Analysis of DNA Methylation Differences during the JIII Formation of Bursaphelenchus xylophilus
Abstract
:1. Introduction
2. Materials and Methods
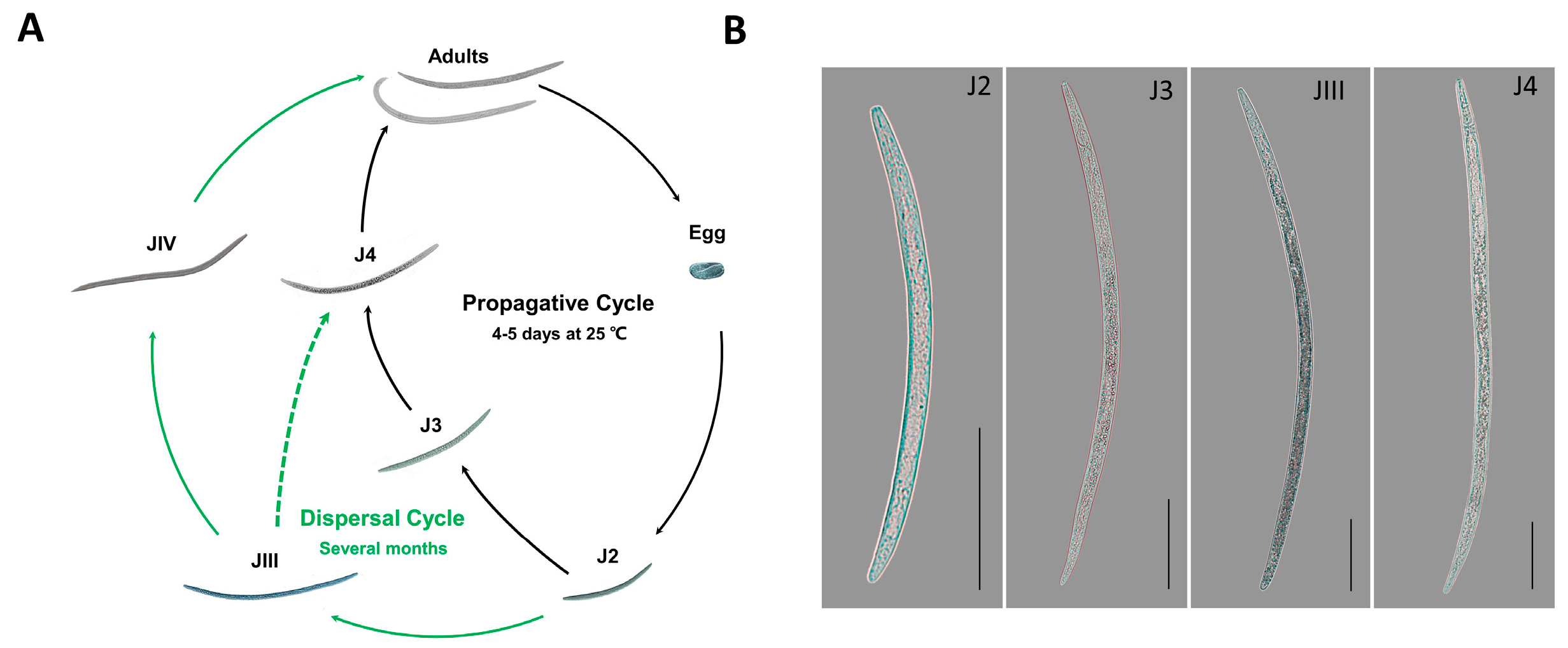
2.1. Nematodes in Different Developmental Stages
2.2. DNA and RNA Extraction
2.3. WGBS Library Preparation
2.4. Sequence Alignment and Detection of Cytosines Methylation Level
2.5. DMC and DMR Detection
2.6. RNA Sequencing
2.7. Validation of RT-qPCR
3. Results
3.1. DNA Methylation Sequencing Data
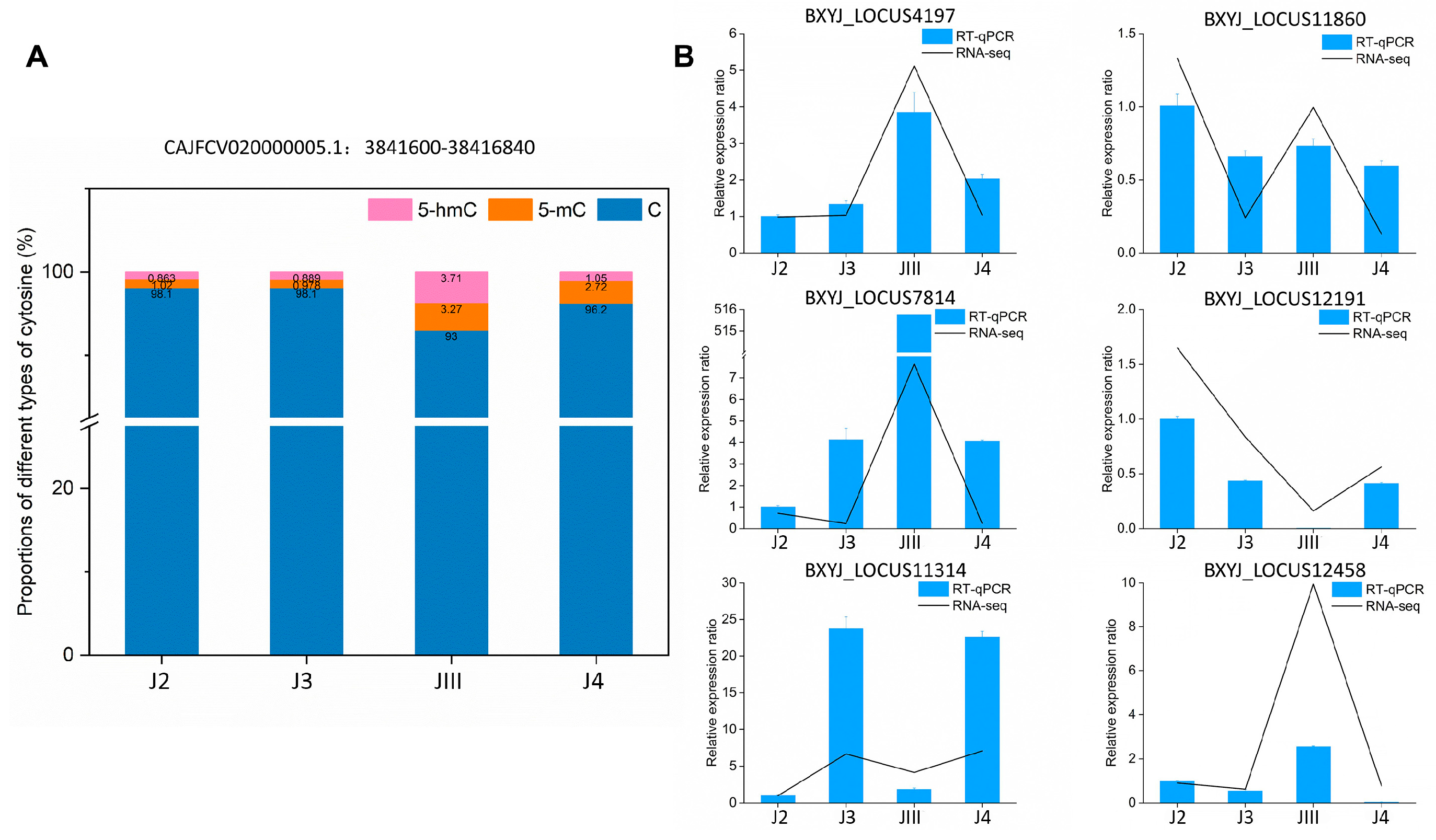
3.2. Distribution and Statistics of Cytosine Methylation
3.3. Identification and Analysis of DMCs and DMRs
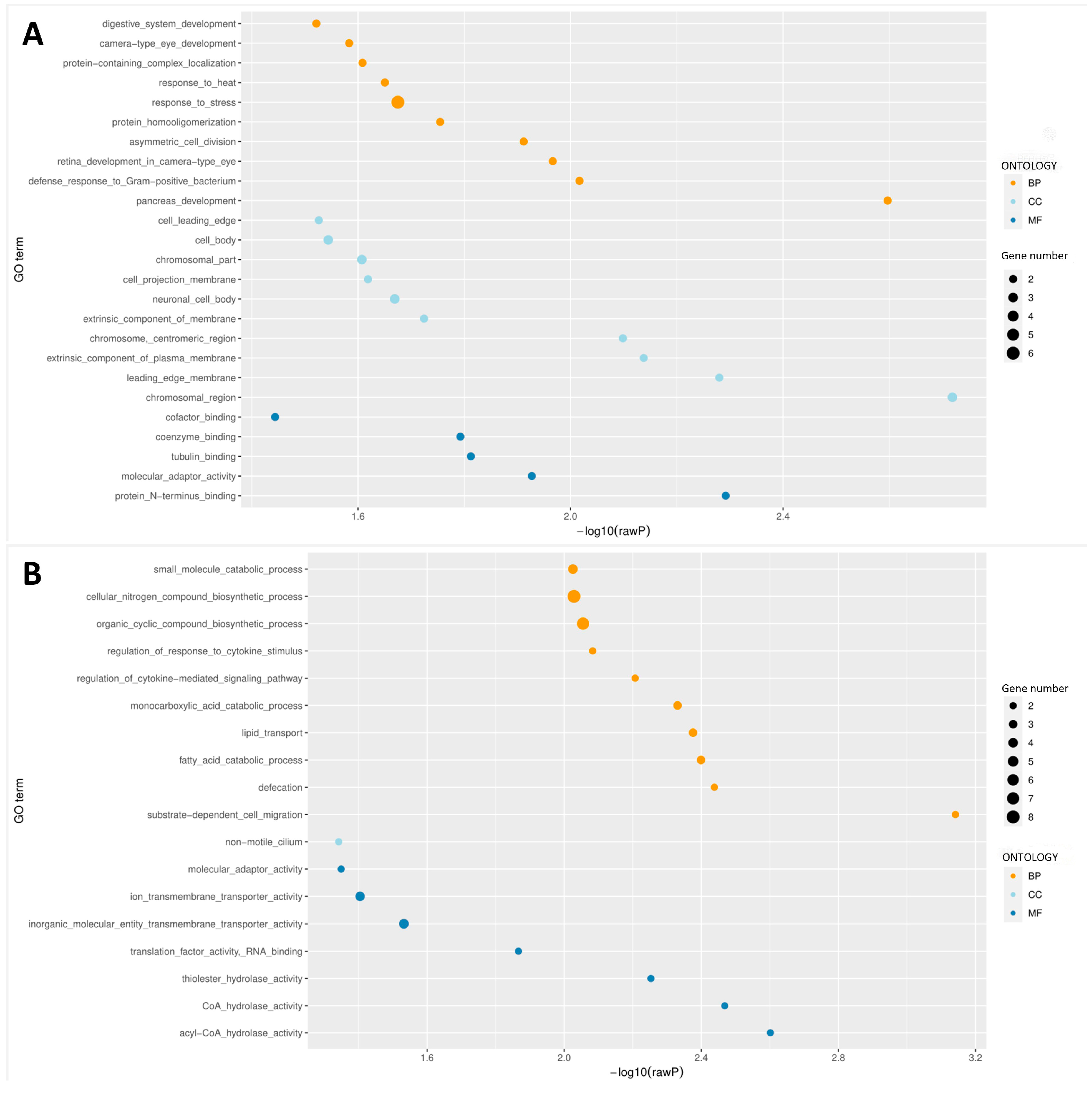
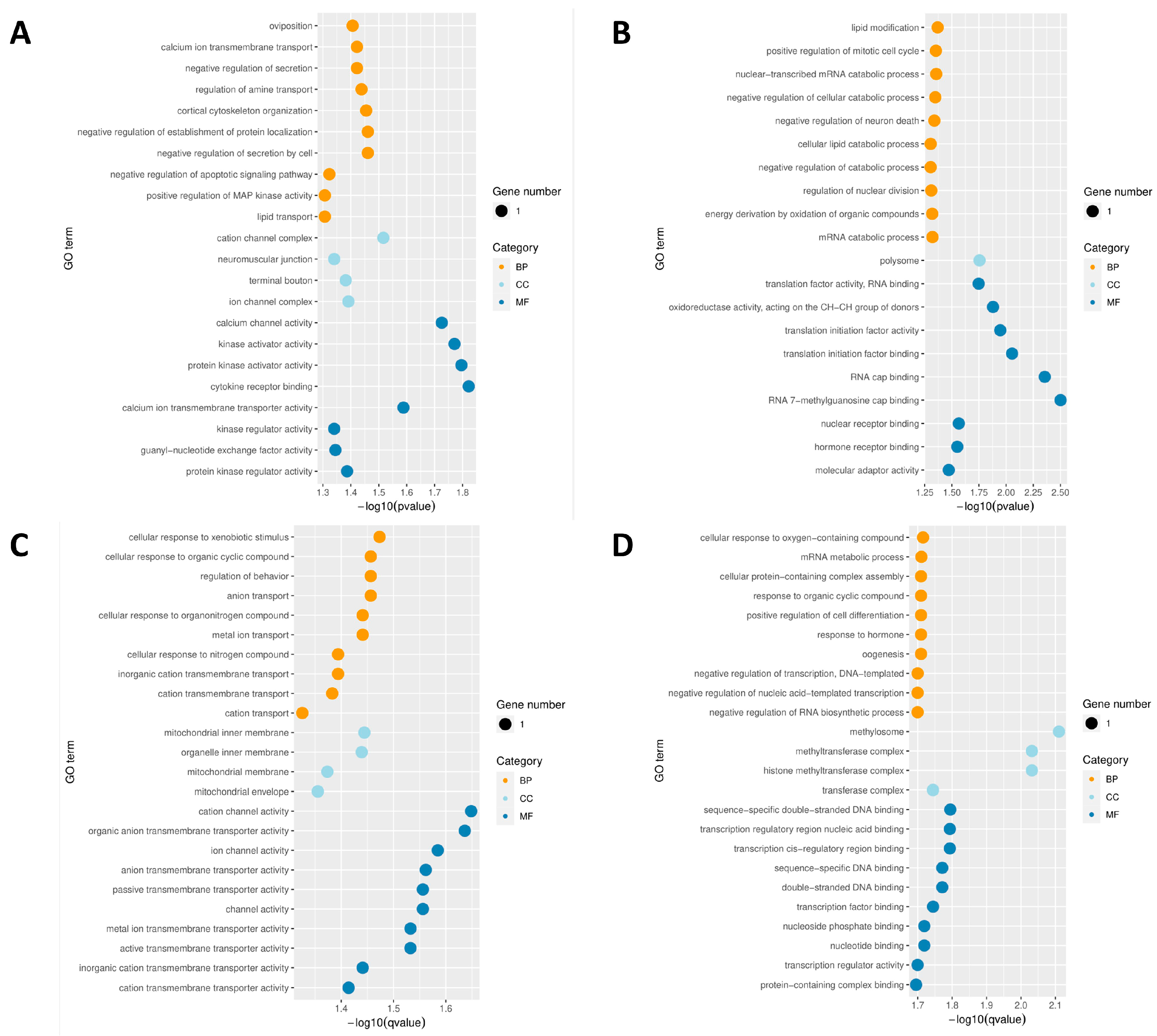
3.4. GO Analysis of DMRs during JIII Formation
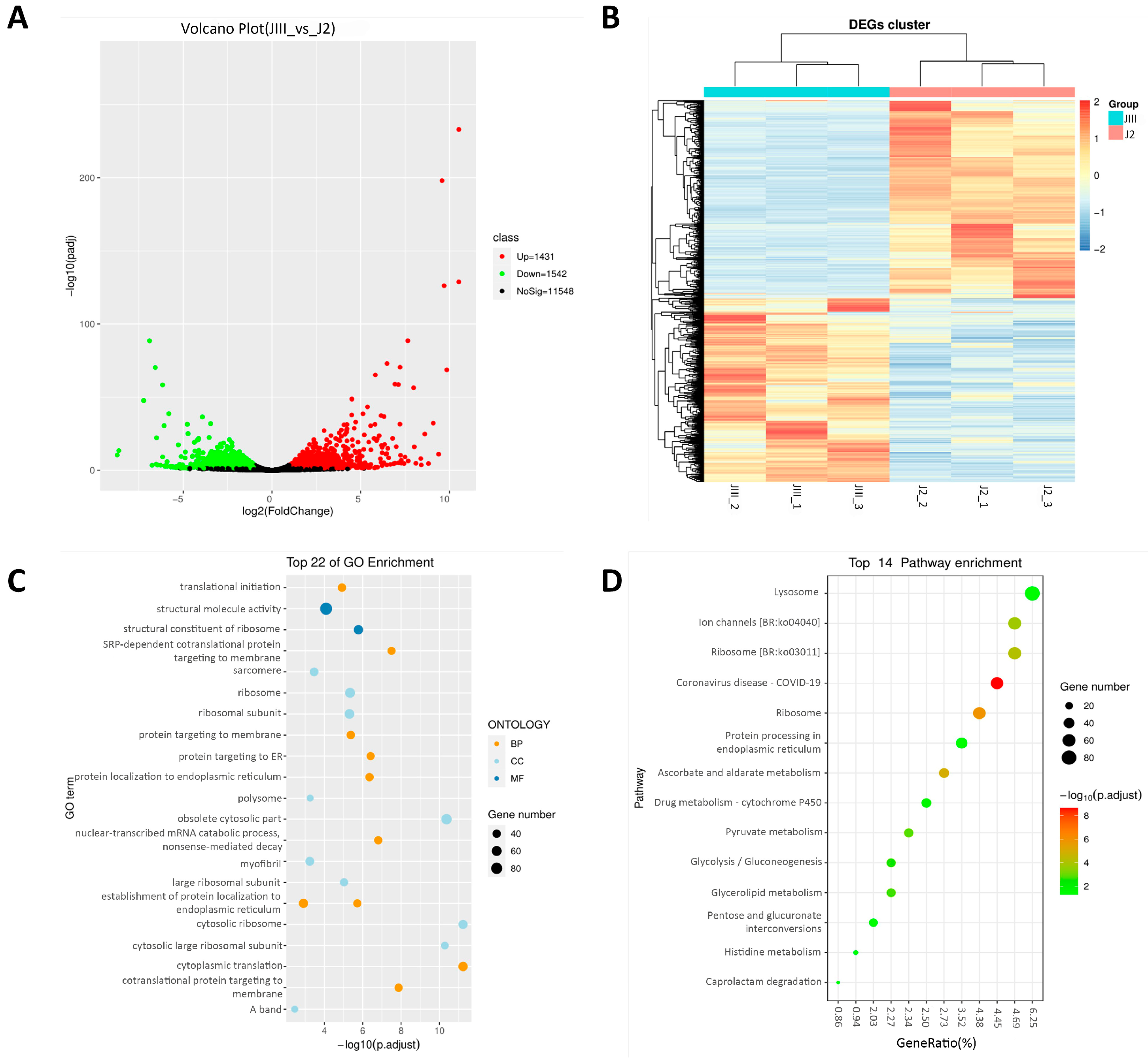
3.5. Summary of Transcriptome Analysis and Pathway Enrichment
3.6. Correlation Analysis of DMRs and Gene Expression during JIII Formation
3.7. Validation of DNA Methylation and Transcriptomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Futai, K. Pine Wood Nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 2013, 51, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 Plant-Parasitic Nematodes in Molecular Plant Pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.T.; Moens, M.; Mota, M.; Li, H.; Kikuchi, T. Bursaphelenchus xylophilus: Opportunities in Comparative Genomics and Molecular Host–Parasite Interactions. Mol. Plant Pathol. 2008, 9, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Nickle, W.R.; Golden, A.M.; Mamiya, Y.; Wergin, W.P. On the Taxonomy and Morphology of the Pine Wood Nematode, Bursaphelenchus xylophilus (Steiner & Buhrer 1934) Nickle 1970. J. Nematol. 1981, 13, 385. [Google Scholar] [PubMed]
- Fukuda, K. Physiological Process of the Symptom Development and Resistance Mechanism in Pine Wilt Disease. J. For. Res. 1997, 2, 171–181. [Google Scholar] [CrossRef]
- Ishibashi, N.; Kondo, E. Occurrence and survival of the dispersal forms of pine wood nematode, Bursaphelenchus lignicolus Mamiya and Kiyohara. Appl. Entomol. Zool. 1977, 12, 293–302. [Google Scholar] [CrossRef]
- Tanaka, S.E.; Aikawa, T.; Takeuchi-Kaneko, Y.; Fukuda, K.; Kanzaki, N. Artificial Induction of Third-Stage Dispersal Juveniles of Bursaphelenchus xylophilus Using Newly Established Inbred Lines. PLoS ONE 2017, 12, e0187127. [Google Scholar] [CrossRef]
- Mamiya, Y. The life history of the pine wood nematode, Bursaphelenchus lignicolus. Jpn. J. Nematol. 1975, 5, 16–25. [Google Scholar]
- Togashi, K. A New Method for Loading Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) on Adult Monochamus alternatus (Coleoptera: Cerambycidae). J. Econ. Entomol. 2004, 97, 941–945. [Google Scholar] [CrossRef]
- Vineis, P.; Chatziioannou, A.; Cunliffe, V.T.; Flanagan, J.M.; Hanson, M.; Kirsch-Volders, M.; Kyrtopoulos, S. Epigenetic Memory in Response to Environmental Stressors. FASEB J. 2017, 31, 2241–2251. [Google Scholar] [CrossRef]
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the Impact of Transgenerational Epigenetic Variation on Complex Traits. PLoS Genet. 2009, 5, e1000530. [Google Scholar] [CrossRef]
- Zhang, Y.; Fischer, M.; Colot, V.; Bossdorf, O. Epigenetic Variation Creates Potential for Evolution of Plant Phenotypic Plasticity. New Phytol. 2013, 197, 314–322. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic Regulation of Gene Expression: How the Genome Integrates Intrinsic and Environmental Signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Anderson, O.S.; Sant, K.E.; Dolinoy, D.C. Nutrition and Epigenetics: An Interplay of Dietary Methyl Donors, One-Carbon Metabolism and DNA Methylation. J. Nutr. Biochem. 2012, 23, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.A. The Impact of Climate Change on Plant Epigenomes. Trends Genet. 2013, 29, 503–505. [Google Scholar] [CrossRef]
- Navarro-Martín, L.; Viñas, J.; Ribas, L.; Díaz, N.; Gutiérrez, A.; Di Croce, L.; Piferrer, F. DNA Methylation of the Gonadal Aromatase (Cyp19a) Promoter Is Involved in Temperature-Dependent Sex Ratio Shifts in the European Sea Bass. PLoS Genet. 2011, 7, e1002447. [Google Scholar] [CrossRef] [PubMed]
- Venegas, D.; Marmolejo-Valencia, A.; Valdes-Quezada, C.; Govenzensky, T.; Recillas-Targa, F.; Merchant-Larios, H. Dimorphic DNA Methylation during Temperature-Dependent Sex Determination in the Sea Turtle Lepidochelys olivacea. Gen. Comp. Endocrinol. 2016, 236, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.; Valente, L.M.P.; Fernandes, J.M.O. Molecular Evolution of Zebrafish Dnmt3 Genes and Thermal Plasticity of Their Expression during Embryonic Development. Gene 2012, 500, 93–100. [Google Scholar] [CrossRef]
- Han, B.; Li, W.; Chen, Z.; Xu, Q.; Luo, J.; Shi, Y.; Li, X.; Yan, X.; Zhang, J. Variation of DNA Methylome of Zebrafish Cells under Cold Pressure. PLoS ONE 2016, 11, e0160358. [Google Scholar] [CrossRef]
- Shi, Y.Y.; Huang, Z.Y.; Zeng, Z.J.; Wang, Z.L.; Wu, X.B.; Yan, W.Y. Diet and Cell Size Both Affect Queen-Worker Differentiation through DNA Methylation in Honey Bees (Apis mellifera, Apidae). PLoS ONE 2011, 6, e18808. [Google Scholar] [CrossRef]
- Arand, J.; Spieler, D.; Karius, T.; Branco, M.R.; Meilinger, D.; Meissner, A.; Jenuwein, T.; Xu, G.; Leonhardt, H.; Wolf, V.; et al. In Vivo Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases. PLoS Genet. 2012, 8, e1002750. [Google Scholar] [CrossRef] [PubMed]
- Rošić, S.; Amouroux, R.; Requena, C.E.; Gomes, A.; Emperle, M.; Beltran, T.; Rane, J.K.; Linnett, S.; Selkirk, M.E.; Schiffer, P.H.; et al. Evolutionary Analysis Indicates That DNA Alkylation Damage Is a Byproduct of Cytosine DNA Methyltransferase Activity. Nat. Genet. 2018, 50, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Takai, D. The Role of DNA Methylation in Mammalian Epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef] [PubMed]
- Loyfer, N.; Magenheim, J.; Peretz, A.; Cann, G.; Bredno, J.; Klochendler, A.; Fox-Fisher, I.; Shabi-Porat, S.; Hecht, M.; Pelet, T.; et al. A DNA Methylation Atlas of Normal Human Cell Types. Nature 2023, 613, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Keller, T.E.; Han, P.; Yi, S.V. Evolutionary Transition of Promoter and Gene Body DNA Methylation across Invertebrate–Vertebrate Boundary. Mol. Biol. Evol. 2016, 33, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.C.; Ruiz-Ferrer, V.; Müller, S.Y.; Pellegrin, C.; Abril-Urías, P.; Martínez-Gómez, Á.; Gómez-Rojas, A.; Berenguer, E.; Testillano, P.S.; Andrés, M.F.; et al. The DNA Methylation Landscape of the Root-Knot Nematode-Induced Pseudo-Organ, the Gall, in Arabidopsis, Is Dynamic, Contrasting over Time, and Critically Important for Successful Parasitism. New Phytol. 2022, 236, 1888–1907. [Google Scholar] [CrossRef]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.-H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. Elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Gao, F.; Liu, X.; Wu, X.-P.; Wang, X.-L.; Gong, D.; Lu, H.; Xia, Y.; Song, Y.; Wang, J.; Du, J.; et al. Differential DNA Methylation in Discrete Developmental Stages of the Parasitic Nematode Trichinella spiralis. Genome Biol. 2012, 13, R100. [Google Scholar] [CrossRef]
- Hadad, N. Early-Life DNA Methylation Profiles Are Indicative of Age-Related Transcriptome Changes. Epigenetics Chromatin 2019, 12, 58. [Google Scholar] [CrossRef]
- Jaiswal, V.; Rawoof, A.; Gahlaut, V.; Ahmad, I.; Chhapekar, S.S.; Dubey, M.; Ramchiary, N. Integrated Analysis of DNA Methylation, Transcriptome, and Global Metabolites in Interspecific Heterotic Capsicum F1 Hybrid. iScience 2022, 25, 105318. [Google Scholar] [CrossRef]
- Beck, D.; Ben Maamar, M.; Skinner, M.K. Genome-Wide CpG Density and DNA Methylation Analysis Method (MeDIP, RRBS, and WGBS) Comparisons. Epigenetics 2022, 17, 518–530. [Google Scholar] [CrossRef]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.A.; et al. Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hu, Q.; Liu, X.; Zou, K.; Sarkodie, E.K.; Liu, X.; Gao, F. AllEnricher: A Comprehensive Gene Set Function Enrichment Tool for Both Model and Non-Model Species. BMC Bioinform. 2020, 21, 106. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Li, D.; Zhang, W.; Wang, X.; Wen, X.; Liu, Z.; Feng, Y.; Zhang, X. Identification of the Extracellular Nuclease Influencing Soaking RNA Interference Efficiency in Bursaphelenchus xylophilus. Int. J. Mol. Sci. 2022, 23, 12278. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, R.; Li, D.; Wang, F. Genetic Characteristics of Bursaphelenchus xylophilus Third-Stage Dispersal Juveniles. Sci. Rep. 2021, 11, 3908. [Google Scholar] [CrossRef]
- Angers, B.; Castonguay, E.; Massicotte, R. Environmentally Induced Phenotypes and DNA Methylation: How to Deal with Unpredictable Conditions until the Next Generation and after. Mol. Ecol. 2010, 19, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, A.; Brickner, J.H. Mechanisms of Epigenetic Memory. Trends Genet. 2014, 30, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Chang, J.-C.; Yen, M.-R.; Huang, Y.-F.; Chen, T.-H.; Chen, L.-H.; Nai, Y.-S. Whole-Genome DNA Methylome Analysis of Different Developmental Stages of the Entomopathogenic Fungus Beauveria bassiana NCHU-157 by Nanopore Sequencing. Front. Genet. 2023, 14, 1085631. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.-H.; Wang, H.-T.; Chen, X.-Q.; Ge, M.-X.; Yan, D.; Yang, X.-L.; Yang, L.-Q.; Lin, R.; Guo, R.-H.; Zhang, W.; et al. Hypermethylation in H3K9me3 Regions Characterizes the Centenarian Methylomes in Healthy Aging. Natl. Sci. Rev. 2023, 10, nwad067. [Google Scholar] [CrossRef] [PubMed]
- Jie, X.; Wu, H.; Yang, M.; He, M.; Zhao, G.; Ling, S.; Huang, Y.; Yue, B.; Yang, N.; Zhang, X. Whole Genome Bisulfite Sequencing Reveals DNA Methylation Roles in the Adaptive Response of Wildness Training Giant Pandas to Wild Environment. Front. Genet. 2022, 13, 995700. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Pan, L.; Meng, F.; Zhang, X. Cold Adaptive Potential of Pine Wood Nematodes Overwintering in Plant Hosts. Biol. Open 2019, 8, bio.041616. [Google Scholar] [CrossRef]
- Massengo-Tiassé, R.P.; Cronan, J.E. Diversity in Enoyl-Acyl Carrier Protein Reductases. Cell. Mol. Life Sci. 2009, 66, 1507–1517. [Google Scholar] [CrossRef]
- Yao, J.; Ericson, M.E.; Frank, M.W.; Rock, C.O. Enoyl-Acyl Carrier Protein Reductase I (FabI) Is Essential for the Intracellular Growth of Listeria monocytogenes. Infect. Immun. 2016, 84, 3597–3607. [Google Scholar] [CrossRef]
- Waterham, H.R.; Ferdinandusse, S.; Wanders, R.J.A. Human Disorders of Peroxisome Metabolism and Biogenesis. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 922–933. [Google Scholar] [CrossRef]
- Lodhi, I.J.; Semenkovich, C.F. Peroxisomes: A Nexus for Lipid Metabolism and Cellular Signaling. Cell Metab. 2014, 19, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-L.; Weigel, A.V.; Ioannou, M.S.; Pasolli, H.A.; Xu, C.S.; Peale, D.R.; Shtengel, G.; Freeman, M.; Hess, H.F.; Blackstone, C.; et al. Spastin Tethers Lipid Droplets to Peroxisomes and Directs Fatty Acid Trafficking through ESCRT-III. J. Cell Biol. 2019, 218, 2583–2599. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, M.; Guo, J.; Hao, Z.; Zhang, Z.; Lu, Z.; Wang, J.; Zhu, X.; Wang, Y.; Chen, J.; et al. The Peroxins BcPex8, BcPex10, and BcPex12 Are Required for the Development and Pathogenicity of Botrytis cinerea. Front. Microbiol. 2022, 13, 962500. [Google Scholar] [CrossRef] [PubMed]


| Gene ID | DMR Genomic Location | DMR Difference | DEG log2 Fold Change |
|---|---|---|---|
| BXYJ_LOCUS13082 | genebody | 0.338333 | −1.104336548 |
| BXYJ_LOCUS11405 (Syt1) | genebody | 0.200167 | −1.268410936 |
| BXYJ_LOCUS7569 | genebody | 0.184889 | 6.037514936 |
| BXYJ_LOCUS7569 | promoter | 0.184889 | 6.037514936 |
| BXYJ_LOCUS7568 | promoter | 0.184889 | 6.477511571 |
| BXYJ_LOCUS8847 | genebody | 0.148556 | −1.069283441 |
| BXYJ_LOCUS12505 | promoter | 0.128333 | 1.891287088 |
| BXYJ_LOCUS15791 (Kmt) | genebody | 0.119778 | 1.718757485 |
| BXYJ_LOCUS10978 | genebody | 0.1175 | −1.451330359 |
| BXYJ_LOCUS8383 | genebody | 0.117333 | 7.439557963 |
| BXYJ_LOCUS8383 | promoter | 0.117333 | 7.439557963 |
| BXYJ_LOCUS8384 | promoter | 0.117333 | 3.671438732 |
| BXYJ_LOCUS10824 | promoter | 0.102778 | −3.842519089 |
| BXYJ_LOCUS8048 (Trpv6) | genebody | −0.100111 | −1.312559644 |
| BXYJ_LOCUS10582 (Enr) | genebody | −0.100778 | 2.43271744 |
| BXYJ_LOCUS6456 (Pex10) | promoter | −0.132444 | 1.010721634 |
| BXYJ_LOCUS8233 (Col-125) | genebody | −0.15 | −4.15283697 |
| BXYJ_LOCUS7390 (Kcnk18) | promoter | −0.161667 | −1.063657134 |
| BXYJ_LOCUS12977 (Srt-41) | genebody | −0.166333 | −1.476196334 |
| BXYJ_LOCUS5952 (Madd) | genebody | −0.171556 | −1.15646022 |
| BXYJ_LOCUS3255 (Nht-62) | genebody | −0.219333 | −1.199834112 |
| BXYJ_LOCUS12921 (Slc25a10) | promoter | −0.227556 | −1.178271087 |
| BXYJ_LOCUS5381 (Mif4gd) | genebody | −0.244333 | 1.390619044 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Li, Y.; Liu, Z.; Zhang, W.; Li, D.; Wang, X.; Wen, X.; Feng, Y.; Zhang, X. Analysis of DNA Methylation Differences during the JIII Formation of Bursaphelenchus xylophilus. Curr. Issues Mol. Biol. 2023, 45, 9656-9673. https://doi.org/10.3390/cimb45120603
Wang P, Li Y, Liu Z, Zhang W, Li D, Wang X, Wen X, Feng Y, Zhang X. Analysis of DNA Methylation Differences during the JIII Formation of Bursaphelenchus xylophilus. Current Issues in Molecular Biology. 2023; 45(12):9656-9673. https://doi.org/10.3390/cimb45120603
Chicago/Turabian StyleWang, Peng, Yongxia Li, Zhenkai Liu, Wei Zhang, Dongzhen Li, Xuan Wang, Xiaojian Wen, Yuqian Feng, and Xingyao Zhang. 2023. "Analysis of DNA Methylation Differences during the JIII Formation of Bursaphelenchus xylophilus" Current Issues in Molecular Biology 45, no. 12: 9656-9673. https://doi.org/10.3390/cimb45120603
APA StyleWang, P., Li, Y., Liu, Z., Zhang, W., Li, D., Wang, X., Wen, X., Feng, Y., & Zhang, X. (2023). Analysis of DNA Methylation Differences during the JIII Formation of Bursaphelenchus xylophilus. Current Issues in Molecular Biology, 45(12), 9656-9673. https://doi.org/10.3390/cimb45120603






