; Mathilde, P.; Ve’ronique, H.; Pascal, S.; Mathieu, G.; et al. Familial Mediterranean fever mutations are hypermorphic mutations that specifically decrease the activation threshold of the Pyrin inflammasome. Rheumatology 2018, 57, 100111. [Google Scholar]
- Mangan, M.; Gorki, F.; Krause, K.; Heinz, A.; Pankow, A.; Ebert, T. Transcriptional licensing is required for Pyrin inflammasome activation in human macrophages and bypassed by mutations causing familial Mediterranean fever. PLoS Biol. 2020, 20, e3001351. [Google Scholar] [CrossRef] [PubMed]
- Heilig, R.; Petr, B. Function and Mechanism of the Pyrin Inflammasome. Eur. J. Immunol. 2018, 48, 230–238. [Google Scholar] [CrossRef]
- Schnappauf, O.; Chae, J.; Kastner, D.; Aksentijevich, I. The PyrinInflammasome in Health and Disease. Front. Immunol. 2019, 10, 1745. [Google Scholar] [CrossRef]
- Sei, S.; Ryusuke, Y.; Yohei, K.; Hideaki, N. The PRY/SPRY domain of pyrin/TRIM20 interacts with β2-microglobulin to promote infammasome formation. Sci. Rep. 2021, 11, 23613. [Google Scholar]
- Sevinç, N.; Erdem, G.; Kireçtepe-Aydın, A.; Altin, S. Increased Pyrin Protein Levels in Patients with Familial Mediterranean Fever (FMF) could be Independent of Mediterranean Fever Gene (MEFV) Exon10 Variations. In Proceedings of the 12th National Congress of Medical Genetics Conference, Turkish Society of Medical Genetics, Turkey, September 2016. [Google Scholar]
- Kirectepe, A.; Kasapcopur, O.; Arisoy, N.; Erdem, G.; Hatemi, G.; Ozdogan, H. Analysis of MEFV Exon Methylation and Expression Patterns in Familial Mediterranean Fever. BMC Med. Genet. 2011, 12, 105. [Google Scholar]
- Samli, H.; Dogru, O.; Bukulmez, A.; Yuksel, E.; Ovali, F.; Solak, M. Relationship of Tel Hashomer Criteria and Mediterranean fever Gene Mutations in a Cohort of Turkish Familial Mediterranean Fever Patients. Saudi Med. J. 2006, 12, 1822–1826. [Google Scholar]
- Pras, E.; Livneh, A.; Balow, J.E., Jr.; Pras, E.; Kastner, D.L.; Pras, M.; Langevitz, P. Clinical Differences between North African and Iraqi Jews with Familial Mediterranean fever. Am. J. Med. Genet. 1998, 75, 216–219. [Google Scholar] [CrossRef]
- Babidge, W.; Butler, L.; Burton, M.; Cowled, P. Methylation of CpG Sites in Exon 2 of the Bcl-2 Gene occurs in Colo-rectal Carcinoma. Anticancer Res. 2001, 21, 2809–2814. [Google Scholar]
- Lo, P.K.; Watanabe, H.; Cheng, P.C.; Teo, W.W.; Liang, X.; Argani, P.; Lee, J.S.; Sukumar, S. Methy SYBR, a Novel Quantitative PCR Assay for the Dual Analysis of DNA Methylation and CpG Methylation Density. J. Mol. Diagn. 2009, 11, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Al-Haggar, M.; Yahia, S.; bdel-Hady, D.; Al-Saied, A.; Al-Kenawy, R.; Abo-El-Kasem, R. Phenotype-Genotype Updates from Familial Mediterranean Fever Database Registry of Mansoura University Children Hospital, Mansoura, Egypt. Indian J. Hum. Genet. 2014, 20, 43. [Google Scholar] [CrossRef]
- Talaat, H.S.; Sheba, M.F.; Mohammed, R.H.; Gomaa, M.A.; El Rifaei, N.; Ibrahim, M.F.M. Genotype Mutations in Egyptian Children with Familial Mediterranean fever: Clinical Profile, and Response to Colchicines. Mediterr. J. Rheumatol. 2020, 31, 206. [Google Scholar] [CrossRef]
- El Gezery, D.; Abou-Zeid, A.; Hashad, D.; El-Sayegh, H. MEFV Gene Mutations in Egyptian Patients with Familial Mediterranean fever. Genet. Test. Mol. Biomark. 2010, 14, 263–268. [Google Scholar] [CrossRef]
- Yomna, F.; Heba, T.; Noha, M.; Diana, F.; Huda, M. Articular manifestations in Egyptian children with familial Mediterranean fever. Egypt. Rheumatol. Rehabil. 2020, 47, 48. [Google Scholar]
- Lofty, H.M.; Marzouk, H.; Farag, Y.; Nabih, M.; Khalifa, I.A.; Mostafa, N.; Salah, A.; Rashed, L.; El Garf, K. Serum Amyloid A level in Egyptian Children with Familial Mediterranean Fever. Int. J. Rheumatol. 2016, 2016, 7354018. [Google Scholar] [CrossRef]
- Mohammed, H.; Omar, E.; Eslam, M.; Aya, M. Clinical and genetic characterization of familial Mediterranean fever among a cohort of Egyptian patients. Gastroenterol. Rev. 2022, 17, 240–244. [Google Scholar]
- Beshlawy, A.; Zekri, A.; Ramadan, M.; Selim, Y.; Abdel-Salam, A.; Hegazy, M.; Ragab, L.; Gaggiano, C.; Cantarini, L.; Ragab, G. Genotype-phenotype associations in familial Mediterranean fever: A study of 500 Egyptian pediatric patients. Clin. Rheumatol. 2022, 41, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Taryma-Leśniak, O.; EwaSokolowska, K.; Kazimierz, T. Current Status of Development of Methylation Biomarkers for in Vitro Diagnostic IVD Applications. Clin. Epigenetics 2020, 12, 100. [Google Scholar] [CrossRef]
- Turanli, E.; Kirectepe, A.; Kasapçopur, Ö. MEFV Methylation Analysis in FMF, and JRA Diseases. Pediatr. Rheumatol. 2013, 11, A67. [Google Scholar] [CrossRef]
- Yepiskoposyan, L.; Harutyunyan, A. Population genetics of familial Mediterranean fever: A review. Eur. J. Hum. Genet. 2007, 15, 91. [Google Scholar]
- Anderson, O.; Sant, E.; Dolinoy, D. Nutrition and epigenetics: An interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutr. Biochem. 2012, 23, 853–859. [Google Scholar]
- Kandi, V.; Vadakedath, S. Effect of DNA methylation in various diseases and the probable protective role of nutrition: A mini-review. Cureus 2015, 7, e309. [Google Scholar] [CrossRef]
- Sharkia, R.; Mahajnah, M.; Zalan, A.; Athamna, M.; Azem, A.; Badarneh, K.; Faris, F. Comparative Screening of FMF Mutations in Various Communities of the Israeli Society. Eur. J. Med. Genet. 2013, 56, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Shahbaznejad, L.; Raeeskarami, S.R.; Assari, R.; Shakoori, A.; Azhideh, H.; Aghighi, Y.; Tahghighi, F.; Ziaee, V. Familial Mediterranean Gene (MEFV) Mutation in Parents of Children with Familial Mediterranean fever: What Are the Exceptions? Int. J. Inflamm. 2018, 2018, 1902791. [Google Scholar] [CrossRef] [PubMed]
- El-Messery, L.; Elhagrasy, H. Study of MEFV Gene R202Q Polymorphism in Egyptian Patients with Familial Mediterranean fever. Egypt. J. Haematol. 2014, 39, 64. [Google Scholar] [CrossRef]
- Kehribar, D.; Özgen, M. The Importance of Mediterranean fever Gene in Familial Mediterranean Fever. Eur. J. Rheumatol. 2020, 7, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zvi, I.; Livneh, A. Chronic Inflammation in FMF: Markers, Risk Factors, Outcomes and Therapy. Nat. Rev. Rheumatol. 2010, 7, 105–112. [Google Scholar] [CrossRef]
- Iwona, G.; Mariusz, K.; Andrzej, C.; Klaudyna, L.; Paweł, J.; Oksana, B.; Bartosz, K.; Andrzej, B.; Lech, C. Polymorphism of Interleukin 1B May Modulate the Risk of Ischemic Stroke in Polish Patients. Medicina 2019, 55, 558. [Google Scholar]
- Panel, C.; Hasmik, A.; Stéphanie, P. Identification of MEFV independent Modifying Genetic factors for FMF. Am. J. Hum. Genet. 2000, 67, 1136–1143. [Google Scholar]
- Duşunsel, R.; Dursun, I.; Gündüz, Z.; Poyrazoğlu, M.; Gürgöze, M.; Dundar, M. Genotype–Phenotype correlation in children with familial Mediterranean fever in a Turkish population. Pediatr. Int. 2008, 50, 208–212. [Google Scholar] [CrossRef]
- Liu, J.; Morgan, M.; Hutchison, K.; Calhoun, V. A Study of the Influence of Sex on Genome Wide Methylation. PLoS ONE 2010, 5, e10028. [Google Scholar] [CrossRef] [PubMed]
- Corsia, A.; Georgin-Lavialle, S.; Hentgen, V.; Hachulla, E.; Grateau, G.; Faye, A.; Quartier, P.; Rossi-Semerano, L.; Koné-Paut, I. A Survey of Resistance to Colchicine Treatment for French Patients with Familial Mediterranean fever. Orphanet J. Rare Dis. 2017, 12, 54. [Google Scholar] [CrossRef]
- Lidar, M.; Scherrmann, J.M.; Shinar, Y.; Chetrit, A.; Niel, E.; Gershoni-Baruch, R.; Langevitz, P.; Livneh, A. Colchicine Non-responsiveness in Familial Mediterranean fever: Clinical, Genetic, Pharmacokinetic, and Socioeconomic Characterization. Semin. Arthritis Rheum. 2004, 33, 273–282. [Google Scholar] [CrossRef]
- Gao, W.; Yang, J.; Liu, W.; Wang, Y.; Shao, F. Site-Specific Phosphorylation and Microtubule Dynamics Control Pyrin Inflammasome Activation. Proc. Natl. Acad. Sci. USA 2016, 113, E4857–E4866. [Google Scholar] [CrossRef]
- Ying, Y.; Laura, L.; Virginia, B. Colchicine—Update on mechanisms of action and therapeutic uses. Semin. Arthritis Rheum. 2015, 45, 341–350. [Google Scholar] [CrossRef]
- Mansfield, E.; Chae, J.J.; Komarow, H.D.; Brotz, T.M.; Frucht, D.M.; Aksentijevich, I.; Kastner, D.L. The Familial Mediterranean Fever Protein, Pyrin, Associates with Microtubules and Colocalizes with Actin Filaments. Blood 2001, 98, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Soylemezoglu, O.; Arga, M.; Fidan, K.; Gonen, S.; Emeksiz, H.; Hasanoglu, E.; Buyan, N. Unresponsiveness to colchicine therapy in patients with familial Mediterranean fever homozygous for the M694V mutation. J. Rheumatol. 2010, 37, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Lidar, M.; Yonath, H.; Shechter, N.; Sikron, F.; Sadetzki, S.; Livneh, A.; Pras, E. Incomplete response to colchicine in M694V homozygote FMF patients. Autoimmun. Rev. 2012, 12, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.; Rania, A.; Eman, B. Genotype Phenotype Correlation of FMF Cases in East Delta of Egypt. Egypt. J. Hosp. Med. 2021, 83, 1149–1154. [Google Scholar]





| Parameter | Patients (n = 63) | Control (n = 19) |
|---|---|---|
| Age (years) | 4–20 | 5–20 |
| Male | 28 (44.4%) | 9 (47.3%) |
| Female (n %) | 35 (55.6%) | 10 (52.6%) |
| Age of onset (mean ± SD) | 5.3 ± 3.7 | 0 |
| Colchicine dose (mg/day) (mean ± SD) | 0.5+2.5 | 0 |
| Consanguineous marriage offspring (n %) | 22 (34.9%) | |
| Similarly affected family members (n %) | 54 (85.72%) | - |
| Colchicine-responders | 37 (58.7%) | |
| Colchicine-non-responders | 26 (41.3%) | 0 |
| Demographic Data Total n = 63 (100%) | Methylation% (Mean + SD) | p-Value | Pyrin Concentration (pg/mL) (Mean + SD) | p-Value |
|---|---|---|---|---|
| Gender: | ||||
| Male n = 28 (44.4%) | 17.13 ± 12.0 | 0.268 | 384.66 ± 73.1 | 0.994 |
| Female n = 35 (55.6%) | 20.41 ± 11.3 | 384.79 ± 73.7 | ||
| Consanguineous marriage: | ||||
| Offspring n = 22 (34.9%) | 23.95 ± 13.6 | 0.011 * | 391.84 ± 85.1 | 0.544 |
| Non-offspring n = 41 (65.1%) | 16.27 ± 9.5 | 380.92 ± 56.6 | ||
| Affected family members: | ||||
| Similarly n = 54 (85.72%) | 19.32 ± 11.7 | 0.539 | 382.93 ± 70.5 | 0.609 |
| Not similarly n = 9 (14.3%) | 16.73 ± 11.1 | 395.51 ± 46.8 |
| Genotyping (Mean+ SD) | Methylation % | p-Value | Pyrin Concentration (pg/mL) | p-Value |
|---|---|---|---|---|
| Mutations n = 53 (77.8%) | 20.2 ± 11.9 | 0.107 | 388.27 ± 72.03 | 0.439 |
| No mutations n = 14 (22.2%) | 14.5 ± 9.8 | 372.30 ± 48.20 | ||
| Exon 2 mutations n = 13 (20.6%) | 21.6 ± 15.6 | 0.169 | 360.0 ± 60 | 0.244 |
| Exon10 mutations n = 40 (63.5%) | 19.8 + 10.5 | 393.3 ± 73.0 | ||
| R202Q n = 2 (3.2%) | 25.1 ± 20.8 | 0.448 | 322.3 ± 36.8 | 0.601 |
| M694V n = 4 (6.3%) | 24.8 ± 9.3 | 416.8 ± 60.9 | ||
| M694I n = 19 (30.2%) | 21.5 ± 11.9 | 387.2 ± 88.8 | ||
| E148Q n = 11 (17.5%) | 20.9 ± 15.7 | 366.9 ± 63.1 | ||
| V726A n = 6 (9.5%) | 18.9 ± 7.9 | 399.5 ± 52.1 | ||
| M680I n = 13 (20.6%) | 18.9 ± 7.9 | 395.6 ± 56.7 | ||
| Heterozygous n = 34 (53.96%) | 19.29 ± 12.06 | 386.54 ± 64.2 | ||
| Homozygous n = 8 (12.6%) | 23.038 ± 12.06 | 0.466 | 413.07 ± 112.2 | 0.469 |
| Compound heterozygous n = 7 (5.55%) | 22.52 ± 15.44 | 354.53 ± 42.965 |
| Parameters/ Disease Severity Total n = 63 (100%) | Methylation% (Mean + SD) | p-Value | Pyrin Concentration (pg/mL) (Mean + SD) | p-Value |
|---|---|---|---|---|
| Mild n = 13 (20.6%) | 11.36 ± 7.8 | 0.016 * | 392.06 ± 51.2 | 0.664 |
| Moderate and severe n = 50 (79.3%) | 20.4 ± 12.1 | 382.82 ± 71.4 |
| Severe n = 32 (50.8%) | Moderate n = 18 (28.6%) | Mild n = 13 (20.6%) | p-Value | |
|---|---|---|---|---|
| Mutations n = 49 (77.8%) | 26 (81.25%) | 14 (77.7%) | 9 (69.23%) | 0.68 |
| No mutations n = 14 (22.2%) | 6 (18.75%) | 4 (22.22%) | 4 (30.7%) | |
| Exon 2 mutations n = 13 (20.6) | 7 (53.8%) | 2 (15.4%) | 4 (30.8%) | 0.31 |
| Exon 10 mutations n = 40 (63.5) | 21 (52.5%) | 13 (32.5%) | 6 (15.0%) | |
| E148Q n = 11 (17.5) | 5 (15.5%) | 2 (10.0%) | 4 (28.5%) | 0.267 |
| R202Q n = 2 (3.2) | 2 (6.0%) | 0 | 0 | |
| M694I n = 19(30.2 | 12 (63.2%) | 5 (25.0%) | 2 (14.25%) | |
| M680I n = 13 (20.6) | 5 (15.15%) | 6 (30.0%) | 2 (14.2%) | |
| V726A = 6 (9.5) | 1 (3.0%) | 3 (15.0%) | 2 (14.2%) | |
| M694V n = 4 (6.3) | 4 (12.12%) | 0 | 0 |
| Phenotype (n%) (Mean + SD) | Methylation% | Pyrin Concentration (pg/mL) | r | p-Value |
|---|---|---|---|---|
| Abdominal pain n = 45 (71.4%) | 19.91 ± 12.1 | 380.91 ± 63.8 | −0.084 | 0.582 |
| Arthritis n = 41 (65.1%) | 19.8 ± 11.7 | 373.49 ± 62.0 | −0.095 | 0.554 |
| Fever n = 37 (58.7%) | 18.64 ± 11.2 | 383.81 ± 63.8 | −0.021 | 0.904 |
| Diarrhea/constipation n = 25 (39.7%) | 20.43 ± 13.7 | 370.46 ± 50.0 | −0.056 | 0.791 |
| Erysipeloid erythema n = 12 (19.0%) | 22.19 ± 12.2 | 377.2 ± 53.5 | 0.01 | 0.976 |
| Vomiting n = 10 (15.9%) | 25.27 ± 12.8 | 390.1 ± 31.3 | 0.576 | 0.082 |
| Chest pain n = 6 (9.5%) | 17.05 ± 6.4 | 441.57 ± 45.9 | −0.087 | 0.869 |
| Parameters | Non-Responder Patients n = 26 (41.3%) | Responder Patients n = 37 (58.7%) | p-Value |
|---|---|---|---|
| Methylation% | 19.54 ± 11.7 | 18.53 ± 11.7 | 0.737 |
| Pyrin concentration (pg/mL) | 378.0 ± 62.4 | 389.5 ± 71.2 | 0.512 |
| Abdominal pain n = 45 (71.4%) | 26 (100.0%) | 19 (51.4%) | <0.001 ** |
| Arthritis n = 41 (65.1%) | 25 (69.2%) | 16 (43.2%) | <0.001 ** |
| Fever n = 37 (58.7) | 20 (76.95%) | 17 (45.9%) | 0.014 * |
| Erysipeloid erythema n = 12 (19.0%) | 9 (34.6%) | 3 (8.1%) | 0.008 ** |
| Vomiting n = 10 (15.9%) | 6 (23.1%) | 4 (10.8%) | 0.19 |
| Diarrhea/constipation n = 25 (39.7%) | 15 (57.7%) | 10 (27.0%) | 0.014 * |
| Chest pain n = 6 (9.5%) | 5 (19.2%) | 1 (2.7%) | 0.028 * |
| FMF severity | <0.001 ** | ||
| Mild n = 13 (20.6) | 0 | 13 (35.1%) | |
| Moderate n = 18 (28.6) | 4 (15.4%) | 14 (37.8%) | |
| Severe n = 32 (50.8) | 22 (84.6%) | 10 (27.0%) | |
| Homozygous n = 8 (12.6%) | 4 (50.0%) | 4 (50.0%) | 0.628 |
| Compound heterozygous n = 7 (5.55%) | 4(70.0%) | 3(30.0%) | |
| Heterozygous n = 34 (53.96%) | 13 (38.2%) | 21(61.8%) | |
| High SAA n = 13 (20.6%) | 8 (30.8%) | 5 (13.5%) | 0.09 |
| High CRP n = 21 (33.3%) | 12 (46.2%) | 9 (24.3%) | 0.07 |
| High ESR n = 28 (44.4%) | 16 (61.5%) | 12 (32.4%) | 0.022 * |
| Anemia n = 32 (50.8%) | 17 (65.4%) | 15 (40.5%) | 0.05 * |
| H-pylori n = 8 (12.7%) | 4 (15.4%) | 4 (10.8%) | 0.591 |
| Renal calculi n = 14 (22.2%) | 8 (30.8%) | 6 (16.2%) | 0.171 |
| Vitamin D deficiency n = 24 (31.7%) | 13 (50.0%) | 11 (29.7%) | 0.103 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zekry, M.E.; Sallam, A.-A.M.; AbdelHamid, S.G.; Zarouk, W.A.; El-Bassyouni, H.T.; El-Mesallamy, H.O. Genetic and Epigenetic Regulation of MEFV Gene and Their Impact on Clinical Outcome in Auto-Inflammatory Familial Mediterranean Fever Patients. Curr. Issues Mol. Biol. 2023, 45, 721-737. https://doi.org/10.3390/cimb45010048
Zekry ME, Sallam A-AM, AbdelHamid SG, Zarouk WA, El-Bassyouni HT, El-Mesallamy HO. Genetic and Epigenetic Regulation of MEFV Gene and Their Impact on Clinical Outcome in Auto-Inflammatory Familial Mediterranean Fever Patients. Current Issues in Molecular Biology. 2023; 45(1):721-737. https://doi.org/10.3390/cimb45010048
Chicago/Turabian StyleZekry, May E., Al-Aliaa M. Sallam, Sherihan G. AbdelHamid, Waheba A. Zarouk, Hala T. El-Bassyouni, and Hala O. El-Mesallamy. 2023. "Genetic and Epigenetic Regulation of MEFV Gene and Their Impact on Clinical Outcome in Auto-Inflammatory Familial Mediterranean Fever Patients" Current Issues in Molecular Biology 45, no. 1: 721-737. https://doi.org/10.3390/cimb45010048
APA StyleZekry, M. E., Sallam, A.-A. M., AbdelHamid, S. G., Zarouk, W. A., El-Bassyouni, H. T., & El-Mesallamy, H. O. (2023). Genetic and Epigenetic Regulation of MEFV Gene and Their Impact on Clinical Outcome in Auto-Inflammatory Familial Mediterranean Fever Patients. Current Issues in Molecular Biology, 45(1), 721-737. https://doi.org/10.3390/cimb45010048






