NRG1 Regulates Proliferation, Migration and Differentiation of Human Limbal Epithelial Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Normal Human Limbus Samples
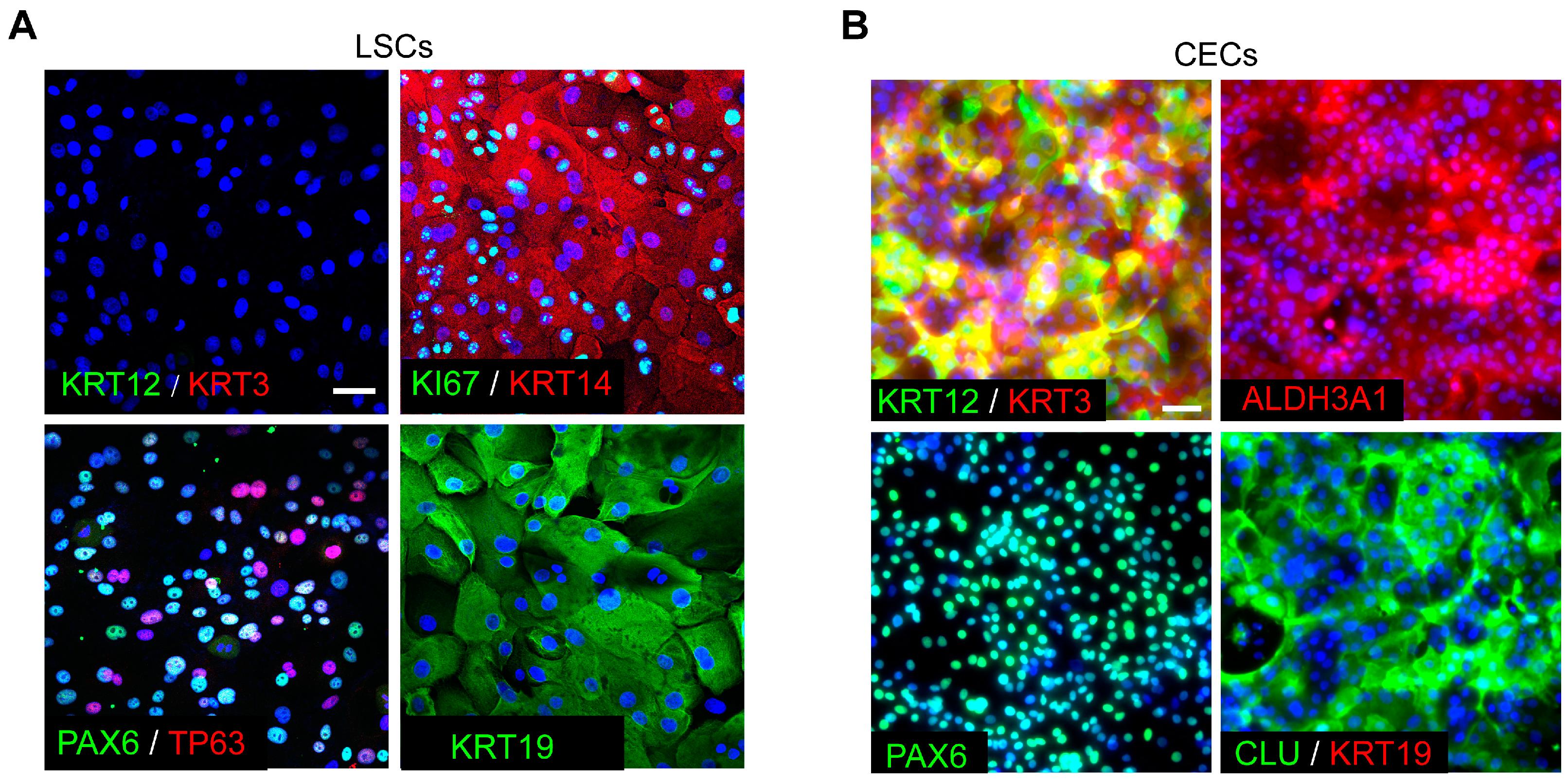
2.2. Isolation and Culture of Human LESCs
2.3. Immunofluorescence Staining
2.4. In Vitro CEC Differentiation
2.5. Cell Proliferation Assay
2.6. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
2.7. Gene Knockdown
2.8. Scratch Wound Closure Assay
2.9. RNA-Seq and Data Analysis
2.10. Statistical Analysis
3. Results
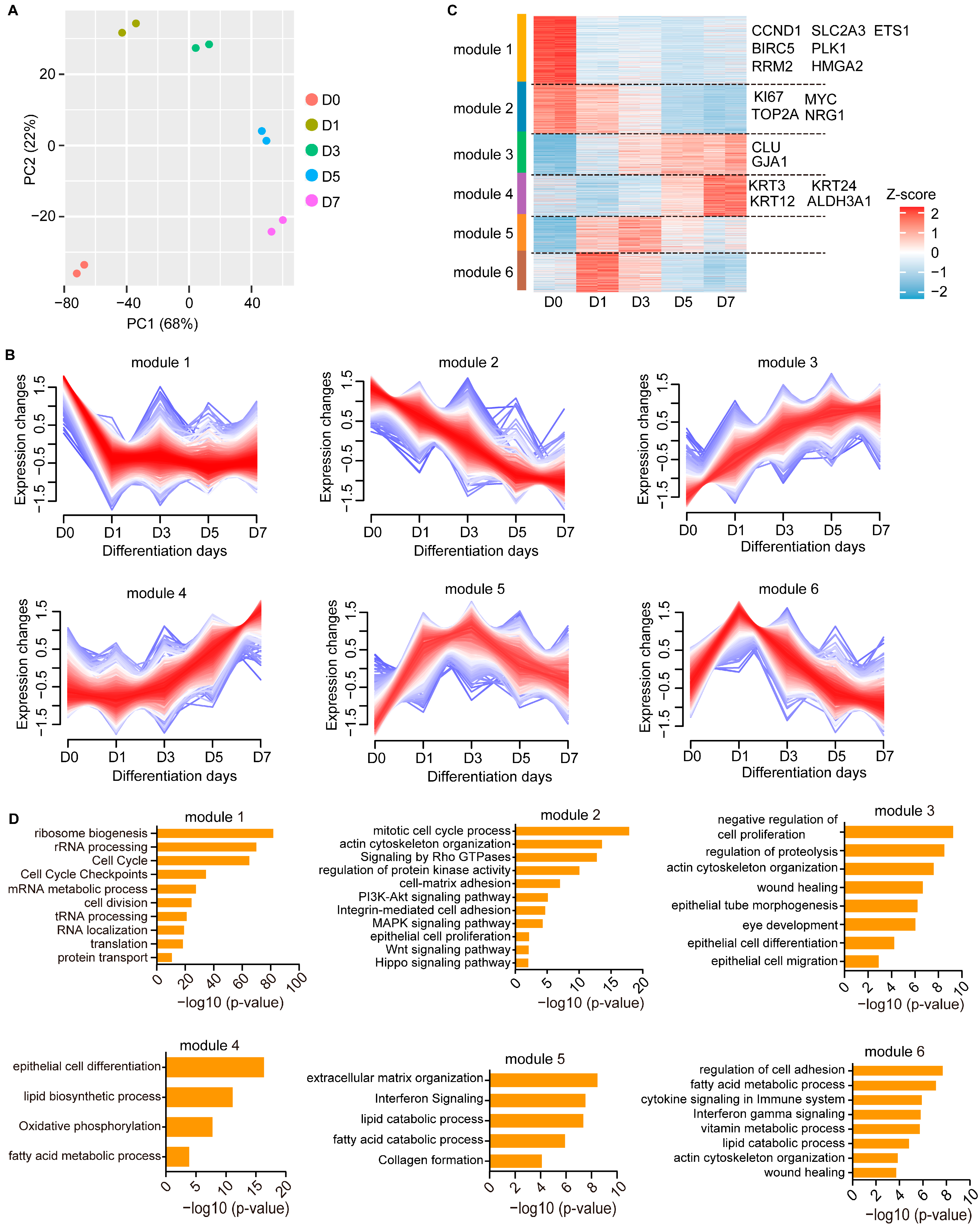
3.1. Gene Expression Profiles during CEC Differentiation
3.2. Temporal Dynamics of Transcriptional Regulation during CEC Differentiation
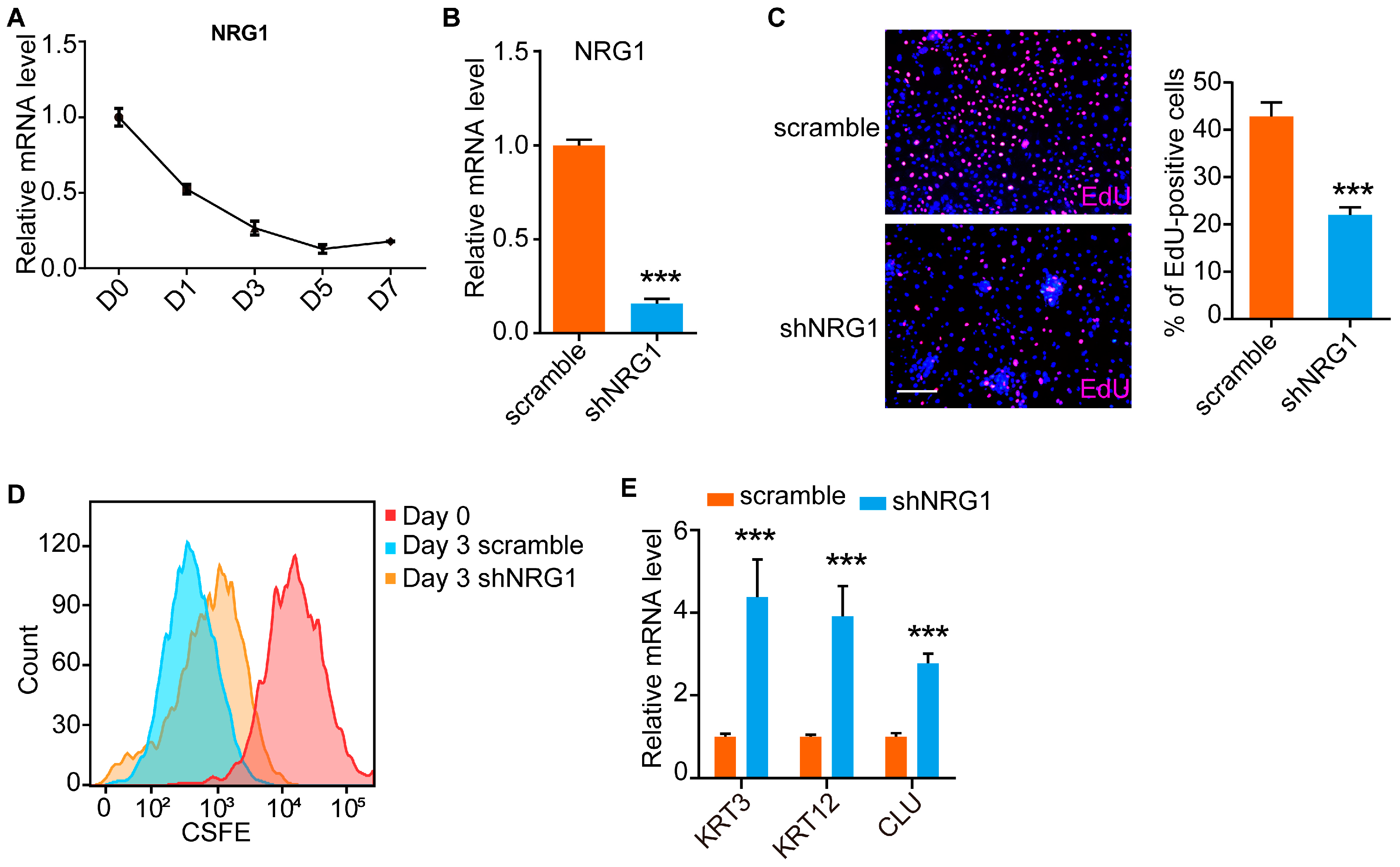
3.3. NRG1 Influences LESC Proliferation and Differentiation
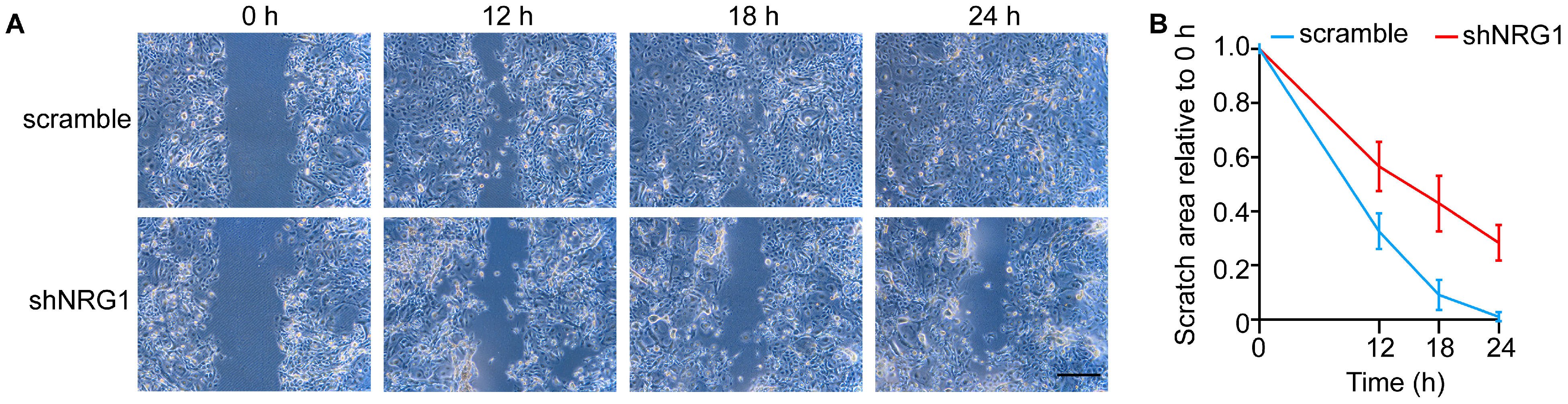
3.4. NRG1 Promotes LESC Migration
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gonzalez, G.; Sasamoto, Y.; Ksander, B.R.; Frank, M.H.; Frank, N.Y. Limbal stem cells: Identity, developmental origin, and therapeutic potential. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e303. [Google Scholar] [CrossRef] [PubMed]
- Nowell, C.S.; Radtke, F. Corneal epithelial stem cells and their niche at a glance. J. Cell Sci. 2017, 130, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Kolli, S.; Lako, M.; Figueiredo, F.; Daniels, J.T. Stem cell therapies for ocular surface disease. Drug Discov. Today 2010, 15, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, G.; De Luca, M. Eyes on the prize: Limbal stem cells and corneal restoration. Cell Stem Cell 2014, 15, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Masood, F.; Chang, J.H.; Akbar, A.; Song, A.; Hu, W.Y.; Azar, D.T.; Rosenblatt, M.I. Therapeutic Strategies for Restoring Perturbed Corneal Epithelial Homeostasis in Limbal Stem Cell Deficiency: Current Trends and Future Directions. Cells 2022, 11, 3247. [Google Scholar] [CrossRef]
- Lee, V.; Rompolas, P. Corneal regeneration: Insights in epithelial stem cell heterogeneity and dynamics. Curr. Opin. Genet. Dev. 2022, 77, 101981. [Google Scholar] [CrossRef] [PubMed]
- Jarde, T.; Chan, W.H.; Rossello, F.J.; Kaur Kahlon, T.; Theocharous, M.; Kurian Arackal, T.; Flores, T.; Giraud, M.; Richards, E.; Chan, E.; et al. Mesenchymal Niche-Derived Neuregulin-1 Drives Intestinal Stem Cell Proliferation and Regeneration of Damaged Epithelium. Cell Stem Cell 2020, 27, 646–662.e647. [Google Scholar] [CrossRef]
- Falls, D.L. Neuregulins: Functions, forms, and signaling strategies. Exp. Cell Res. 2003, 284, 14–30. [Google Scholar] [CrossRef]
- Kataria, H.; Alizadeh, A.; Karimi-Abdolrezaee, S. Neuregulin-1/ErbB network: An emerging modulator of nervous system injury and repair. Prog. Neurobiol. 2019, 180, 101643. [Google Scholar] [CrossRef]
- Van Ho, A.T.; Hayashi, S.; Brohl, D.; Aurade, F.; Rattenbach, R.; Relaix, F. Neural crest cell lineage restricts skeletal muscle progenitor cell differentiation through Neuregulin1-ErbB3 signaling. Dev. Cell 2011, 21, 273–287. [Google Scholar] [CrossRef]
- Trombetta, D.; Rossi, A.; Fabrizio, F.P.; Sparaneo, A.; Graziano, P.; Fazio, V.M.; Muscarella, L.A. NRG1-ErbB Lost in Translation: A New Paradigm for Lung Cancer? Curr. Med. Chem. 2017, 24, 4213–4228. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Chen, J.; Khan, R.A.; Song, Z.; Wang, M.; Li, Z.; Shen, J.; Li, W.; Shi, Y. Genetic association between NRG1 and schizophrenia, major depressive disorder, bipolar disorder in Han Chinese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2016, 171B, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.J.; Lin, B.; Holguin, B. Expression of neuregulin 1, a member of the epidermal growth factor family, is expressed as multiple splice variants in the adult human cornea. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3021–3029. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Wang, B.; Guo, H.; Liu, D.; Wu, S.; Liu, J.; Lan, X.; Huang, H.; An, F.; Zhu, J.; Ji, J.; et al. ETS1-HMGA2 Axis Promotes Human Limbal Epithelial Stem Cell Proliferation. Investig. Ophthalmol. Vis. Sci. 2023, 64, 12. [Google Scholar] [CrossRef]
- Li, J.M.; Kim, S.; Zhang, Y.; Bian, F.; Hu, J.; Lu, R.; Pflugfelder, S.C.; Chen, R.; Li, D.Q. Single-Cell Transcriptomics Identifies a Unique Entity and Signature Markers of Transit-Amplifying Cells in Human Corneal Limbus. Investig. Ophthalmol. Vis. Sci. 2021, 62, 36. [Google Scholar] [CrossRef]
- Portal, C.; Wang, Z.; Scott, D.K.; Wolosin, J.M.; Iomini, C. The c-Myc Oncogene Maintains Corneal Epithelial Architecture at Homeostasis, Modulates p63 Expression, and Enhances Proliferation During Tissue Repair. Investig. Ophthalmol. Vis. Sci. 2022, 63, 3. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, L.Y.; Li, C.Y.; Wu, J.Y.; Zhang, Y.T.; Pang, K.P.; Wei, Y.; Du, L.Q.; Liu, M.; Wu, X.Y. SPARC promotes self-renewal of limbal epithelial stem cells and ocular surface restoration through JNK and p38-MAPK signaling pathways. Stem Cells 2020, 38, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, M.N.; Ding, Z.; Ng, M.Y.; Truong, T.T.; Yu, F.; Deng, S.X. Wnt/beta-catenin signaling regulates proliferation of human cornea epithelial stem/progenitor cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4734–4741. [Google Scholar] [CrossRef]
- Chen, K.; Li, Y.; Zhang, X.; Ullah, R.; Tong, J.; Shen, Y. The role of the PI3K/AKT signalling pathway in the corneal epithelium: Recent updates. Cell Death Dis. 2022, 13, 513. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Fu, W.; Liu, Y.; Wang, Q.; Wang, L.; Huang, Y. Agrin Promotes Limbal Stem Cell Proliferation and Corneal Wound Healing Through Hippo-Yap Signaling Pathway. Investig. Ophthalmol. Vis. Sci. 2020, 61, 7. [Google Scholar] [CrossRef] [PubMed]
- Osei-Bempong, C.; Figueiredo, F.C.; Lako, M. The limbal epithelium of the eye--a review of limbal stem cell biology, disease and treatment. Bioessays 2013, 35, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Serror, L.; Nir, E.; Dhiraj, D.; Altshuler, A.; Khreish, M.; Tiosano, B.; Hasson, P.; Panman, L.; Luxenburg, C.; et al. SOX2 Regulates P63 and Stem/Progenitor Cell State in the Corneal Epithelium. Stem Cells 2019, 37, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, V.; Testa, A.; Di Iorio, E.; Mavilio, F.; Pellegrini, G.; De Luca, M. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J. Cell Biol. 2007, 177, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Gautam, P.; Hamashima, K.; Chen, Y.; Zeng, Y.; Makovoz, B.; Parikh, B.H.; Lee, H.Y.; Lau, K.A.; Su, X.; Wong, R.C.B.; et al. Multi-species single-cell transcriptomic analysis of ocular compartment regulons. Nat. Commun. 2021, 12, 5675. [Google Scholar] [CrossRef]
- Klein, R.H.; Hu, W.; Kashgari, G.; Lin, Z.; Nguyen, T.; Doan, M.; Andersen, B. Characterization of enhancers and the role of the transcription factor KLF7 in regulating corneal epithelial differentiation. J. Biol. Chem. 2017, 292, 18937–18950. [Google Scholar] [CrossRef]
- Li, G.; Xu, F.; Zhu, J.; Krawczyk, M.; Zhang, Y.; Yuan, J.; Patel, S.; Wang, Y.; Lin, Y.; Zhang, M.; et al. Transcription Factor PAX6 (Paired Box 6) Controls Limbal Stem Cell Lineage in Development and Disease. J. Biol. Chem. 2015, 290, 20448–20454. [Google Scholar] [CrossRef]
- Zhao, S.; Wan, X.; Dai, Y.; Gong, L.; Le, Q. WNT16B enhances the proliferation and self-renewal of limbal epithelial cells via CXCR4/MEK/ERK signaling. Stem Cell Rep. 2022, 17, 864–878. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.; Uhm, H.; Deng, S.X. Notch Inhibition Prevents Differentiation of Human Limbal Stem/Progenitor Cells in vitro. Sci. Rep. 2019, 9, 10373. [Google Scholar] [CrossRef] [PubMed]
- Vauclair, S.; Majo, F.; Durham, A.D.; Ghyselinck, N.B.; Barrandon, Y.; Radtke, F. Corneal epithelial cell fate is maintained during repair by Notch1 signaling via the regulation of vitamin A metabolism. Dev. Cell 2007, 13, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Nowell, C.S.; Odermatt, P.D.; Azzolin, L.; Hohnel, S.; Wagner, E.F.; Fantner, G.E.; Lutolf, M.P.; Barrandon, Y.; Piccolo, S.; Radtke, F. Chronic inflammation imposes aberrant cell fate in regenerating epithelia through mechanotransduction. Nat. Cell Biol. 2016, 18, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Chen, S.Y.; Zhu, Y.T.; Tseng, S.C. Integration of BMP/Wnt signaling to control clonal growth of limbal epithelial progenitor cells by niche cells. Stem Cell Res. 2014, 12, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.Y.T.; Roberts, J.S.; Deng, S.X. Regulation of Limbal Epithelial Stem Cells: Importance of the Niche. Int. J. Mol. Sci. 2021, 22, 11975. [Google Scholar] [CrossRef]
- Britsch, S. The neuregulin-I/ErbB signaling system in development and disease. Adv. Anat. Embryol. Cell Biol. 2007, 190, 1–65. [Google Scholar]
- Yoon, D.; Yoon, D.; Cha, H.J.; Lee, J.S.; Chun, W. Enhancement of wound healing efficiency mediated by artificial dermis functionalized with EGF or NRG1. Biomed. Mater. 2018, 13, 045007. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Guo, H.; Han, Z.; Wu, S.; Liu, J.; Lin, Z.; An, F.; Zhu, J.; Li, M. NRG1 Regulates Proliferation, Migration and Differentiation of Human Limbal Epithelial Stem Cells. Curr. Issues Mol. Biol. 2023, 45, 10121-10130. https://doi.org/10.3390/cimb45120632
Wang B, Guo H, Han Z, Wu S, Liu J, Lin Z, An F, Zhu J, Li M. NRG1 Regulates Proliferation, Migration and Differentiation of Human Limbal Epithelial Stem Cells. Current Issues in Molecular Biology. 2023; 45(12):10121-10130. https://doi.org/10.3390/cimb45120632
Chicago/Turabian StyleWang, Bofeng, Huizhen Guo, Zhuo Han, Siqi Wu, Jiafeng Liu, Zesong Lin, Fengjiao An, Jin Zhu, and Mingsen Li. 2023. "NRG1 Regulates Proliferation, Migration and Differentiation of Human Limbal Epithelial Stem Cells" Current Issues in Molecular Biology 45, no. 12: 10121-10130. https://doi.org/10.3390/cimb45120632
APA StyleWang, B., Guo, H., Han, Z., Wu, S., Liu, J., Lin, Z., An, F., Zhu, J., & Li, M. (2023). NRG1 Regulates Proliferation, Migration and Differentiation of Human Limbal Epithelial Stem Cells. Current Issues in Molecular Biology, 45(12), 10121-10130. https://doi.org/10.3390/cimb45120632





