Enhanced UV-B Radiation in Potato Stems and Leaves Promotes the Accumulation of Anthocyanins in Tubers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Grafting Experiment
2.3. UV-B Treatment
2.4. Determination of Total Anthocyanin Content (TAC) and Monomeric Anthocyanidin Content (MAC)
2.5. Transcriptome Sequencing and Bioinformatics Analysis
2.6. Verification of the qRT-PCR (Quantitative Real-Time Polymerase Chain Reaction)
2.7. Statistical Analysis
3. Results
3.1. Anthocyanins in Colored Potato Tubers Are Synthesized In Situ
3.2. Enhanced UV-B Radiation in Colored Potato Stems and Leaves Increases the Anthocyanin Content in Tubers
3.3. Effects of Enhanced UV-B Exposure of Potato Stems and Leaves on the Gene Expression in Tubers
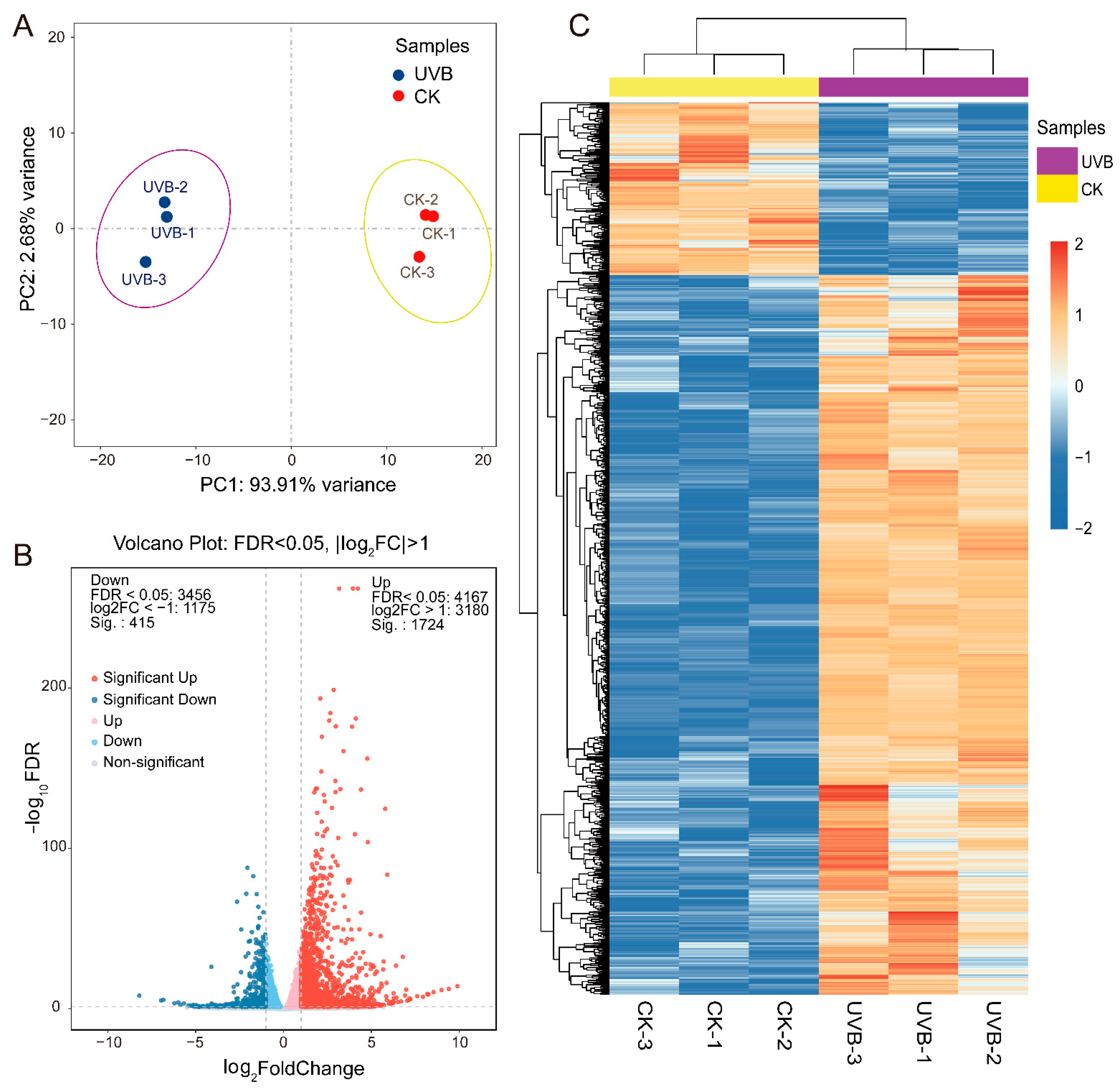
3.3.1. Transcriptome Overview
3.3.2. Analysis of Anthocyanin Biosynthetic Genes in UVB and CK Tubers
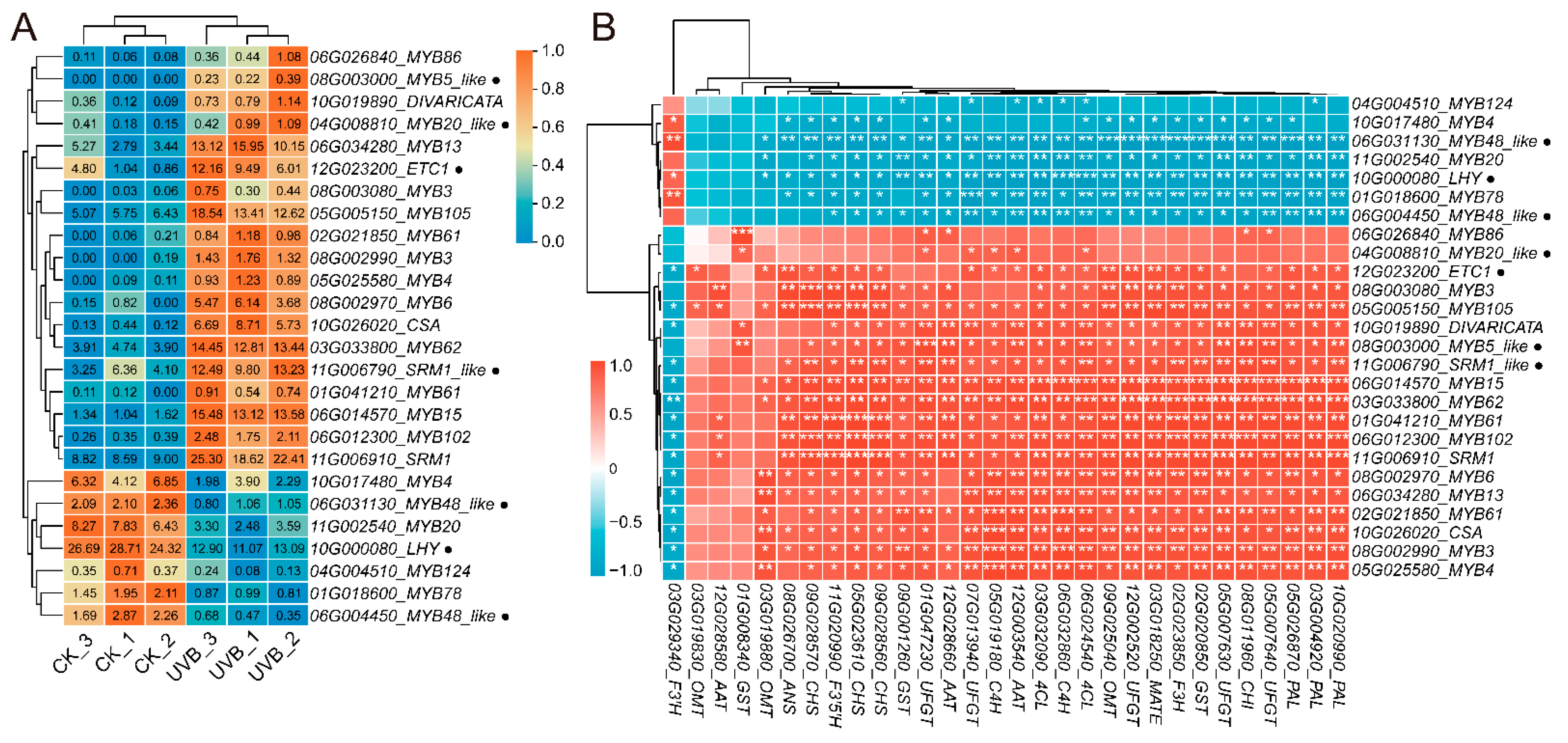
3.3.3. Identification of Transcription Factors in UVB and CK Tubers
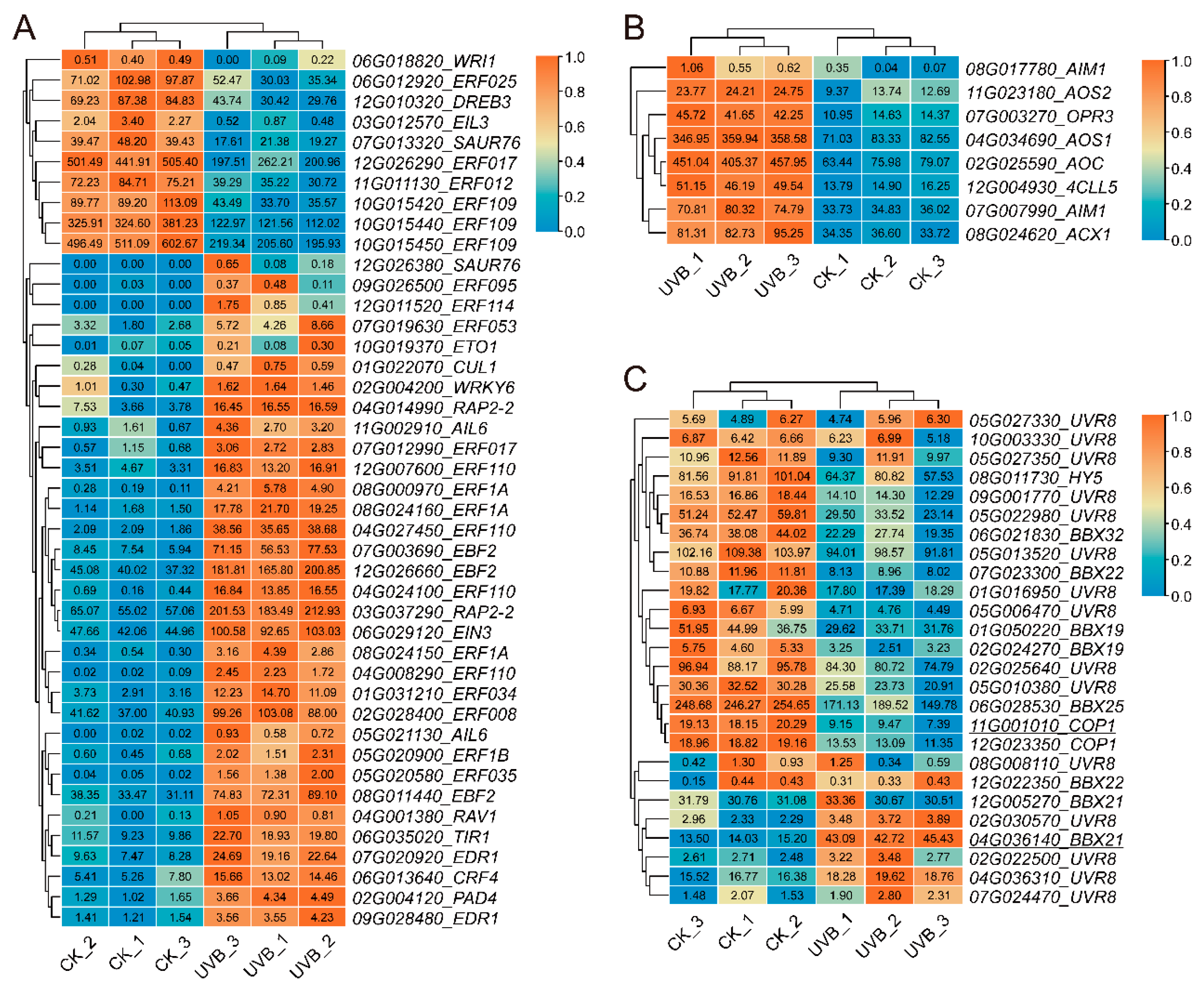
3.3.4. Identification of Hormone-Related Genes and UV-B-Related Genes in UVB and CK Tubers
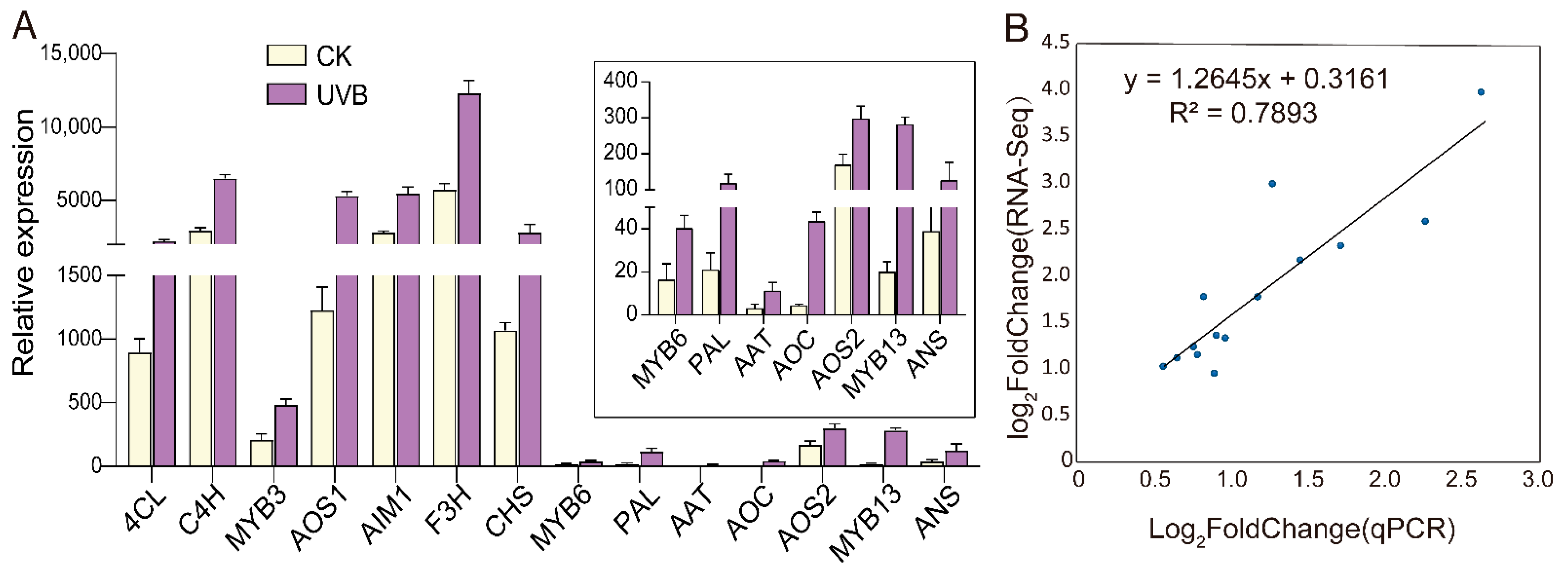
3.3.5. qRT-PCR Verification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, C.R.; Wrolstad, R.; Durst, R.; Yang, C.P.; Clevidence, B. Breeding studies in potatoes containing high concentrations of anthocyanins. Am. J. Potato Res. 2003, 80, 241–249. [Google Scholar] [CrossRef]
- Giusti, M.M.; Polit, M.F.; Ayvaz, H.; Tay, D.; Manrique, I. Characterization and Quantitation of Anthocyanins and Other Phenolics in Native Andean Potatoes. J. Agric. Food Chem. 2014, 62, 4408–4416. [Google Scholar] [CrossRef]
- Brown, C.R. Antioxidants in potato. Am. J. Potato Res. 2005, 82, 163–172. [Google Scholar] [CrossRef]
- Han, K.; Sekikawa, M.; Shimada, K.; Hashimoto, M.; Hashimoto, N.; Noda, T.; Tanaka, H.; Fukushima, M. Anthocyanin-rich purple potato flake extract has antioxidant capacity and improves antioxidant potential in rats. Br. J. Nutr. 2007, 96, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Camire, M.E.; Kubow, S.; Donnelly, D.J. Potatoes and Human Health. Crit. Rev. Food Sci. Nutr. 2009, 49, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, R.; Jayathilake, C.; Chaminda Jayawardana, B.; Liyanage, R. Health-beneficial properties of potato and compounds of interest. J. Sci. Food Agric. 2016, 96, 4850–4860. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Strygina, K.V.; Kochetov, A.V.; Khlestkina, E.K. Genetic control of anthocyanin pigmentation of potato tissues. BMC Genet. 2019, 20 (Suppl. S1), 35–43. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Liu, J.; Guo, D.; Hou, J.; Chen, S.; Song, B.; Xie, C. Analysis of structural genes and key transcription factors related to anthocyanin biosynthesis in potato tubers. Sci. Hortic. 2017, 225, 310–316. [Google Scholar] [CrossRef]
- Kaur, S.; Sharma, N.; Kapoor, P.; Chunduri, V.; Pandey, A.K.; Garg, M. Spotlight on the overlapping routes and partners for anthocyanin transport in plants. Physiol. Plant. 2021, 171, 868–881. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Liu, Z.; Wang, L.; Bi, Z.; Sun, C.; Yao, P.; Zhang, J.; Bai, J.; Zeng, Y. Integrated transcriptomic and metabolomic analysis revealed altitude-related regulatory mechanisms on flavonoid accumulation in potato tubers. Food Res. Int. 2023, 170, 112997. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.L.F.; Serra, P.; Casati, P. Recent advances on the roles of flavonoids as plant protective molecules after UV and high light exposure. Physiol. Plant. 2021, 173, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Huang, J.; Guo, H.; Yang, C.; Li, Y.; Shen, S.; Zhan, C.; Qu, L.; Liu, X.; Wang, S.; et al. OsRLCK160 contributes to flavonoid accumulation and UV-B tolerance by regulating OsbZIP48 in rice. Sci. China Life Sci. 2022, 65, 1380–1394. [Google Scholar] [CrossRef] [PubMed]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Changing scenario in plant UV-B research:UV-B from a generic stressor to a specific regulator. J. Photochem. Photobiol. B Biol. 2015, 153, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Wang, F.; Li, X.; Chu, J.; Huang, J.; et al. Brassinosteroid-Activated BRI1-EMS-SUPPRESSOR 1 Inhibits Flavonoid Biosynthesis and Coordinates Growth and UV-B Stress Responses in Plants. Plant Cell 2020, 32, 3224–3239. [Google Scholar] [CrossRef]
- Henry-Kirk, R.A.; Plunkett, B.; Hall, M.; McGhie, T.; Allan, A.C.; Wargent, J.J.; Espley, R.V. Solar UV light regulates flavonoid metabolism in apple (Malus x domestica). Plant Cell Environ. 2018, 41, 675–688. [Google Scholar] [CrossRef]
- Nguyen, C.T.T.; Lim, S.; Lee, J.G.; Lee, E.J. VcBBX, VcMYB21, and VcR2R3MYB Transcription Factors Are Involved in UV–B-Induced Anthocyanin Biosynthesis in the Peel of Harvested Blueberry Fruit. J. Agric. Food Chem. 2017, 65, 2066–2073. [Google Scholar] [CrossRef]
- Martínez-Lüscher, J.; Sánchez-Díaz, M.; Delrot, S.; Aguirreolea, J.; Pascual, I.; Gomès, E. Ultraviolet-B radiation and water deficit interact to alter flavonol and anthocyanin profiles in grapevine berries through transcriptomic regulation. Plant Cell Physiol. 2014, 55, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.; Lütken, H.; Barba-Espin, G.; Hernández, J.A.; Acosta-Motos, J.R.; Großkinsky, D.K. UV-B exposure of black carrot (Daucus carota ssp. sativus var. atrorubens) plants promotes growth, accumulation of anthocyanin, and phenolic compounds. Agronomy 2019, 9, 323. [Google Scholar] [CrossRef]
- Wu, X.; Chen, B.; Xiao, J.; Guo, H. Different doses of UV-B radiation affect pigmented potatoes’ growth and quality during the whole growth period. Front. Plant Sci. 2023, 14, 1101172. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, Q.; Xu, X.; Li, J.; Li, K.; Li, M.; Guo, H. Physiological and biochemical responses of potato to enhanced UV-B radiation. J. Shanxi Agric. Univ. (Nat. Sci. Ed.) 2022, 42, 93–101. (In Chinese) [Google Scholar] [CrossRef]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and Functional Analysis of a MYB Transcription Factor Gene that is a Key Regulator for the Development of Red Coloration in Apple Skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Gao-Takai, M.; Katayama-Ikegami, A.; Matsuda, K.; Shindo, H.; Uemae, S.; Oyaizu, M. A low temperature promotes anthocyanin biosynthesis but does not accelerate endogenous abscisic acid accumulation in red-skinned grapes. Plant Sci. 2019, 283, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Guo, H. Effects of tomato and potato heterografting on photosynthesis, quality and yield of grafted parents. Hortic. Environ. Biotechnol. 2018, 60, 9–18. [Google Scholar] [CrossRef]
- Zhang, Z.; Kou, X.; Fugal, K.; McLaughlin, J. Comparison of HPLC Methods for Determination of Anthocyanins and Anthocyanidins in Bilberry Extracts. J. Agric. Food Chem. 2004, 52, 688–691. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using teal-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nie, T.; Wang, D.; Ma, X.; Chen, Y.; Chen, Q. Analysis of Key Genes Involved in Potato Anthocyanin Biosynthesis Based on Genomics and Transcriptomics Data. Front. Plant Sci. 2019, 10, 603. [Google Scholar] [CrossRef]
- Bao, Y.; Nie, T.; Wang, D.; Chen, Q. Anthocyanin regulatory networks in Solanum tuberosum L. leaves elucidated via integrated metabolomics, transcriptomics, and StAN1 overexpression. BMC Plant Biol. 2022, 22, 228. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.E.; Walker, J.R.L.; Lancaster, J.E.; Conner, A.J. Light regulation of anthocyanin, flavonoid and phenolic acid biosynthesis in potato minitubers in vitro. Aust. J. Plant Physiol. 1998, 25, 915–922. [Google Scholar] [CrossRef]
- Araguirang, G.E.; Richter, A.S. Activation of anthocyanin biosynthesis in high light—What is the initial signal? New Phytol. 2022, 236, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Apoorva; Jaiswal, D.; Pandey-Rai, S.; Agrawal, S.B. Untangling the UV-B radiation-induced transcriptional network regulating plant morphogenesis and secondary metabolite production. Environ. Exp. Bot. 2021, 192, 104655. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Wang, H.; Zhang, Y.; Li, W.; Liu, J.; Cheng, Q.; Sun, L.; Shen, H. Identification of the regulatory genes of UV-B-induced anthocyanin biosynthesis in pepper fruit. Int. J. Mol. Sci. 2022, 23, 1960. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Ma, J.; Zhang, J.; Gui, M.; Li, J.; Zhang, L. Effects of low doses of UV-B radiation supplementation on tuber quality in purple potato (Solanum tuberosum L.). Plant Signal. Behav. 2020, 15, 1783490. [Google Scholar] [CrossRef] [PubMed]
- Lachman, J.; Hamouz, K.; Šulc, M.; Orsák, M.; Pivec, V.; Hejtmánková, A.; Dvořák, P.; Čepl, J. Cultivar differences of total anthocyanins and anthocyanidins in red and purple-fleshed potatoes and their relation to antioxidant activity. Food Chem. 2009, 114, 836–843. [Google Scholar] [CrossRef]
- Oertel, A.; Matros, A.; Hartmann, A.; Arapitsas, P.; Dehmer, K.; Martens, S.; Mock, H. Metabolite profiling of red and blue potatoes revealed cultivar and tissue specific patterns for anthocyanins and other polyphenols. Planta 2017, 246, 281–297. [Google Scholar] [CrossRef]
- Song, Y.; Ma, B.; Guo, Q.; Zhou, L.; Lv, C.; Liu, X.; Wang, J.; Zhou, X.; Zhang, C. UV-B induces the expression of flavonoid biosynthetic pathways in blueberry (Vaccinium corymbosum) calli. Front. Plant Sci. 2022, 13, 1079087. [Google Scholar] [CrossRef]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Castellano Moreno, M.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Zhang, J.; Yu, B.; Wang, J.; Wang, D. The MYB transcription factor StMYBA1 from potato requires light to activate anthocyanin biosynthesis in transgenic tobacco. J. Plant Biol. 2017, 60, 93–101. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, H.; Liu, T.; Zhao, Y.; Hu, X.; Liu, S.; Lin, Y.; Song, B.; He, C. Transcriptome analysis provides StMYBA1 gene that regulates potato anthocyanin biosynthesis by activating structural genes. Front. Plant Sci. 2023, 14, 1087121. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, M.; Brendolise, C.; Dare, A.P.; Lin-Wang, K.; Davies, K.M.; Hellens, R.P.; Allan, A.C. In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 2015, 66, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, B.; Wang, M.; Chen, M.; Yin, J.M.; Kaleri, G.M.; Zhang, R.J.; Zuo, T.N.; You, X.; Yang, Q. Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation. J. Integr. Plant Biol. 2014, 56, 364–372. [Google Scholar] [CrossRef]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Nie, S.; Xu, C.; Liu, H.; Jia, K.; Zhou, S.; Zhao, W.; Zhou, X.; El-Kassaby, Y.; Wang, X.; et al. UV-B-induced molecular mechanisms of stress physiology responses in the major northern Chinese conifer Pinus tabuliformis Carr. Tree Physiol. 2021, 41, 1247–1263. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Vanhaelewyn, L.; Prinsen, E.; Van Der Straeten, D.; Vandenbussche, F. Hormone-controlled UV-B responses in plants. J. Exp. Bot. 2016, 67, 4469–4482. [Google Scholar] [CrossRef]
- Conconi, A.; Smerdon, M.J.; Howe, G.A.; Ryan, C.A. The octadecanoid signalling pathway in plants mediates a response to ultraviolet radiation. Nature 1996, 383, 826–829. [Google Scholar] [CrossRef]
- Demkura, P.V.; Abdala, G.; Baldwin, I.T.; Ballaré, C.L. Jasmonate-dependent and -independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant Physiol. 2010, 152, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Love, A.; Laval, V.; Geri, C.; Laird, J.; Tomos, A.; Hooks, M.; Milner, J. Components of Arabidopsis defense- and ethylene-signaling pathways regulate susceptibility to Cauliflower mosaic virus by restricting long-distance movement. Mol. Plant-Microbe Interact. 2007, 20, 659–670. [Google Scholar] [CrossRef]
- Tamogami, S.; Noge, K.; Abe, M.; Agrawal, G.K.; Rakwal, R. Methyl jasmonate is transported to distal leaves via vascular process metabolizing itself into JA-Ile and triggering VOCs emission as defensive metabolites. Plant Signal. Behav. 2012, 7, 1378–1381. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, W.T.; Kim, Y.B.; Seo, J.M.; Kim, S.-J.; Chung, E.; Lee, J.-H.; Park, S.U. Accumulation of Anthocyanin and Associated Gene Expression in Radish Sprouts Exposed to Light and Methyl Jasmonate. J. Agric. Food Chem. 2013, 61, 4127–4132. [Google Scholar] [CrossRef] [PubMed]
- Gaillochet, C.; Burko, Y.; Platre, M.P.; Zhang, L.; Simura, J.; Willige, B.C.; Kumar, S.V.; Ljung, K.; Chory, J.; Busch, W. HY5 and phytochrome activity modulate shoot-to-root coordination during thermomorphogenesis in Arabidopsis. Development 2020, 147, dev192625. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Burko, Y.; Gaillochet, C.; Seluzicki, A.; Chory, J.; Busch, W. Local HY5 Activity Mediates Hypocotyl Growth and Shoot-to-Root Communication. Plant Commun. 2020, 1, 100078. [Google Scholar] [CrossRef]
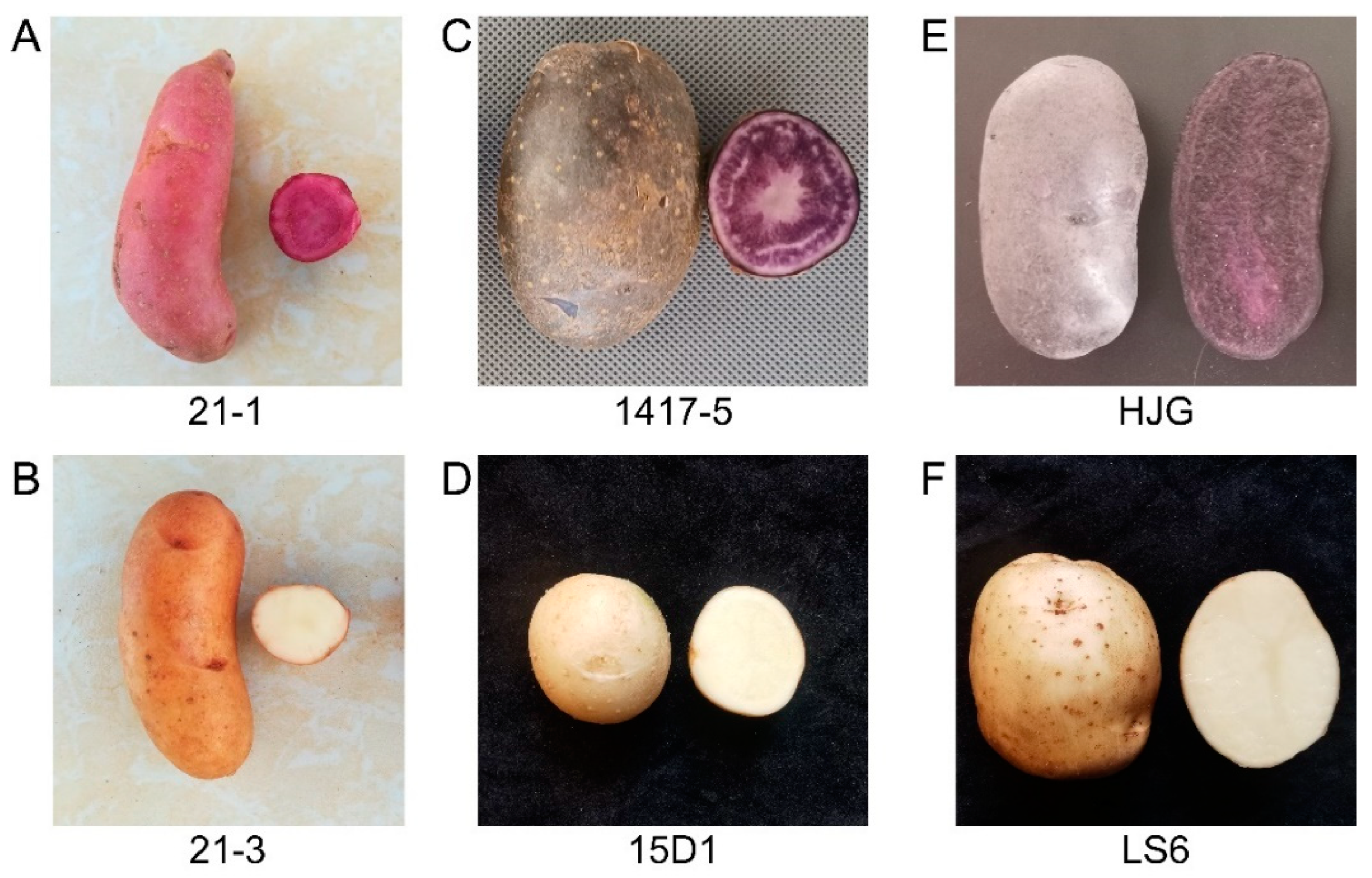



| Potato Clones | Tuber Color (Skin, Flesh) | TAC (mg/100 g FW) | Grafting Plants (Rootstock/Scion) | Tuber Color (Skin, Flesh) | TAC (mg/100 g FW) |
|---|---|---|---|---|---|
| 21-1 | Red, Red | 26.77 ± 2.34 | 21-3/21-1 | Red, Red | 35.53 ± 2.03 |
| 21-3 | Pale Yellow, White | nd | 21-1/21-3 | Pale Yellow, White | nd |
| 1417-5 | Purple, Purple | 9.71 ± 0.72 | 15D1/1417-5 | Purple, Purple | 18.68 ± 1.83 |
| 15D1 | White, White | nd | 1417-5/15D1 | White, White | nd |
| HJG | Purple, Purple | 39.79 ± 0.90 | LS6/HJG | Purple, Purple | 68.92 ± 4.04 |
| LS6 | White, White | nd | HJG/LS6 | White, White | nd |
| Gene ID (Soltu.DM.) | Gene Name | Gene Function Annotation | Fold Change (UVB/CK) | Regulation |
|---|---|---|---|---|
| 03G004920 | PAL | Phenylalanine ammonia-lyase | 2.118 | Up |
| 05G026870 | PAL | 5.041 | Up | |
| 10G020990 | PAL | 2.957 | Up | |
| 05G019180 | C4H | Trans-cinnamate 4-monooxygenase | 12.765 | Up |
| 06G032860 | C4H | 2.229 | Up | |
| 03G032090 | 4CL | 4-coumarate-CoA ligase | 2.046 | Up |
| 06G024540 | 4CL | 2.576 | Up | |
| 05G023610 | CHS | Chalcone synthase | 2.526 | Up |
| 09G028560 | CHS | 4.110 | Up | |
| 09G028570 | CHS | 3.845 | Up | |
| 08G011960 | CHI | Chalcone isomerase | 3.561 | Up |
| 02G023850 | F3H | Flavanone 3-hydroxylase | 2.366 | Up |
| 03G029340 | F3’H | Flavonoid 3’-hydroxylase | 0.333 | Down |
| 11G020990 | F3’5’H | Flavonoid 3’,5’-hydroxylase | 3.169 | Up |
| 08G026700 | ANS | Anthocyanidin synthase | 3.437 | Up |
| 01G047230 | UFGT | UDP-glucose:flavonoid 3-O-glucosyltransferase | 7.760 | Up |
| 05G007630 | UFGT | 6.472 | Up | |
| 05G007640 | UFGT | 6.909 | Up | |
| 07G013940 | UFGT | 2.384 | Up | |
| 12G002520 | UFGT | 3.848 | Up | |
| 03G019830 | OMT | O-methyltransferase | 4.838 | Up |
| 03G019880 | OMT | 9.946 | Up | |
| 09G025040 | OMT | 2.589 | Up | |
| 12G003540 | AAT | Anthocyanin acyltransferase | 3.158 | Up |
| 12G028580 | AAT | 14.592 | Up | |
| 12G028660 | AAT | 8.012 | Up | |
| 01G008340 | GST | Glutathione S-transferase | 14.654 | Up |
| 02G020850 | GST | 2.594 | Up | |
| 09G001260 | GST | 2.820 | Up | |
| 03G018250 | MATE | Multidrug and toxic compound extrusion | 2.443 | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Li, M.; Zhang, X.; Guo, Z.; Li, K.; Shi, Y.; Wang, Q.; Guo, H. Enhanced UV-B Radiation in Potato Stems and Leaves Promotes the Accumulation of Anthocyanins in Tubers. Curr. Issues Mol. Biol. 2023, 45, 9943-9960. https://doi.org/10.3390/cimb45120621
Cui L, Li M, Zhang X, Guo Z, Li K, Shi Y, Wang Q, Guo H. Enhanced UV-B Radiation in Potato Stems and Leaves Promotes the Accumulation of Anthocyanins in Tubers. Current Issues in Molecular Biology. 2023; 45(12):9943-9960. https://doi.org/10.3390/cimb45120621
Chicago/Turabian StyleCui, Lingyan, Maoxing Li, Xing Zhang, Zongming Guo, Kaifeng Li, Yuhan Shi, Qiong Wang, and Huachun Guo. 2023. "Enhanced UV-B Radiation in Potato Stems and Leaves Promotes the Accumulation of Anthocyanins in Tubers" Current Issues in Molecular Biology 45, no. 12: 9943-9960. https://doi.org/10.3390/cimb45120621
APA StyleCui, L., Li, M., Zhang, X., Guo, Z., Li, K., Shi, Y., Wang, Q., & Guo, H. (2023). Enhanced UV-B Radiation in Potato Stems and Leaves Promotes the Accumulation of Anthocyanins in Tubers. Current Issues in Molecular Biology, 45(12), 9943-9960. https://doi.org/10.3390/cimb45120621





