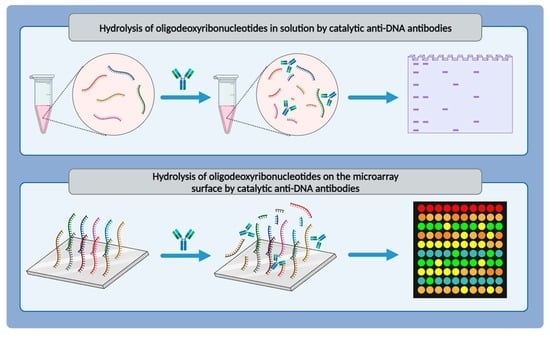
Hydrolysis of Oligodeoxyribonucleotides on the Microarray Surface and in Solution by Catalytic Anti-DNA Antibodies in Systemic Lupus Erythematosus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants and Biological Material
2.2. Purification of Antibodies from the Serum
2.3. Model Oligodeoxyribonucleotides Used for Analysis
2.4. Microarray Slide Production
2.5. Analysis of Hydrolysis of Immobilized ODNs on Microarray Surface by Antibodies
2.6. Analysis of ODN Hydrolysis in Solution by Antibodies Using Electrophoresis
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Patients and Healthy Individuals
3.2. Hydrolysis of Immobilized ODNs on Microarray Surface by Antibodies
3.3. Hydrolysis of ODN by Antibodies in Solution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaul, A.; Gordon, C.; Crow, M.K.; Touma, Z.; Urowitz, M.B.; Van Vollenhoven, R.; Ruiz-Irastorza, G.; Hughes, G. Systemic Lupus Erythematosus. Nat. Rev. Dis. Primers 2016, 2, 16039. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, G.C.; Lo, M.S.; Reis, P.C.; Sullivan, K.E. New Insights into the Immunopathogenesis of Systemic Lupus Erythematosus. Nat. Rev. Rheumatol. 2016, 12, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S. Anti-DNA Antibodies—Quintessential Biomarkers of SLE. Nat. Rev. Rheumatol. 2016, 12, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S.; Lipsky, P.E. New Insights into the Role of Antinuclear Antibodies in Systemic Lupus Erythematosus. Nat. Rev. Rheumatol. 2020, 16, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S.; Rovin, B.H.; Lipsky, P.E. New Perspectives in Rheumatology: Biomarkers as Entry Criteria for Clinical Trials of New Therapies for Systemic Lupus Erythematosus: The Example of Antinuclear Antibodies and Anti-DNA. Arthritis Rheumatol. 2017, 69, 487–493. [Google Scholar] [CrossRef]
- Aringer, M.; Costenbader, K.; Daikh, D.; Brinks, R.; Mosca, M.; Ramsey-Goldman, R.; Smolen, J.S.; Wofsy, D.; Boumpas, D.T.; Kamen, D.L.; et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Ann. Rheum. Dis. 2019, 78, 1151–1159. [Google Scholar] [CrossRef]
- Ho, A.; Magder, L.S.; Barr, S.G.; Petri, M. Decreases in Anti-Double-Stranded DNA Levels Are Associated with Concurrent Flares in Patients with Systemic Lupus Erythematosus. Arthritis Rheum. 2001, 44, 2342–2349. [Google Scholar] [CrossRef]
- Yeo, A.L.; Kandane-Rathnayake, R.; Koelmeyer, R.; Golder, V.; Louthrenoo, W.; Chen, Y.-H.; Cho, J.; Lateef, A.; Hamijoyo, L.; Luo, S.-F.; et al. SMART-SLE: Serology Monitoring and Repeat Testing in Systemic Lupus Erythematosus—An Analysis of Anti-Double-Stranded DNA Monitoring. Rheumatology 2023, kead231. [Google Scholar] [CrossRef]
- Hamilton, K.J.; Schett, G.; Reich, C.F.; Smolen, J.S.; Pisetsky, D.S. The Binding of Sera of Patients with SLE to Bacterial and Mammalian DNA. Clin. Immunol. 2006, 118, 209–218. [Google Scholar] [CrossRef]
- Rekvig, O.P.; Van Der Vlag, J.; Seredkina, N. Review: Antinucleosome Antibodies: A Critical Reflection on Their Specificities and Diagnostic Impact. Arthritis Rheumatol. 2014, 66, 1061–1069. [Google Scholar] [CrossRef]
- Wang, X.; Xia, Y. Anti-Double Stranded DNA Antibodies: Origin, Pathogenicity, and Targeted Therapies. Front. Immunol. 2019, 10, 1667. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Song, Y.; Chang, J.; Zhou, X.; Qi, Q.; Tian, X.; Li, M.; Zeng, X.; Xu, M.; Zhang, W.; et al. High Levels of Circulating Cell-free DNA Are a Biomarker of Active SLE. Eur. J. Clin. Investig. 2018, 48, e13015. [Google Scholar] [CrossRef] [PubMed]
- Duvvuri, B.; Lood, C. Cell-Free DNA as a Biomarker in Autoimmune Rheumatic Diseases. Front. Immunol. 2019, 10, 502. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kaplan, M.J. The Role of Neutrophils and NETosis in Autoimmune and Renal Diseases. Nat. Rev. Nephrol. 2016, 12, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S.; Wenk, K.S.; Reich, C.F. The Role of CpG Sequences in the Induction of Anti-DNA Antibodies. Clin. Immunol. 2001, 100, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Rekvig, O.P. The Anti-DNA Antibody: Origin and Impact, Dogmas and Controversies. Nat. Rev. Rheumatol. 2015, 11, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Spencer, D.M.; Svenungsson, E.; Gunnarsson, I.; Caricchio, R.; Pisetsky, D.S. The Expression of Antibodies to Z-DNA in the Blood of Patients with Systemic Lupus Erythematosus: Relationship to Autoantibodies to B-DNA. Clin. Immunol. 2023, 255, 109763. [Google Scholar] [CrossRef]
- Casperson, G.F.; Voss, E.W. Specificity of Anti-Dna Antibodies in SLE—II. Relative Contribution of Backbone, Secondary Structure and Nucleotide Sequence to DNA Binding. Mol. Immunol. 1983, 20, 581–588. [Google Scholar] [CrossRef]
- Richardson, C.; Chida, A.S.; Adlowitz, D.; Silver, L.; Fox, E.; Jenks, S.A.; Palmer, E.; Wang, Y.; Heimburg-Molinaro, J.; Li, Q.-Z.; et al. Molecular Basis of 9G4 B Cell Autoreactivity in Human Systemic Lupus Erythematosus. J. Immunol. 2013, 191, 4926–4939. [Google Scholar] [CrossRef]
- Pisetsky, D.S.; Shaffer, R.; Armstrong, D.D.; Spencer, D.M. The Interaction of Anti-DNA Antibodies with DNA Antigen: Evidence for Hysteresis for High Avidity Binding. Clin. Immunol. 2021, 231, 108848. [Google Scholar] [CrossRef]
- Tanner, J.J.; Komissarov, A.A.; Deutscher, S.L. Crystal Structure of an Antigen-Binding Fragment Bound to Single-Stranded DNA. J. Mol. Biol. 2001, 314, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mi, J.; Cao, X. Anti-DNA Antibodies Exhibit Different Binding Motif Preferences for Single Stranded or Double Stranded DNA. Immunol. Lett. 2000, 73, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Swanson, P.C.; Ackroyd, C.; Glick, G.D. Ligand Recognition by Anti-DNA Autoantibodies. Affinity, Specificity, and Mode of Binding. Biochemistry 1996, 35, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Böröcz, K.; Simon, D.; Erdő-Bonyár, S.; Kovács, K.T.; Tuba, É.; Czirják, L.; Németh, P.; Berki, T. Relationship between Natural and Infection-Induced Antibodies in Systemic Autoimmune Diseases (SAD): SLE, SSc and RA. Clin. Exp. Immunol. 2020, 203, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Ding, J.L. Natural Antibodies Bridge Innate and Adaptive Immunity. J. Immunol. 2015, 194, 13–20. [Google Scholar] [CrossRef]
- Maddur, M.S.; Lacroix-Desmazes, S.; Dimitrov, J.D.; Kazatchkine, M.D.; Bayry, J.; Kaveri, S.V. Natural Antibodies: From First-Line Defense Against Pathogens to Perpetual Immune Homeostasis. Clin. Rev. Allerg. Immunol. 2020, 58, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Nevinsky, G.A.; Buneva, V.N. Catalytic Antibodies in Healthy Humans and Patients with Autoimmune and Viral Diseases. J. Cell. Mol. Med. 2003, 7, 265–276. [Google Scholar] [CrossRef]
- Ermakov, E.A.; Nevinsky, G.A.; Buneva, V.N. Immunoglobulins with Non-Canonical Functions in Inflammatory and Autoimmune Disease States. Int. J. Mol. Sci. 2020, 21, 5392. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, J.; Hu, X.; Zhang, S. Catalytic Antibodies: Design, Expression, and Their Applications in Medicine. Appl. Biochem. Biotechnol. 2023, 195, 1514–1540. [Google Scholar] [CrossRef]
- Shuster, A.M.; Gololobov, G.V.; Kvashuk, O.A.; Bogomolova, A.E.; Smirnov, I.V.; Gabibov, A.G. DNA Hydrolyzing Autoantibodies. Science 1992, 256, 665–667. [Google Scholar] [CrossRef]
- Botvinovskaya, A.V.; Kostrikina, I.A.; Buneva, V.N.; Nevinsky, G.A. Systemic Lupus Erythematosus: Molecular Cloning of Several Recombinant DNase Monoclonal Kappa Light Chains with Different Catalytic Properties. J. Mol. Recognit. 2013, 26, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, V.; Pandit, P.; Surve, P.; Lecerf, M.; Rajadhyaksha, A.; Nadkar, M.; Khadilkar, P.V.; Chougule, D.A.; Naigaonkar, A.A.; Lacroix-Desmazes, S.; et al. Catalytic Antibodies in Patients with Systemic Lupus Erythematosus. Eur. J. Rheumatol. 2018, 5, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Gabibov, A.G.; Ponomarenko, N.A.; Tretyak, E.B.; Paltsev, M.A.; Suchkov, S.V. Catalytic Autoantibodies in Clinical Autoimmunity and Modern Medicine. Autoimmun. Rev. 2006, 5, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Wootla, B.; Lacroix-Desmazes, S.; Warrington, A.E.; Bieber, A.J.; Kaveri, S.V.; Rodriguez, M. Autoantibodies with Enzymatic Properties in Human Autoimmune Diseases. J. Autoimmun. 2011, 37, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Nygren, H.; Werthen, M.; Stenberg, M. Kinetics of Antibody Binding to Solid-Phase-Immobilised Antigen. J. Immunol. Methods 1987, 101, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Werthén, M.; Nygren, H. Effect of Antibody Affinity on the Isotherm of Antibody Binding to Surface-Immobilized Antigen. J. Immunol. Methods 1988, 115, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Laurent, N.; Haddoub, R.; Flitsch, S.L. Enzyme Catalysis on Solid Surfaces. Trends Biotechnol. 2008, 26, 328–337. [Google Scholar] [CrossRef]
- Sinyakov, A.N.; Ryabinin, V.A.; Kostina, E.V. Application of Array-Based Oligonucleotides for Synthesis of Genetic Designs. Mol. Biol. 2021, 55, 487–500. [Google Scholar] [CrossRef]
- Tamirou, F.; Arnaud, L.; Talarico, R.; Scirè, C.A.; Alexander, T.; Amoura, Z.; Avcin, T.; Bortoluzzi, A.; Cervera, R.; Conti, F.; et al. Systemic Lupus Erythematosus: State of the Art on Clinical Practice Guidelines. RMD Open 2019, 4, e000793. [Google Scholar] [CrossRef]
- Petri, M.; Kim, M.Y.; Kalunian, K.C.; Grossman, J.; Hahn, B.H.; Sammaritano, L.R.; Lockshin, M.; Merrill, J.T.; Belmont, H.M.; Askanase, A.D.; et al. Combined Oral Contraceptives in Women with Systemic Lupus Erythematosus. N. Engl. J. Med. 2005, 353, 2550–2558. [Google Scholar] [CrossRef]
- Melamud, M.M.; Ermakov, E.A.; Boiko, A.S.; Parshukova, D.A.; Sizikov, A.E.; Ivanova, S.A.; Nevinsky, G.A.; Buneva, V.N. Serum Cytokine Levels of Systemic Lupus Erythematosus Patients in the Presence of Concomitant Cardiovascular Diseases. EMIDDT 2022, 22, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, E.A.; Kabirova, E.M.; Sizikov, A.E.; Buneva, V.N.; Nevinsky, G.A. IgGs-Abzymes from the Sera of Patients with Systemic Lupus Erythematosus Hydrolyzed miRNAs. JIR 2020, 13, 681–699. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.M.; Sedykh, S.E.; Ermakov, E.A.; Matveev, A.L.; Odegova, E.I.; Sedykh, T.A.; Shcherbakov, D.N.; Merkuleva, I.A.; Volosnikova, E.A.; Nesmeyanova, V.S.; et al. Natural IgG against S-Protein and RBD of SARS-CoV-2 Do Not Bind and Hydrolyze DNA and Are Not Autoimmune. Int. J. Mol. Sci. 2022, 23, 13681. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, E.A.; Ivanova, S.A.; Buneva, V.N.; Nevinsky, G.A. Hydrolysis by Catalytic IgGs of microRNA Specific for Patients with Schizophrenia: DIFFERENT MECHANISMS OF SCZ DEVELOPMENT. IUBMB Life 2018, 70, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Kostina, E.V.; Ryabinin, V.A.; Ternovoi, V.A.; Sinyakov, A.N. On-Microchip PCR for Detection of Subtypes of Influenza a Viruses Circulating in the Human Population. Russ. J. Bioorg. Chem. 2015, 41, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.E.; Chaires, J.B. Characterization of Preferred Deoxyribonuclease I Cleavage Sites. J. Mol. Biol. 1994, 236, 405–411. [Google Scholar] [CrossRef]
- Halling, P.J.; Ulijn, R.V.; Flitsch, S.L. Understanding Enzyme Action on Immobilised Substrates. Curr. Opin. Biotechnol. 2005, 16, 385–392. [Google Scholar] [CrossRef]
- Zhu, Q.; Uttamchandani, M.; Li, D.; Lesaicherre, M.L.; Yao, S.Q. Enzymatic Profiling System in a Small-Molecule Microarray. Org. Lett. 2003, 5, 1257–1260. [Google Scholar] [CrossRef]
- Fang, S.; Lee, H.J.; Wark, A.W.; Kim, H.M.; Corn, R.M. Determination of Ribonuclease H Surface Enzyme Kinetics by Surface Plasmon Resonance Imaging and Surface Plasmon Fluorescence Spectroscopy. Anal. Chem. 2005, 77, 6528–6534. [Google Scholar] [CrossRef]
- Salisbury, C.M.; Maly, D.J.; Ellman, J.A. Peptide Microarrays for the Determination of Protease Substrate Specificity. J. Am. Chem. Soc. 2002, 124, 14868–14870. [Google Scholar] [CrossRef]
- Ulijn, R.V.; Baragaña, B.; Halling, P.J.; Flitsch, S.L. Protease-Catalyzed Peptide Synthesis on Solid Support. J. Am. Chem. Soc. 2002, 124, 10988–10989. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Wark, A.W.; Goodrich, T.T.; Fang, S.; Corn, R.M. Surface Enzyme Kinetics for Biopolymer Microarrays: A Combination of Langmuir and Michaelis−Menten Concepts. Langmuir 2005, 21, 4050–4057. [Google Scholar] [CrossRef] [PubMed]
- Gaspers, P.B.; Robertson, C.R.; Gast, A.P. Enzymes on Immobilized Substrate Surfaces: Diffusion. Langmuir 1994, 10, 2699–2704. [Google Scholar] [CrossRef]
- Anne, A.; Demaille, C. Kinetics of Enzyme Action on Surface-Attached Substrates: A Practical Guide to Progress Curve Analysis in Any Kinetic Situation. Langmuir 2012, 28, 14665–14671. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, O.A.; Chavez, M.; Lissi, E. A Theoretical Approach to Some Analytical Properties of Heterogeneous Enzymatic Assays. Anal. Chem. 2004, 76, 2664–2668. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S.; Garza Reyna, A.; Belina, M.E.; Spencer, D.M. The Interaction of Anti-DNA Antibodies with DNA: Evidence for Unconventional Binding Mechanisms. Int. J. Mol. Sci. 2022, 23, 5227. [Google Scholar] [CrossRef] [PubMed]
- Andreev, S.L.; Buneva, V.N.; Nevinsky, G.A. How Human IgGs against DNA Recognize Oligonucleotides and DNA. J Mol. Recognit. 2016, 29, 596–610. [Google Scholar] [CrossRef]
- Arbuckle, M.R.; James, J.A.; Kohlhase, K.F.; Rubertone, M.V.; Dennis, G.J.; Harley, J.B. Development of Anti-dsDNA Autoantibodies Prior to Clinical Diagnosis of Systemic Lupus Erythematosus. Scand. J. Immunol. 2001, 54, 211–219. [Google Scholar] [CrossRef]
- Tseng, C.; Buyon, J.P.; Kim, M.; Belmont, H.M.; Mackay, M.; Diamond, B.; Marder, G.; Rosenthal, P.; Haines, K.; Ilie, V.; et al. The Effect of Moderate-dose Corticosteroids in Preventing Severe Flares in Patients with Serologically Active, but Clinically Stable, Systemic Lupus Erythematosus: Findings of a Prospective, Randomized, Double-blind, Placebo-controlled Trial. Arthritis Rheum. 2006, 54, 3623–3632. [Google Scholar] [CrossRef]
- Böhm, I. Decrease of B-Cells and Autoantibodies after Low-Dose Methotrexate. Biomed. Pharmacother. 2003, 57, 278–281. [Google Scholar] [CrossRef]
- Han, D.S.C.; Ni, M.; Chan, R.W.Y.; Chan, V.W.H.; Lui, K.O.; Chiu, R.W.K.; Lo, Y.M.D. The Biology of Cell-Free DNA Fragmentation and the Roles of DNASE1, DNASE1L3, and DFFB. Am. J. Hum. Genet. 2020, 106, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.; Toong, C.; Adelstein, S. Clearing the Complexity: Immune Complexes and Their Treatment in Lupus Nephritis. IJNRD 2011, 4, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Yung, S.; Chan, T.M. Anti-dsDNA Antibodies and Resident Renal Cells—Their Putative Roles in Pathogenesis of Renal Lesions in Lupus Nephritis. Clin. Immunol. 2017, 185, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Abdelal, I.T.; Zakaria, M.A.; Sharaf, D.M.; Elakad, G.M. Levels of Plasma Cell-Free DNA and Its Correlation with Disease Activity in Rheumatoid Arthritis and Systemic Lupus Erythematosus Patients. Egypt. Rheumatol. 2016, 38, 295–300. [Google Scholar] [CrossRef]
- Hendy, O.M.; Motalib, T.A.; El Shafie, M.A.; Khalaf, F.A.; Kotb, S.E.; Khalil, A.; Ali, S.R. Circulating Cell Free DNA as a Predictor of Systemic Lupus Erythematosus Severity and Monitoring of Therapy. Egypt. J. Med. Hum. Genet. 2016, 17, 79–85. [Google Scholar] [CrossRef]
- Tamkovich, S.; Laktionov, P. Cell-surface-bound Circulating DNA in the Blood: Biology and Clinical Application. IUBMB Life 2019, 71, 1201–1210. [Google Scholar] [CrossRef]
- Rykova, E.; Sizikov, A.; Roggenbuck, D.; Antonenko, O.; Bryzgalov, L.; Morozkin, E.; Skvortsova, K.; Vlassov, V.; Laktionov, P.; Kozlov, V. Circulating DNA in Rheumatoid Arthritis: Pathological Changes and Association with Clinically Used Serological Markers. Arthritis Res. Ther. 2017, 19, 85. [Google Scholar] [CrossRef]








| ODN Name | Sequence | Number of Nucleotides | Sequence Description |
|---|---|---|---|
| A10 | ROX-5′-AAAAAAAAAA 1 | 10 | consists entirely of adenosines |
| C10 | ROX-5′-СССССССССС 1 | 10 | consists entirely of cytidines |
| T10 | ROX-5′-TTTTTTTTTT 1 | 10 | consists entirely of thymidines |
| (AC)5 | ROX-5′-ACACACACAC 1 | 10 | consists of alternating purine and pyrimidine nucleobases |
| (AT)5 | ROX-5′-ATATATATAT | 10 | consists of alternating purine and pyrimidine nucleobases |
| (GT)5 | ROX-5′-GTGTGTGTGT 1 | 10 | consists of alternating purine and pyrimidine nucleobases |
| (AG)5 | ROX-5′-AGAGAGAGAG 1 | 10 | composed of alternating purine bases |
| (CT)5 | ROX-5′-CTCTCTCTCT 1 | 10 | consists of alternating pyrimidine bases |
| (CT)3 | ROX-5′-CTCTCT-linker-NH2 2 | 6 | ODNs consisting of alternating pyrimidine bases varying in length |
| (CT)5 | ROX-5′-CTCTCTCTCT-linker-NH2 2 | 10 | |
| (GT)3 | ROX-5′-GTGTGT-linker-NH2 2 | 6 | ODNs consisting of alternating purine and pyrimidine nucleobases varying in length |
| (GT)5 | ROX-5′-GTGTGTGTGT-linker-NH2 2 | 10 | |
| C14T10 | ROX-5′-CCCCCCCCCCCCCCTTTTTTTTTT-linker-NH2 2 | 24 | Long ODN consisting of 24 nucleotides |
| Group | No. of IgG Samples | Sex | Age, Years | Disease Duration, Years | Course of SLE | SELENA-SLEDAI Score |
|---|---|---|---|---|---|---|
| SLE | SLE22 | F | 34 | 4 | subacute | 10 |
| SLE23 | F | 25 | 5 | subacute | 8 | |
| SLE24 | F | 47 | 5 | subacute | 9 | |
| SLE33 | F | 58 | 2 | chronic | 9 | |
| SLE44 | F | 71 | 1 | subacute | 10 | |
| Mean ± SD | - | - | 47.0 ± 18.4 | 3.4 ± 1.8 | - | 9.2 ± 0.8 |
| Healthy subjects | HS74 | F | 61 | - | - | - |
| HS81 | M | 25 | - | - | - | |
| HS45 | M | 43 | - | - | - | |
| HS20 | F | 42 | - | - | - | |
| HS40 | M | 27 | - | - | - | |
| Mean ± SD | - | - | 39.6 ± 14.6 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novikova, T.S.; Ermakov, E.A.; Kostina, E.V.; Sinyakov, A.N.; Sizikov, A.E.; Nevinsky, G.A.; Buneva, V.N. Hydrolysis of Oligodeoxyribonucleotides on the Microarray Surface and in Solution by Catalytic Anti-DNA Antibodies in Systemic Lupus Erythematosus. Curr. Issues Mol. Biol. 2023, 45, 9887-9903. https://doi.org/10.3390/cimb45120617
Novikova TS, Ermakov EA, Kostina EV, Sinyakov AN, Sizikov AE, Nevinsky GA, Buneva VN. Hydrolysis of Oligodeoxyribonucleotides on the Microarray Surface and in Solution by Catalytic Anti-DNA Antibodies in Systemic Lupus Erythematosus. Current Issues in Molecular Biology. 2023; 45(12):9887-9903. https://doi.org/10.3390/cimb45120617
Chicago/Turabian StyleNovikova, Tatiana S., Evgeny A. Ermakov, Elena V. Kostina, Alexander N. Sinyakov, Alexey E. Sizikov, Georgy A. Nevinsky, and Valentina N. Buneva. 2023. "Hydrolysis of Oligodeoxyribonucleotides on the Microarray Surface and in Solution by Catalytic Anti-DNA Antibodies in Systemic Lupus Erythematosus" Current Issues in Molecular Biology 45, no. 12: 9887-9903. https://doi.org/10.3390/cimb45120617
APA StyleNovikova, T. S., Ermakov, E. A., Kostina, E. V., Sinyakov, A. N., Sizikov, A. E., Nevinsky, G. A., & Buneva, V. N. (2023). Hydrolysis of Oligodeoxyribonucleotides on the Microarray Surface and in Solution by Catalytic Anti-DNA Antibodies in Systemic Lupus Erythematosus. Current Issues in Molecular Biology, 45(12), 9887-9903. https://doi.org/10.3390/cimb45120617