Exploring Plant Meiosis: Insights from the Kinetochore Perspective
Abstract
1. Introduction
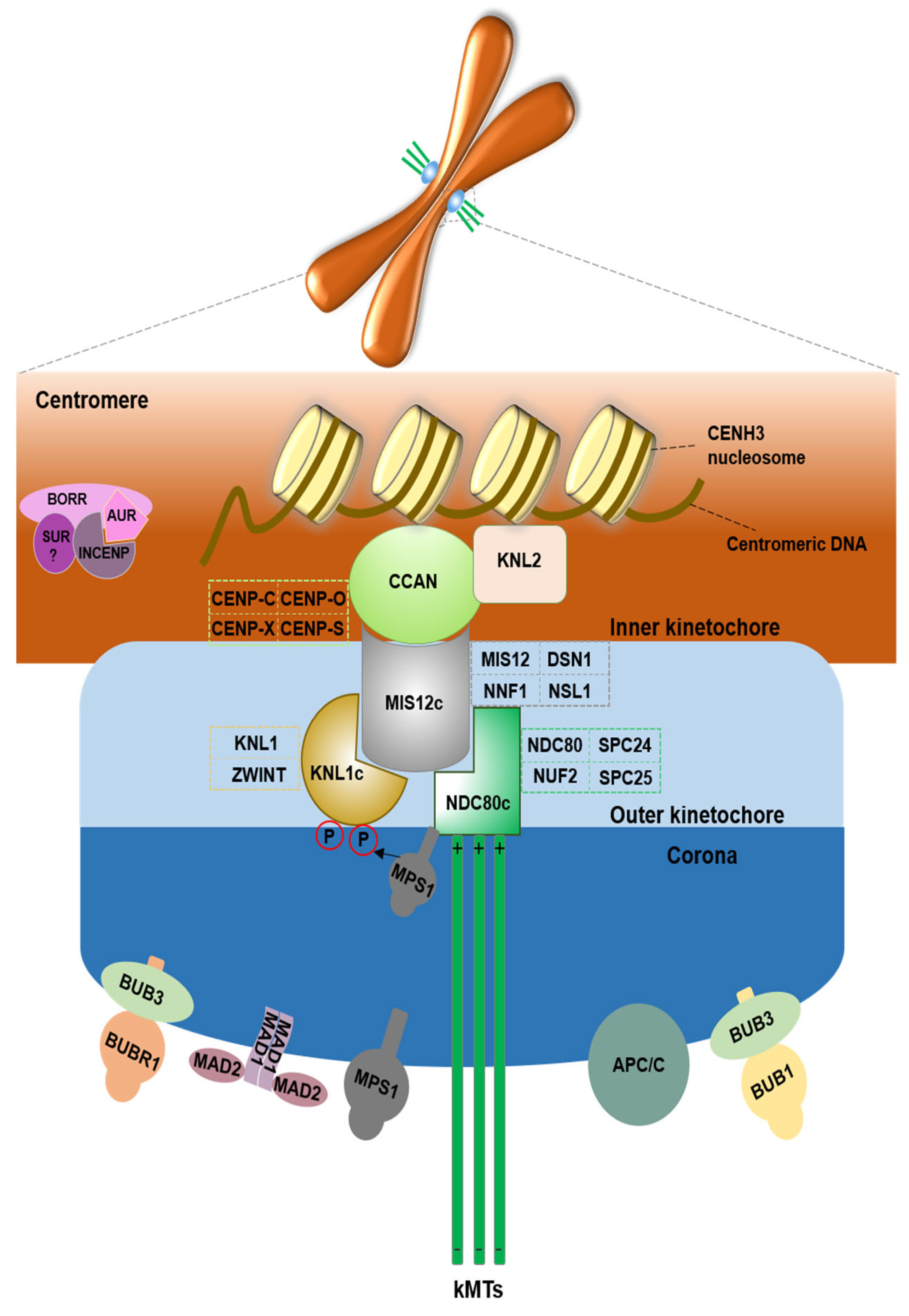
2. Kinetochore Structural Proteins
2.1. CENH3 Protein
2.2. Inner Kinetochore
2.3. Outer Kinetochore and KMN Network
3. Spindle Assembly Checkpoint (SAC) Proteins
3.1. MPS1 Protein
3.2. BUB Proteins
3.3. MAD Proteins
4. Chromosomal Passenger Complex (CPC) Proteins
4.1. Aurora Kinase
4.2. Other CPC Components
5. Centromeric Cohesion in Meiosis
5.1. Sister Chromatid Cohesion
5.2. Mono-Orientation
5.3. Cohesin Protectors
6. Concluding Remarks and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Earnshaw, W.C. Discovering centromere proteins: From cold white hands to the A, B, C of CENPs. Nat. Rev. Mol. Cell Biol. 2015, 16, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Zafarullah, M. An Investigation of Functional Dependency of CENP-C and CENH3 in Arabidopsis thaliana. Master’s Thesis, University of California, Davis, CA, USA, 2017. [Google Scholar]
- Heckmann, S.; Jankowska, M.; Schubert, V.; Kumke, K.; Ma, W.; Houben, A. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat. Commun. 2014, 5, 4979. [Google Scholar] [CrossRef] [PubMed]
- Cabral, G.; Marques, A.; Schubert, V.; Pedrosa-Harand, A.; Schlögelhofer, P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Commun. 2014, 5, 5070. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Li, C.; Li, H.; Wang, Z.; Jiang, Y.; Guo, Y.; Sun, P.; Chen, X.; Li, Q.; Tian, H. Disruption of REC8 in Meiosis I led to watermelon seedless. Plant Sci. 2022, 323, 111394. [Google Scholar] [CrossRef]
- Watts, A.; Kumar, V.; Raipuria, R.K.; Bhattacharya, R. In vivo haploid production in crop plants: Methods and challenges. Plant Mol. Biol. Rep. 2018, 36, 685–694. [Google Scholar] [CrossRef]
- Wang, K. Fixation of hybrid vigor in rice: Synthetic apomixis generated by genome editing. aBIOTECH 2020, 1, 15–20. [Google Scholar] [CrossRef]
- Xie, E.; Li, Y.; Tang, D.; Lv, Y.; Shen, Y.; Cheng, Z. A strategy for generating rice apomixis by gene editing. J. Integr. Plant Biol. 2019, 61, 911–916. [Google Scholar] [CrossRef]
- Meng, D.; Liu, C.; Chen, S.; Jin, W. Haploid induction and its application in maize breeding. Mol. Breed. 2021, 41, 20. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Rothfield, N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 1985, 91, 313–321. [Google Scholar] [CrossRef]
- Malik, H.S.; Henikoff, S. Phylogenomics of the nucleosome. Nat. Struct. Mol. Biol. 2003, 10, 882–891. [Google Scholar] [CrossRef]
- Schubert, V.; Lermontova, I.; Schubert, I. Loading of the centromeric histone H3 variant during meiosis–how does it differ from mitosis? Chromosoma 2014, 123, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Lermontova, I.; Koroleva, O.; Rutten, T.; Fuchs, J.; Schubert, V.; Moraes, I.; Koszegi, D.; Schubert, I. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 2011, 68, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Jokelainen, P.T. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 1967, 19, 19–44. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Chan, S.W. Haploid plants produced by centromere-mediated genome elimination. Nature 2010, 464, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Silva, N.; Ulmer, J.; Kiuchi, T.; Hsieh, E.; Cornilleau, G.; Ladid, I.; Dingli, F.; Loew, D.; Katsuma, S.; Drinnenberg, I.A. CenH3-independent kinetochore assembly in Lepidoptera requires CCAN, including CENP-T. Curr. Biol. 2020, 30, 561–572.e510. [Google Scholar] [CrossRef]
- Perpelescu, M.; Fukagawa, T. The abcs of cenps. Chromosoma 2011, 120, 425–446. [Google Scholar] [CrossRef]
- Kozgunova, E.; Nishina, M.; Goshima, G. Kinetochore protein depletion underlies cytokinesis failure and somatic polyploidization in the moss Physcomitrella patens. eLife 2019, 8, e43652. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Osman, F.; Feeney, L.; Lorenz, A.; Bryer, C.; Whitby, M.C. MHF1–2/CENP-SX performs distinct roles in centromere metabolism and genetic recombination. Open Biol. 2013, 3, 130102. [Google Scholar] [CrossRef]
- Dawe, R.K.; Reed, L.M.; Yu, H.-G.; Muszynski, M.G.; Hiatt, E.N. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 1999, 11, 1227–1238. [Google Scholar] [CrossRef]
- Ogura, Y.; Shibata, F.; Sato, H.; Murata, M. Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet. Syst. 2004, 79, 139–144. [Google Scholar] [CrossRef]
- Tanaka, K.; Chang, H.L.; Kagami, A.; Watanabe, Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 2009, 17, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Unhavaithaya, Y.; Orr-Weaver, T.L. Centromere proteins CENP-C and CAL1 functionally interact in meiosis for centromere clustering, pairing, and chromosome segregation. Proc. Natl. Acad. Sci. USA 2013, 110, 19878–19883. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Liu, Y.; Zhong, Y. Immunization with CENP-C causes aberrant chromosome segregation during oocyte meiosis in mice. J. Immunol. Res. 2021, 2021, 4610494. [Google Scholar] [CrossRef] [PubMed]
- Kral, L.G. Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. F1000Research 2016, 4, 474. [Google Scholar] [CrossRef]
- Fellmeth, J.E.; McKim, K.S. Meiotic CENP-C is a shepherd: Bridging the space between the centromere and the kinetochore in time and space. Essays Biochem. 2020, 64, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Sullivan, B.A.; Trazzi, S.; Della Valle, G.; Robertson, K.D. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum. Mol. Genet. 2009, 18, 3178–3193. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Shang, W.-H.; Hara, M.; Ariyoshi, M.; Arimura, Y.; Fujita, R.; Kurumizaka, H.; Fukagawa, T. Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates. Dev. Cell 2017, 42, 181–189. [Google Scholar] [CrossRef]
- Sandmann, M.; Talbert, P.; Demidov, D.; Kuhlmann, M.; Rutten, T.; Conrad, U.; Lermontova, I. Targeting of Arabidopsis KNL2 to centromeres depends on the conserved CENPC-k motif in its C terminus. Plant Cell 2017, 29, 144–155. [Google Scholar] [CrossRef]
- Lermontova, I.; Kuhlmann, M.; Friedel, S.; Rutten, T.; Heckmann, S.; Sandmann, M.; Demidov, D.; Schubert, V.; Schubert, I. Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell 2013, 25, 3389–3404. [Google Scholar] [CrossRef]
- Hayashi, T.; Fujita, Y.; Iwasaki, O.; Adachi, Y.; Takahashi, K.; Yanagida, M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 2004, 118, 715–729. [Google Scholar] [CrossRef]
- De Rop, V.; Padeganeh, A.; Maddox, P.S. CENP-A: The key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma 2012, 121, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Yadala, R.; Yang, F.; Talbert, P.; Fuchs, J.; Schubert, V.; Ahmadli, U.; Rutten, T.; Pecinka, A.; Lysak, M.A. Recurrent plant-specific duplications of KNL2 and its conserved function as a kinetochore assembly factor. Mol. Biol. Evol. 2022, 39, msac123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Martyniuk, C.J.; Trudeau, V.L. SANTA domain: A novel conserved protein module in Eukaryota with potential involvement in chromatin regulation. Bioinformatics 2006, 22, 2459–2462. [Google Scholar] [CrossRef] [PubMed]
- Dambacher, S.; Deng, W.; Hahn, M.; Sadic, D.; Fröhlich, J.; Nuber, A.; Hoischen, C.; Diekmann, S.; Leonhardt, H.; Schotta, G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 2012, 3, 101–110. [Google Scholar] [CrossRef]
- Stellfox, M.E.; Nardi, I.K.; Knippler, C.M.; Foltz, D.R. Differential binding partners of the Mis18α/β YIPPEE domains regulate Mis18 complex recruitment to centromeres. Cell Rep. 2016, 15, 2127–2135. [Google Scholar] [CrossRef]
- Fujita, Y.; Hayashi, T.; Kiyomitsu, T.; Toyoda, Y.; Kokubu, A.; Obuse, C.; Yanagida, M. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev. Cell 2007, 12, 17–30. [Google Scholar] [CrossRef]
- Su, H.; Liu, Y.; Wang, C.; Liu, Y.; Feng, C.; Sun, Y.; Yuan, J.; Birchler, J.A.; Han, F. Knl1 participates in spindle assembly checkpoint signaling in maize. Proc. Natl. Acad. Sci. USA 2021, 118, e2022357118. [Google Scholar] [CrossRef]
- Neumann, P.; Oliveira, L.C.; Jang, T.S.; Novak, P.; Koblizkova, A.; Schubert, V.; Houben, A.; Macas, J. Disruption of the standard kinetochore in holocentric Cuscuta species. Proc. Natl. Acad. Sci. USA 2023, 120, e2300877120. [Google Scholar] [CrossRef]
- Du, Y.; Dawe, R.K. Maize NDC80 is a constitutive feature of the central kinetochore. Chromosome Res. 2007, 15, 767–775. [Google Scholar] [CrossRef]
- Li, X.; Dawe, R.K. Fused sister kinetochores initiate the reductional division in meiosis I. Nat. Cell Biol. 2009, 11, 1103–1108. [Google Scholar] [CrossRef]
- Cheeseman, I.M.; Chappie, J.S.; Wilson-Kubalek, E.M.; Desai, A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006, 127, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, A.; Mosalaganti, S.; Keller, J.; Mattiuzzo, M.; Overlack, K.; Krenn, V.; De Antoni, A.; Wohlgemuth, S.; Cecatiello, V.; Pasqualato, S. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell 2014, 53, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, A.; Pasqualato, S.; Dube, P.; Krenn, V.; Santaguida, S.; Cittaro, D.; Monzani, S.; Massimiliano, L.; Keller, J.; Tarricone, A. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 2010, 190, 835–852. [Google Scholar] [CrossRef] [PubMed]
- Allipra, S.; Anirudhan, K.; Shivanandan, S.; Raghunathan, A.; Maruthachalam, R. The kinetochore protein NNF1 has a moonlighting role in the vegetative development of Arabidopsis thaliana. Plant J. 2022, 109, 1064–1085. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zou, W.; Jian, L.; Fu, Y.; Zhao, J. AtNUF2 modulates spindle microtubule organization and chromosome segregation during mitosis. Plant J. 2021, 107, 801–816. [Google Scholar] [CrossRef]
- Shin, J.; Jeong, G.; Park, J.Y.; Kim, H.; Lee, I. MUN (MERISTEM UNSTRUCTURED), encoding a SPC24 homolog of NDC80 kinetochore complex, affects development through cell division in Arabidopsis thaliana. Plant J. 2018, 93, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, Y.; Guo, X.; Birchler, J.A.; Han, F.; Su, H. Centromeres: From chromosome biology to biotechnology applications and synthetic genomes in plants. Plant Biotechnol. J. 2022, 20, 2051–2063. [Google Scholar] [CrossRef]
- London, N.; Biggins, S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 2014, 15, 736–748. [Google Scholar] [CrossRef]
- van Hooff, J.J.; Tromer, E.; van Wijk, L.M.; Snel, B.; Kops, G.J. Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 2017, 18, 1559–1571. [Google Scholar] [CrossRef]
- Sun, S.-C.; Kim, N.-H. Spindle assembly checkpoint and its regulators in meiosis. Hum. Reprod. Update 2012, 18, 60–72. [Google Scholar] [CrossRef]
- Liu, B.; Lee, Y.-R.J. Spindle assembly and mitosis in plants. Annu. Rev. Plant Biol. 2022, 73, 227–254. [Google Scholar] [CrossRef] [PubMed]
- Marston, A.L.; Wassmann, K. Multiple duties for spindle assembly checkpoint kinases in meiosis. Front. Cell Dev. Biol. 2017, 5, 109. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, F.F.; Wu, Y.T.; Zhou, X.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Dong, R.B.; Cao, K.M.; Yang, Z.N. MULTIPOLAR SPINDLE 1 (MPS1), a novel coiled-coil protein of Arabidopsis thaliana, is required for meiotic spindle organization. Plant J. 2009, 59, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- De Muyt, A.; Pereira, L.; Vezon, D.; Chelysheva, L.; Gendrot, G.; Chambon, A.; Lainé-Choinard, S.; Pelletier, G.; Mercier, R.; Nogue, F. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000654. [Google Scholar] [CrossRef]
- Xue, Z.; Liu, C.; Shi, W.; Miao, Y.; Shen, Y.; Tang, D.; Li, Y.; You, A.; Xu, Y.; Chong, K. OsMTOPVIB is required for meiotic bipolar spindle assembly. Proc. Natl. Acad. Sci. USA 2019, 116, 15967–15972. [Google Scholar] [CrossRef]
- Wang, C.; Qu, S.; Zhang, J.; Fu, M.; Chen, X.; Liang, W. OsPRD2 is essential for double-strand break formation, but not spindle assembly during rice meiosis. Front. Plant Sci. 2022, 13, 1122202. [Google Scholar] [CrossRef]
- Wang, M.; Tang, D.; Luo, Q.; Jin, Y.; Shen, Y.; Wang, K.; Cheng, Z. BRK1, a Bub1-related kinase, is essential for generating proper tension between homologous kinetochores at metaphase I of rice meiosis. Plant Cell 2012, 24, 4961–4973. [Google Scholar] [CrossRef]
- Vaur, S.; Cubizolles, F.; Plane, G.; Genier, S.; Rabitsch, P.K.; Gregan, J.; Nasmyth, K.; Vanoosthuyse, V.; Hardwick, K.G.; Javerzat, J.-P. Control of Shugoshin function during fission-yeast meiosis. Curr. Biol. 2005, 15, 2263–2270. [Google Scholar] [CrossRef]
- Nicklas, R.B.; Waters, J.C.; Salmon, E.; Ward, S.C. Checkpoint signals in grasshopper meiosis are sensitive to microtubule attachment, but tension is still essential. J. Cell Sci. 2001, 114, 4173–4183. [Google Scholar] [CrossRef]
- Weimer, A.K.; Demidov, D.; Lermontova, I.; Beeckman, T.; Van Damme, D. Aurora kinases throughout plant development. Trends Plant Sci. 2016, 21, 69–79. [Google Scholar] [CrossRef]
- Demidov, D.; Van Damme, D.; Geelen, D.; Blattner, F.R.; Houben, A. Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 2005, 17, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Jeyaprakash, A.A.; Klein, U.R.; Lindner, D.; Ebert, J.; Nigg, E.A.; Conti, E. Structure of a Survivin–Borealin–INCENP core complex reveals how chromosomal passengers travel together. Cell 2007, 131, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Kaitna, S.; Mendoza, M.; Jantsch-Plunger, V.; Glotzer, M. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 2000, 10, 1172–1181. [Google Scholar] [CrossRef]
- Winey, M.; Goetsch, L.; Baum, P.; Byers, B. MPS1 and MPS2: Novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 1991, 114, 745–754. [Google Scholar] [CrossRef]
- Lauze, E.Y.; Stoelcker, B.; Luca, F.; Weiss, E.; Schutz, A.; Winey, M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995, 14, 1655–1663. [Google Scholar] [CrossRef]
- Espeut, J.; Lara-Gonzalez, P.; Sassine, M.; Shiau, A.K.; Desai, A.; Abrieu, A. Natural loss of Mps1 kinase in nematodes uncovers a role for polo-like kinase 1 in spindle checkpoint initiation. Cell Rep. 2015, 12, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Maure, J.-F.; Kitamura, E.; Tanaka, T.U. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 2017, 17, 2175–2182. [Google Scholar] [CrossRef]
- Hewitt, L.; Tighe, A.; Santaguida, S.; White, A.M.; Jones, C.D.; Musacchio, A.; Green, S.; Taylor, S.S. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex. J. Cell Biol. 2010, 190, 25–34. [Google Scholar] [CrossRef]
- Maciejowski, J.; George, K.A.; Terret, M.-E.; Zhang, C.; Shokat, K.M.; Jallepalli, P.V. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 2010, 190, 89–100. [Google Scholar] [CrossRef]
- Santaguida, S.; Tighe, A.; D’Alise, A.M.; Taylor, S.S.; Musacchio, A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010, 190, 73–87. [Google Scholar] [CrossRef]
- Tighe, A.; Staples, O.; Taylor, S. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 2008, 181, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Benzi, G.; Camasses, A.; Atsunori, Y.; Katou, Y.; Shirahige, K.; Piatti, S. A common molecular mechanism underlies the role of Mps1 in chromosome biorientation and the spindle assembly checkpoint. EMBO Rep. 2020, 21, e50257. [Google Scholar] [CrossRef] [PubMed]
- London, N.; Ceto, S.; Ranish, J.A.; Biggins, S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 2012, 22, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Gao, H.; Jia, L.; Li, B.; Yu, H. A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. eLife 2017, 6, e22513. [Google Scholar] [CrossRef] [PubMed]
- Tipton, A.R.; Ji, W.; Sturt-Gillespie, B.; Bekier, M.E.; Wang, K.; Taylor, W.R.; Liu, S.-T. Monopolar spindle 1 (MPS1) kinase promotes production of closed MAD2 (C-MAD2) conformer and assembly of the mitotic checkpoint complex. J. Biol. Chem. 2013, 288, 35149–35158. [Google Scholar] [CrossRef] [PubMed]
- Straight, P.D.; Giddings, T.H., Jr.; Winey, M. Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell. 2000, 11, 3525–3537. [Google Scholar] [CrossRef]
- Gilliland, W.D.; Wayson, S.M.; Hawley, R.S. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr. Biol. 2005, 15, 672–677. [Google Scholar] [CrossRef]
- Gilliland, W.D.; Hughes, S.E.; Cotitta, J.L.; Takeo, S.; Xiang, Y.; Hawley, R.S. The multiple roles of Mps1 in Drosophila female meiosis. PLoS Genet. 2007, 3, e113. [Google Scholar] [CrossRef]
- Hached, K.; Xie, S.Z.; Buffin, E.; Cladière, D.; Rachez, C.; Sacras, M.; Sorger, P.K.; Wassmann, K. Mps1 at kinetochores is essential for female mouse meiosis I. Development 2011, 138, 2261–2271. [Google Scholar] [CrossRef]
- Paganelli, L.; Caillaud, M.C.; Quentin, M.; Damiani, I.; Govetto, B.; Lecomte, P.; Karpov, P.A.; Abad, P.; Chabouté, M.E.; Favery, B. Retracted: Three BUB 1 and BUBR 1/MAD 3-related spindle assembly checkpoint proteins are required for accurate mitosis in Arabidopsis. New Phytol. 2015, 205, 202–215. [Google Scholar] [CrossRef]
- Su, H.; Liu, Y.; Dong, Q.; Feng, C.; Zhang, J.; Liu, Y.; Birchler, J.A.; Han, F. Dynamic location changes of Bub1-phosphorylated-H2AThr133 with CENH3 nucleosome in maize centromeric regions. New Phytol. 2017, 214, 682–694. [Google Scholar] [CrossRef]
- Miyazaki, S.; Kim, J.; Yamagishi, Y.; Ishiguro, T.; Okada, Y.; Tanno, Y.; Sakuno, T.; Watanabe, Y. Meikin-associated polo-like kinase specifies Bub1 distribution in meiosis I. Genes Cells 2017, 22, 552–567. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Decottignies, A.; Nurse, P. Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint. EMBO J. 2003, 22, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, S.; Yuan, J.; Wang, Z.-B.; Sun, S.-C.; Schatten, H.; Sun, Q.-Y. Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. PLoS ONE 2009, 4, e7701. [Google Scholar] [CrossRef] [PubMed]
- Fraschini, R.; Beretta, A.; Sironi, L.; Musacchio, A.; Lucchini, G.; Piatti, S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001, 20, 6648–6659. [Google Scholar] [CrossRef] [PubMed]
- Lermontova, I.; Fuchs, J.; Schubert, I. The Arabidopsis checkpoint protein Bub3. 1 is essential for gametophyte development. Front. Biosci. 2008, 13, 5202–5211. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Komaki, S.; Schnittger, A. The spindle assembly checkpoint in Arabidopsis is rapidly shut off during severe stress. Dev. Cell 2017, 43, 172–185.e175. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, X.; Sun, B.; Lee Van, S.; Kang, Z.; Lin, H.; Lee, Y.-R.J.; Liu, B. Role of the BUB3 protein in phragmoplast microtubule reorganization during cytokinesis. Nat. Plants 2018, 4, 485–494. [Google Scholar] [CrossRef]
- Rancati, G.; Crispo, V.; Lucchini, G.; Piatti, S. Mad3/BubR1 phosphorylation during spindle checkpoint activation depends on both Polo and Aurora kinases in budding yeast. Cell Cycle 2005, 4, 972–980. [Google Scholar] [CrossRef]
- Pérez-Mongiovi, D.; Malmanche, N.; Bousbaa, H.; Sunkel, C. Maternal expression of the checkpoint protein BubR1 is required for synchrony of syncytial nuclear divisions and polar body arrest in Drosophila melanogaster. Development 2005, 132, 4509–4520. [Google Scholar] [CrossRef][Green Version]
- Wei, L.; Liang, X.-W.; Zhang, Q.-H.; Li, M.; Yuan, J.; Li, S.; Sun, S.-C.; Ouyang, Y.-C.; Schatten, H.; Sun, Q.-Y. BubR1 is a spindle assembly checkpoint protein regulating meiotic cell cycle progression of mouse oocyte. Cell Cycle 2010, 9, 1112–1121. [Google Scholar] [CrossRef]
- Komaki, S.; Schnittger, A. The spindle checkpoint in plants—A green variation over a conserved theme? Curr. Opin. Plant Biol. 2016, 34, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Parra, M.T.; Gomez, R.; Viera, A.; Llano, E.; Pendas, A.M.; Rufas, J.S.; Suja, J.A. Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet. 2009, 5, e1000417. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, K.B.; Van Deursen, J. Differential mitotic checkpoint protein requirements in somatic and germ cells. Biochem. Soc. Trans. 2006, 34, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Heng-Yu, F. BubR1, a spindle assembly checkpoint protein in mammalian oocyte meiosis. Cell Cycle 2010, 9, 1456–1465. [Google Scholar] [CrossRef]
- Caillaud, M.-C.; Paganelli, L.; Lecomte, P.; Deslandes, L.; Quentin, M.; Pecrix, Y.; Le Bris, M.; Marfaing, N.; Abad, P.; Favery, B. Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS ONE 2009, 4, e6757. [Google Scholar] [CrossRef] [PubMed]
- Kimbara, J.; Endo, T.R.; Nasuda, S. Characterization of the genes encoding for MAD2 homologues in wheat. Chromosome Res. 2004, 12, 703–714. [Google Scholar] [CrossRef]
- Chen, R.-H.; Brady, D.M.; Smith, D.; Murray, A.W.; Hardwick, K.G. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell. 1999, 10, 2607–2618. [Google Scholar] [CrossRef]
- Cheslock, P.S.; Kemp, B.J.; Boumil, R.M.; Dawson, D.S. The roles of MAD1, MAD2 and MAD3 in meiotic progression and the segregation of nonexchange chromosomes. Nat. Genet. 2005, 37, 756–760. [Google Scholar] [CrossRef]
- Kitagawa, R.; Rose, A.M. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1999, 1, 514–521. [Google Scholar] [CrossRef]
- Zhang, D.; Li, M.; Ma, W.; Hou, Y.; Li, Y.-H.; Li, S.-W.; Sun, Q.-Y.; Wang, W.-H. Localization of mitotic arrest deficient 1 (MAD1) in mouse oocytes during the first meiosis and its functions as a spindle checkpoint protein. Biol. Reprod. 2005, 72, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Muthuswamy, S.; Meier, I. Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol. Biol. 2012, 79, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Zhang, N.; Hua, J. Endopolyploidization and flowering time are antagonistically regulated by checkpoint component MAD1 and immunity modulator MOS1. Nat. Commun. 2014, 5, 5628. [Google Scholar] [CrossRef]
- Kallio, M.; Eriksson, J.E.; Gorbsky, G.J. Differences in spindle association of the mitotic checkpoint protein Mad2 in mammalian spermatogenesis and oogenesis. Dev. Biol. 2000, 225, 112–123. [Google Scholar] [CrossRef]
- Zhang, D.; Ma, W.; Li, Y.-H.; Hou, Y.; Li, S.-W.; Meng, X.-Q.; Sun, X.-F.; Sun, Q.-Y.; Wang, W.-H. Intra-oocyte localization of MAD2 and its relationship with kinetochores, microtubules, and chromosomes in rat oocytes during meiosis. Biol. Reprod. 2004, 71, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-G.; Muszynski, M.G.; Kelly Dawe, R. The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 1999, 145, 425–435. [Google Scholar] [CrossRef]
- Niault, T.; Hached, K.; Sotillo, R.; Sorger, P.K.; Maro, B.; Benezra, R.; Wassmann, K. Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PLoS ONE 2007, 2, e1165. [Google Scholar] [CrossRef]
- Wang, J.Y.; Lei, Z.L.; Nan, C.L.; Yin, S.; Liu, J.; Hou, Y.; Li, Y.L.; Chen, D.Y.; Sun, Q.Y. RNA interference as a tool to study the function of MAD2 in mouse oocyte meiotic maturation. Mol. Reprod. Dev. 2007, 74, 116–124. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kitamura, K.; Hihara, D.; Hirose, Y.; Katsuyama, S.; Hiraoka, Y. Spindle checkpoint activation at meiosis I advances anaphase II onset via meiosis-specific APC/C regulation. J. Cell Biol. 2008, 182, 277–288. [Google Scholar] [CrossRef]
- Ruchaud, S.; Carmena, M.; Earnshaw, W.C. Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 2007, 8, 798–812. [Google Scholar] [CrossRef]
- Vader, G.; Cruijsen, C.W.; Van Harn, T.; Vromans, M.J.; Medema, R.H.; Lens, S.M. The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule–kinetochore interactions. Mol. Biol. Cell 2007, 18, 4553–4564. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Barrera, M.; Monje-Casas, F. Increased Aurora B activity causes continuous disruption of kinetochore–microtubule attachments and spindle instability. Proc. Natl. Acad. Sci. USA 2014, 111, E3996–E4005. [Google Scholar] [CrossRef] [PubMed]
- Landino, J.; Ohi, R. The timing of midzone stabilization during cytokinesis depends on myosin II activity and an interaction between INCENP and actin. Curr. Biol. 2016, 26, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Lee, S.H. The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front. Cell Dev. Biol. 2015, 3, 14. [Google Scholar] [CrossRef]
- Giet, R.; Prigent, C. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci. 1999, 112, 3591–3601. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Paris, J.; Willer, M.; Philippe, M.; Hagan, I.M. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 2001, 114, 4371–4384. [Google Scholar] [CrossRef]
- Ducat, D.; Zheng, Y. Aurora kinases in spindle assembly and chromosome segregation. Exp. Cell Res. 2004, 301, 60–67. [Google Scholar] [CrossRef]
- Goldenson, B.; Crispino, J.D. The aurora kinases in cell cycle and leukemia. Oncogene 2015, 34, 537–545. [Google Scholar] [CrossRef]
- Carmena, M.; Earnshaw, W.C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003, 4, 842–854. [Google Scholar] [CrossRef]
- Yu, H.-G.; Koshland, D. The Aurora kinase Ipl1 maintains the centromeric localization of PP2A to protect cohesin during meiosis. J. Cell Biol. 2007, 176, 911–918. [Google Scholar] [CrossRef]
- Monje-Casas, F.; Prabhu, V.R.; Lee, B.H.; Boselli, M.; Amon, A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 2007, 128, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Balboula, A.Z.; Schindler, K. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet. 2014, 10, e1004194. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-J.C.; Lin, C.-Y.; Tang, T.K. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev. Biol. 2006, 290, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Demidov, D.; Lermontova, I.; Weiss, O.; Fuchs, J.; Rutten, T.; Kumke, K.; Sharbel, T.F.; Van Damme, D.; De Storme, N.; Geelen, D. Altered expression of Aurora kinases in Arabidopsis results in aneu-and polyploidization. Plant J. 2014, 80, 449–461. [Google Scholar] [CrossRef]
- Niu, B.; Wang, L.; Zhang, L.; Ren, D.; Ren, R.; Copenhaver, G.P.; Ma, H.; Wang, Y. Arabidopsis cell division cycle 20.1 is required for normal meiotic spindle assembly and chromosome segregation. Plant Cell 2015, 27, 3367–3382. [Google Scholar] [CrossRef]
- Zamariola, L.; Tiang, C.L.; De Storme, N.; Pawlowski, W.; Geelen, D. Chromosome segregation in plant meiosis. Front. Plant Sci. 2014, 5, 279. [Google Scholar] [CrossRef]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef]
- Komaki, S.; Takeuchi, H.; Hamamura, Y.; Heese, M.; Hashimoto, T.; Schnittger, A. Functional analysis of the plant chromosomal passenger complex. Plant Physiol. 2020, 183, 1586–1599. [Google Scholar] [CrossRef]
- Kirioukhova, O.; Johnston, A.J.; Kleen, D.; Kägi, C.; Baskar, R.; Moore, J.M.; Bäumlein, H.; Groß-Hardt, R.; Grossniklaus, U. Female gametophytic cell specification and seed development require the function of the putative Arabidopsis INCENP ortholog WYRD. Development 2011, 138, 3409–3420. [Google Scholar] [CrossRef]
- Gassmann, R.; Carvalho, A.; Henzing, A.J.; Ruchaud, S.; Hudson, D.F.; Honda, R.; Nigg, E.A.; Gerloff, D.L.; Earnshaw, W.C. Borealin: A novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004, 166, 179–191. [Google Scholar] [CrossRef]
- Komaki, S.; Tromer, E.C.; De Jaeger, G.; De Winne, N.; Heese, M.; Schnittger, A. Molecular convergence by differential domain acquisition is a hallmark of chromosomal passenger complex evolution. Proc. Natl. Acad. Sci. USA 2022, 119, e2200108119. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-C.; Wei, L.; Li, M.; Lin, S.-L.; Xu, B.-Z.; Liang, X.-W.; Kim, N.-H.; Schatten, H.; Lu, S.-S.; Sun, Q.-Y. Perturbation of survivin expression affects chromosome alignment and spindle checkpoint in mouse oocyte meiotic maturation. Cell Cycle 2009, 8, 3365–3372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, K.; Jiang, G.-J.; Wei, L.; Liang, X.-W.; Miao, D.-Q.; Sun, S.-C.; Guo, L.; Wang, Z.-B.; Lu, S.-S. Survivin is a critical regulator of spindle organization and chromosome segregation during rat oocyte meiotic maturation. Zygote 2011, 19, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Chelysheva, L.; Diallo, S.; Vezon, D.; Gendrot, G.; Vrielynck, N.; Belcram, K.; Rocques, N.; Márquez-Lema, A.; Bhatt, A.M.; Horlow, C. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 2005, 118, 4621–4632. [Google Scholar] [CrossRef]
- Cuacos, M.; H. Franklin, F.C.; Heckmann, S. Atypical centromeres in plants—What they can tell us. Front. Plant Sci. 2015, 6, 913. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Haering, C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005, 74, 595–648. [Google Scholar] [CrossRef]
- Haering, C.H.; Löwe, J.; Hochwagen, A.; Nasmyth, K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 2002, 9, 773–788. [Google Scholar] [CrossRef]
- Hirano, T. SMC proteins and chromosome mechanics: From bacteria to humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 507–514. [Google Scholar] [CrossRef]
- Schubert, V. SMC proteins and their multiple functions in higher plants. Cytogenet. Genome Res. 2009, 124, 202–214. [Google Scholar] [CrossRef]
- Cai, X.; Dong, F.; Edelmann, R.E.; Makaroff, C.A. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 2003, 116, 2999–3007. [Google Scholar] [CrossRef]
- Kudo, N.R.; Anger, M.; Peters, A.H.; Stemmann, O.; Theussl, H.-C.; Helmhart, W.; Kudo, H.; Heyting, C.; Nasmyth, K. Role of cleavage by separase of the Rec8 kleisin subunit of cohesin during mammalian meiosis I. J. Cell Sci. 2009, 122, 2686–2698. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Nurse, P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 1999, 400, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Sakuno, T.; Hiraoka, Y. Rec8 cohesin: A structural platform for shaping the meiotic chromosomes. Genes 2022, 13, 200. [Google Scholar] [CrossRef]
- Kudo, N.R.; Wassmann, K.; Anger, M.; Schuh, M.; Wirth, K.G.; Xu, H.; Helmhart, W.; Kudo, H.; Mckay, M.; Maro, B. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell 2006, 126, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Perez, E.; Schvarzstein, M.; Barroso, C.; Lightfoot, J.; Dernburg, A.F.; Villeneuve, A.M. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008, 22, 2886–2901. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Beasley, M.D.; Warren, W.D.; van der Horst, G.T.; McKay, M.J. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 2005, 8, 949–961. [Google Scholar] [CrossRef]
- Doll, E.; Molnar, M.; Cuanoud, G.; Octobre, G.; Latypov, V.; Ludin, K.; Kohli, J.R. Cohesin and recombination proteins influence the G1-to-S transition in azygotic meiosis in Schizosaccharomyces pombe. Genetics 2008, 180, 727–740. [Google Scholar] [CrossRef]
- Peirson, B.N.; Bowling, S.E.; Makaroff, C.A. A defect in synapsis causes male sterility in a T-DNA-tagged Arabidopsis thaliana mutant. Plant J. 1997, 11, 659–669. [Google Scholar] [CrossRef]
- Lam, W.S.; Yang, X.; Makaroff, C.A. Characterization of Arabidopsis thaliana SMC1 and SMC3: Evidence that AtSMC3 may function beyond chromosome cohesion. J. Cell Sci. 2005, 118, 3037–3048. [Google Scholar] [CrossRef]
- Lhuissier, F.G.; Offenberg, H.H.; Wittich, P.E.; Vischer, N.O.; Heyting, C. The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell 2007, 19, 862–876. [Google Scholar] [CrossRef]
- Mercier, R.; Grelon, M. Meiosis in plants: Ten years of gene discovery. Cytogenet. Genome Res. 2008, 120, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Mercier, R.; Mézard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015, 66, 297–327. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Ma, H.; Cande, W.Z. Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 2006, 57, 267–302. [Google Scholar] [CrossRef] [PubMed]
- Shonn, M.A.; McCarroll, R.; Murray, A.W. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 2002, 16, 1659–1671. [Google Scholar] [CrossRef]
- Yokobayashi, S.; Watanabe, Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 2005, 123, 803–817. [Google Scholar] [CrossRef]
- Kim, J.; Ishiguro, K.-i.; Nambu, A.; Akiyoshi, B.; Yokobayashi, S.; Kagami, A.; Ishiguro, T.; Pendas, A.M.; Takeda, N.; Sakakibara, Y. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 2015, 517, 466–471. [Google Scholar] [CrossRef]
- Bonner, A.M.; Hughes, S.E.; Hawley, R.S. Regulation of polo kinase by matrimony is required for cohesin maintenance during Drosophila melanogaster female meiosis. Curr. Biol. 2020, 30, 715–722.e713. [Google Scholar] [CrossRef]
- Yokobayashi, S.; Yamamoto, M.; Watanabe, Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 2003, 23, 3965–3973. [Google Scholar] [CrossRef]
- Ferrandiz, N.; Barroso, C.; Telecan, O.; Shao, N.; Kim, H.-M.; Testori, S.; Faull, P.; Cutillas, P.; Snijders, A.P.; Colaiácovo, M.P. Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat. Commun. 2018, 9, 834. [Google Scholar] [CrossRef]
- McKee, B.D.; Yan, R.; Tsai, J.-H. Meiosis in male Drosophila. Spermatogenesis 2012, 2, 167–184. [Google Scholar] [CrossRef]
- Yu, H.-G.; Dawe, R.K. Functional redundancy in the maize meiotic kinetochore. J. Cell Biol. 2000, 151, 131–142. [Google Scholar] [CrossRef]
- Shao, T.; Tang, D.; Wang, K.; Wang, M.; Che, L.; Qin, B.; Yu, H.; Li, M.; Gu, M.; Cheng, Z. OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 2011, 156, 1386–1396. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Peirson, B.N.; Dong, F.; Xue, C.; Makaroff, C.A. Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 1999, 11, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.M.; Lister, C.; Page, T.; Fransz, P.; Findlay, K.; Jones, G.H.; Dickinson, H.G.; Dean, C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999, 19, 463–472. [Google Scholar] [CrossRef]
- Zhang, L.; Tao, J.; Wang, S.; Chong, K.; Wang, T. The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol. Biol. 2006, 60, 533. [Google Scholar] [CrossRef] [PubMed]
- Moens, P.B.; Kolas, N.K.; Tarsounas, M.; Marcon, E.; Cohen, P.E.; Spyropoulos, B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 2002, 115, 1611–1622. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y. Modifying sister chromatid cohesion for meiosis. J. Cell Sci. 2004, 117, 4017–4023. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kerrebrock, A.W.; Moore, D.P.; Wu, J.S.; Orr-Weaver, T.L. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 1995, 83, 247–256. [Google Scholar] [CrossRef]
- Yao, Y.; Dai, W. Shugoshins function as a guardian for chromosomal stability in nuclear division. Cell Cycle 2012, 11, 2631–2642. [Google Scholar] [CrossRef]
- Katis, V.L.; Galova, M.; Rabitsch, K.P.; Gregan, J.; Nasmyth, K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004, 14, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, T.S.; Kawashima, S.A.; Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 2004, 427, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Marston, A.L.; Tham, W.-H.; Shah, H.; Amon, A. A genome-wide screen identifies genes required for centromeric cohesion. Science 2004, 303, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Rabitsch, K.P.; Gregan, J.; Schleiffer, A.; Javerzat, J.-P.; Eisenhaber, F.; Nasmyth, K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004, 14, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Golubovskaya, I.; Meeley, R.; Fiume, E.; Timofejeva, L.; Schleiffer, A.; Nasmyth, K.; Cande, W.Z. A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr. Biol. 2005, 15, 948–954. [Google Scholar] [CrossRef]
- Gómez, R.; Valdeolmillos, A.; Parra, M.T.; Viera, A.; Carreiro, C.; Roncal, F.; Rufas, J.S.; Barbero, J.L.; Suja, J.A. Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep. 2007, 8, 173–180. [Google Scholar] [CrossRef]
- Lee, J.; Kitajima, T.S.; Tanno, Y.; Yoshida, K.; Morita, T.; Miyano, T.; Miyake, M.; Watanabe, Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 2008, 10, 42–52. [Google Scholar] [CrossRef]
- Xu, Z.; Cetin, B.; Anger, M.; Cho, U.S.; Helmhart, W.; Nasmyth, K.; Xu, W. Structure and function of the PP2A-shugoshin interaction. Mol. Cell 2009, 35, 426–441. [Google Scholar] [CrossRef]
- Macy, B.; Wang, M.; Yu, H.-G. The many faces of shugoshin, the “guardian spirit,” in chromosome segregation. Cell Cycle 2009, 8, 35–37. [Google Scholar] [CrossRef]
- Tanno, Y.; Kitajima, T.S.; Honda, T.; Ando, Y.; Ishiguro, K.-I.; Watanabe, Y. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010, 24, 2169–2179. [Google Scholar] [CrossRef]
- Clift, D.; Marston, A. The role of shugoshin in meiotic chromosome segregation. Cytogenet. Genome Res. 2011, 133, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Zamariola, L.; De Storme, N.; Tiang, C.; Armstrong, S.; Franklin, F.; Geelen, D. SGO1 but not SGO2 is required for maintenance of centromere cohesion in Arabidopsis thaliana meiosis. Plant Reprod. 2013, 26, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Zamariola, L.; De Storme, N.; Vannerum, K.; Vandepoele, K.; Armstrong, S.J.; Franklin, F.C.H.; Geelen, D. SHUGOSHIN s and PATRONUS protect meiotic centromere cohesion in Arabidopsis thaliana. Plant J. 2014, 77, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Cromer, L.; Jolivet, S.; Horlow, C.; Chelysheva, L.; Heyman, J.; De Jaeger, G.; Koncz, C.; De Veylder, L.; Mercier, R. Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr. Biol. 2013, 23, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tang, D.; Wang, K.; Shen, Y.; Qin, B.; Miao, C.; Li, M.; Cheng, Z. OsSGO1 maintains synaptonemal complex stabilization in addition to protecting centromeric cohesion during rice meiosis. Plant J. 2011, 67, 583–594. [Google Scholar] [CrossRef]
- Mahjoubi, H.; Tamari, Y.; Takeda, S.; Bouchabké-Coussa, O.; Hanin, M.; Herzog, E.; Schmit, A.-C.; Chabouté, M.-E.; Ebel, C. The wheat TdRL1 is the functional homolog of the rice RSS1 and promotes plant salt stress tolerance. Plant Cell Rep. 2018, 37, 1625–1637. [Google Scholar] [CrossRef]
- Cromer, L.; Jolivet, S.; Singh, D.K.; Berthier, F.; De Winne, N.; De Jaeger, G.; Komaki, S.; Prusicki, M.A.; Schnittger, A.; Guérois, R. Patronus is the elusive plant securin, preventing chromosome separation by antagonizing separase. Proc. Natl. Acad. Sci. USA 2019, 116, 16018–16027. [Google Scholar] [CrossRef]
- Ogawa, D.; Abe, K.; Miyao, A.; Kojima, M.; Sakakibara, H.; Mizutani, M.; Morita, H.; Toda, Y.; Hobo, T.; Sato, Y. RSS1 regulates the cell cycle and maintains meristematic activity under stress conditions in rice. Nat. Com. 2011, 2, 278. [Google Scholar] [CrossRef]
- Ravi, M.; Bondada, R. Genome elimination by tailswap CenH3: In vivo haploid production in Arabidopsis thaliana. In Chromosome and Genomic Engineering in Plants: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2016; pp. 77–99. [Google Scholar] [CrossRef]
- Marimuthu, M.P.; Jolivet, S.; Ravi, M.; Pereira, L.; Davda, J.N.; Cromer, L.; Wang, L.; Nogue, F.; Chan, W.L.; Siddiqi, I.; et al. Synthetic clonal reproduction through seeds. Science 2011, 331, 876. [Google Scholar] [CrossRef]
- Wang, N.; Gent, J.I.; Dawe, R.K. Haploid induction by a maize cenh3 null mutant. Sci. Adv. 2021, 7, eabe2299. [Google Scholar] [CrossRef]
- Lv, J.; Yu, K.; Wei, J.; Gui, H.; Liu, C.; Liang, D.; Wang, Y.; Zhou, H.; Carlin, R.; Rich, R. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat. Biotechnol 2020, 38, 1397–1401. [Google Scholar] [CrossRef] [PubMed]
- Sanei, M.; Pickering, R.; Kumke, K.; Nasuda, S.; Houben, A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. USA 2011, 108, E498–E505. [Google Scholar] [CrossRef] [PubMed]

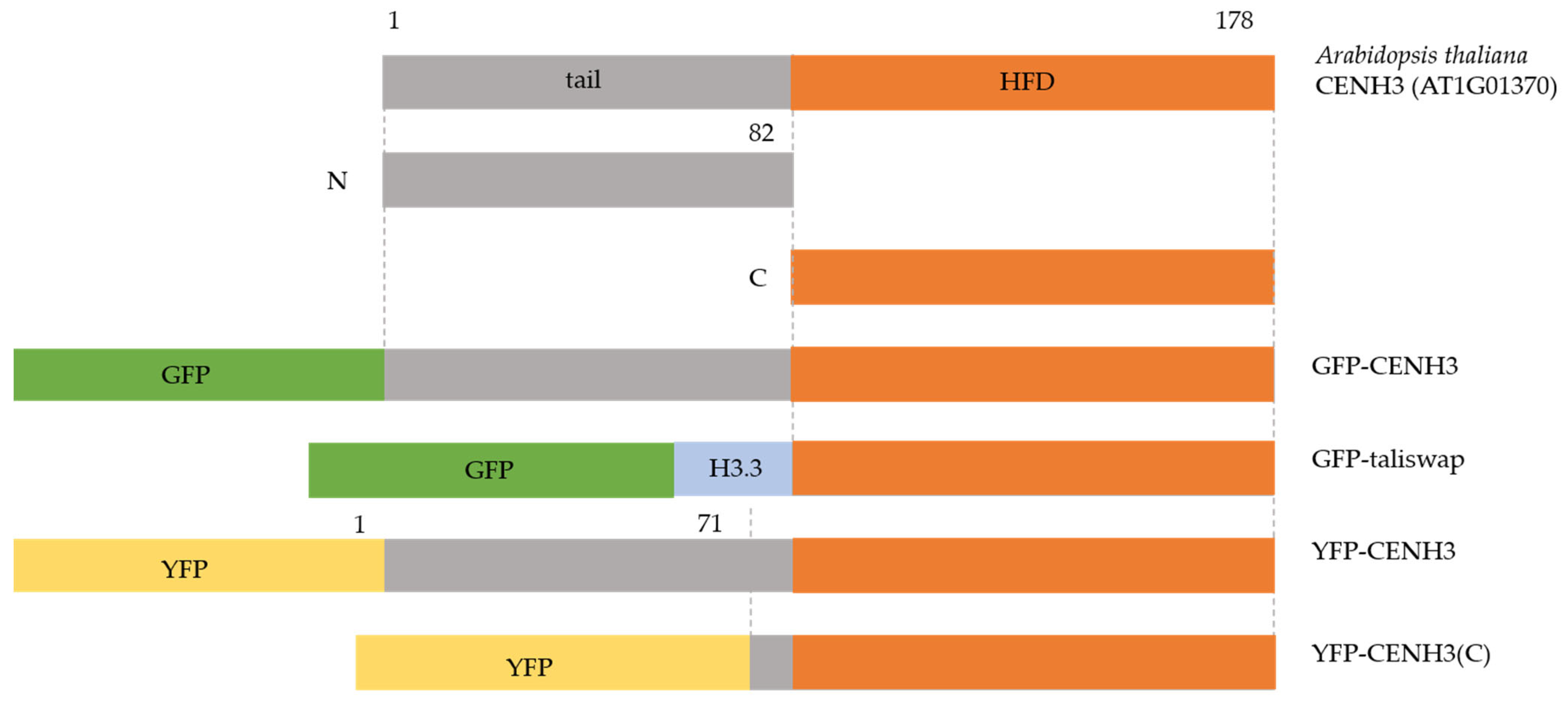

| Name | Homologs | Protein Function or Feature | Mutant Phenotype | Reference |
|---|---|---|---|---|
| SAC proteins | ||||
| MPS1 | AtMPS1 (Arabidopsis) | required for faithful chromosome segregation | chromosome mis-segregation; aneuploidy; precocious into anaphase I | [54] |
| AtPRD2 (Arabidopsis) | involves the formation of DSB and spindle structure | gametophytes aborted; abnormal meiosis products | [55] | |
| OsPRD2 (Rice) | meiotic DSB formation | male and female completely sterile; twenty-four univalent | [56,57] | |
| BUB1 | BRK1 (Rice) | proper tension between homologous kinetochores | precocious separations of sister chromatids; sterile tetrad | [58] |
| ZmBUB1 (Maize) | Bub1-mediated phosphorylation of H2AThr133 | decline of anther fertility | [59] | |
| BUB3 | ZmBUB3 (Maize) | located at the kinetochore | [59] | |
| MAD2 | MAD2 (Maize) | at centromere; relates to the distance between kinetochores | [60] | |
| CPC proteins | ||||
| Aurora | α-Aurora β-Aurora (Arabidopsis) | catalytic subunit of the CPC | microsporogenesis and defects in polyploid and aneuploid offspring | [61,62] |
| INCENP | WYR (Arabidopsis) | involved in cell cycle control | defects in gametophyte cell division and development | [63] |
| Borealis | BORR (Arabidopsis) | required for proper chromosome segregation and cell division | undeveloped ovules, aborted seeds and embryonic defects | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, K.-D.; Zhang, C.-X.; Niu, F.-R.; Bai, H.-C.; Wu, D.-D.; Deng, J.-C.; Qian, H.-Y.; Jiang, Y.-L.; Ma, W. Exploring Plant Meiosis: Insights from the Kinetochore Perspective. Curr. Issues Mol. Biol. 2023, 45, 7974-7995. https://doi.org/10.3390/cimb45100504
Zhou K-D, Zhang C-X, Niu F-R, Bai H-C, Wu D-D, Deng J-C, Qian H-Y, Jiang Y-L, Ma W. Exploring Plant Meiosis: Insights from the Kinetochore Perspective. Current Issues in Molecular Biology. 2023; 45(10):7974-7995. https://doi.org/10.3390/cimb45100504
Chicago/Turabian StyleZhou, Kang-Di, Cai-Xia Zhang, Fu-Rong Niu, Hao-Chen Bai, Dan-Dan Wu, Jia-Cheng Deng, Hong-Yuan Qian, Yun-Lei Jiang, and Wei Ma. 2023. "Exploring Plant Meiosis: Insights from the Kinetochore Perspective" Current Issues in Molecular Biology 45, no. 10: 7974-7995. https://doi.org/10.3390/cimb45100504
APA StyleZhou, K.-D., Zhang, C.-X., Niu, F.-R., Bai, H.-C., Wu, D.-D., Deng, J.-C., Qian, H.-Y., Jiang, Y.-L., & Ma, W. (2023). Exploring Plant Meiosis: Insights from the Kinetochore Perspective. Current Issues in Molecular Biology, 45(10), 7974-7995. https://doi.org/10.3390/cimb45100504





