Abstract
Interferons (IFNs) and IFN-stimulated genes (ISGs) play essential roles for the control of viral infections. Their expression in infants with respiratory syncytial virus (RSV) bronchiolitis is poorly defined. Human endogenous retroviruses (HERVs) represent 8% of our genome and modulate inflammatory and immune reactions. TRIM28 and SETDB1 participate in the epigenetic regulation of genes involved in the immune response, including IFNs and HERVs. No study has explored the expression of HERVs, TRIM28, and SETDB1 during RSV bronchiolitis. We assessed, through a PCR real-time Taqman amplification assay, the transcription levels of six IFN-I ISGs, four IFNλs, the pol genes of HERV-H, -K, and -W families, the env genes of Syncytin (SYN)1 and SYN2, and of TRIM28/SETDB1 in whole blood from 37 children hospitalized for severe RSV bronchiolitis and in healthy children (HC). The expression of most IFN-I ISGs was significantly higher in RSV+ patients than in age-matched HC, but it was inhibited by steroid therapy. The mRNA concentrations of IFN-λs were comparable between patients and age-matched HC. This lack of RSV-driven IFN-III activation may result in the defective protection of the airway mucosal surface leading to severe bronchiolitis. The expression of IFN-III showed a positive correlation with age in HC, that could account for the high susceptibility of young children to viral respiratory tract infections. The transcription levels of every HERV gene were significantly lower in RSV+ patients than in HC, while the expressions of TRIM28/SETDB1 were overlapping. Given the negative impact of HERVs and the positive effects of TRIM28/SETDB1 on innate and adaptive immune responses, the downregulation of the former and the normal expression of the latter may contribute to preserving immune functions against infection.
1. Introduction
Respiratory syncytial virus (RSV) is one of the most frequent causes of infection during the first 2 years of life, with approximately two thirds of children infected by the end of their first year [1]. Infants and young children may develop acute bronchiolitis upon RSV infection. Most patients do not require hospitalization; however, about 3% are hospitalized because of severe clinical pictures, sometimes with admission to a pediatric intensive care unit (PICU) and requiring the support of invasive mechanical ventilation [1,2]. Prematurity, younger age, and co-morbidity were identified as risk factors for increased morbidity and mortality [3,4]. However, nearly half of the children admitted to the PICU were previously healthy [2]. Severe RSV bronchiolitis may predispose children to childhood asthma [5,6]. With increasing age, RSV episodes decrease in severity and in adult severe manifestations occur only in individuals with previous pulmonary problems and in the elderly [7]. Most hypotheses about the particular susceptibility of infants to bronchiolitis are based on structural elements, such as small airway size, different innervation patterns, and lack of interalveolar pores and channels; however, biological factors also appear to be involved, such as an altered inflammatory response with increased mucus production, massive neutrophil accumulation, and prevalent Th2 responses in a genetically susceptible host [8].
Viral recognition elicits the production of interferons (IFNs), which in turn induce the transcription of IFN-stimulated genes (ISGs). These exert potent antiviral activities and modulate a large array of innate and adaptive immune responses. Three types of IFNs have been identified. Type I IFNs encompass several subtypes [9], which through their binding to a ubiquitous receptor (IFNAR) are able to induce a rapid transcription of thousands of genes [9,10]. The direct detection of type I interferons in biologic samples has proven to be challenging. Thus, expressions of ISGs are commonly used to calculate type I IFN signature scores [11,12]. In contrast to the type II IFN, which is mostly a lately produced T-cell cytokine, the type III interferon (IFN-III) is also involved in the early phases of the antiviral response. It encompasses four members, also referred to as lambdas (IFNλs). They bind to a unique receptor expressed by epithelial cells and a subset of immune cells [13]. IFNλs represent the front-line antiviral defense of the respiratory mucosa [14,15]; they have less inflammatory activity than type I IFNs and fine-tune their antiviral effects [16]. Given the potent IFN-driven antiviral functions, many viruses have developed mechanisms to escape their inhibitory effects [16,17,18], including RSV [19,20].
Human endogenous retroviruses (HERVs) constitute about 8% of the human genome. They originate from ancestral infections of somatic cells with subsequent stable integration into the DNA of germinal cells [21]. Following deletions, multiplications, and mutations, they cannot produce infectious virions. Some viral sequences are, however, transcribed, and a few encode proteins, such as Syncytin-1 (SYN1) [22] and Syncytin-2 (SYN2) [23], that are involved in crucial physiologic functions, such as placental syncytiotrophoblast formation and materno-fetal immune tolerance [24]. HERVs share the typical structure of retroviruses, with three major genes: group-associated antigens (gag), polymerase (pol), and envelope (env), flanked between two regulatory long terminal repeats (LTRs). HERVs can regulate the transcription of adjacent cellular genes [25,26]; their RNAs through retro-transposition may generate novel insertions into DNA or be sensed as non-self via pattern recognition receptors (PRRs) can elicit inflammatory and immune responses [26,27,28,29]. Some retroviral proteins can trigger autoimmunity [30,31], while others, such as syncytins, have intrinsic immunomodulatory properties [32,33,34,35]. Aberrant HERV expressions are associated with immune mediated and inflammatory diseases, supporting their pathogenetic role in these pathologies [25,30,31,36,37]. Exogenous viral infections can induce abnormal HERV transcription. These include human herpes simplex virus 1 [38,39] and 6 [39,40], varicella-zoster virus [40], Epstein–Barr virus [41,42], human cytomegalovirus [43], HIV [44,45], hepatitis B [46] and C [47] viruses, influenza virus [48], and SARS-CoV-2 [12,49,50,51]. Furthermore, IFNs and inflammatory cytokines lead to the independent and synergistic activation of retroviral sequences [52]. The impact of virus-induced aberrant HERV expressions on the antiviral immune response remains poorly understood. Protective effects have been reported [50,53], but growing evidence suggests a negative action on viral control and disease progression [33,34,49,51,54,55,56].
HERV expression may be modulated by environmental factors through epigenetic mechanisms, such as DNA methylation and histone modifications. Krüppel-associated box domain zinc finger proteins (KRAB-ZFPs) represent the largest family of transcription regulators in our genome. The tripartite motif-containing 28 (TRIM28), also called KAP1 or TIF1-β, is a nuclear corepressor of KRAB-ZFPs [57]. SET domain bifurcated histone lysine methyltransferase 1 (SETDB1), also called ESET, is a histone H3K9 methyltransferase that participates with TRIM28 and KRAB-ZFPs to form heterochromatin [58]. TRIM28 and SETDB1 were uncovered mainly through investigations on host factors regulating the transcription of endogenous retroviruses [59,60]. Recent studies have, however, drawn attention to the crucial roles of TRIM28 and SETDB1 in the epigenetic control of the immune response [60,61,62,63], including the antiviral response and IFN production [64,65,66].
Despite these premises, the importance of IFN signature scores in infants hospitalized with severe RSV bronchiolitis remains elusive. Furthermore, in light of the potential cross-talks between RSV infection, HERVs, and TRIM28/SETDB1, no data on their expressions in infected patients are available. To obtain targeted information on the relevance of these variables during severe RSV bronchiolitis, we compared the transcriptional levels of six IFN-I ISGs and of type III IFNs, of HERV sequences, in particular of env genes of SYN1 and SYN2, and of pol genes of HERV-H, -K, and -W (the three retroviral families most widely studied), as well as of TRIM28 and SETDB1 in children hospitalized for acute RSV bronchiolitis versus healthy children (HC).
2. Materials and Methods
2.1. Study Populations
Two groups of children were enrolled in the study. Group A included children who were admitted at the Regina Margherita Children’s Hospital, Turin, Italy, with acute bronchiolitis due to RSV infection. Group B included HC tested at the same hospital for various reasons, such as planned surgical interventions and the exclusion of genetic, metabolic, endocrinological, or nephrological diseases, whose laboratory examinations, including white blood cell count and inflammatory markers, were all within normal ranges for age and whose results were all within normal limits. Subjects with any confirmed or suspected diseases associated with abnormal IFN production and/or HERV expression were excluded from the study. Exclusion criteria included children with confirmed or suspected immune defects (e.g., primary immunodeficiencies, born to HIV+ mothers), cancers, viral coinfections, autoimmune disorders [31], food allergies [37], prematurity [67], or autism spectrum disorder [68,69].
2.2. Total RNA Extraction
Total RNA was extracted from whole blood using the automated extractor Maxwell following the RNA Blood Kit protocol without modification (Promega, Madison, WI). This kit provides treatment with DNase during the RNA extraction process. To further exclude any contamination of genomic DNA, RNA extracts were directly amplified without reverse transcription to validate the RNA extraction protocol. RNA concentration and purity were assessed via traditional UV spectroscopy with absorbance at 260 and 280 nm. The nucleic acid concentration was calculated using the Beer–Lambert law, which predicts a linear change in absorbance with concentration. The RNA concentration range was within manufacturer specifications for the NanoDrop (Thermo Fisher Scientific, Waltham, MA USA). UV absorbance measurements were acquired using 1 µL of RNA sample in an ND-1000 spectrophotometer under the RNA-40 settings at room temperature (RT). Using this equation, an A260 reading of 1.0 is equivalent to ~40 µg/mL single-stranded RNA. The A260/A280 ratio was used to define RNA purity. An A260/A280 ratio of 1.8/2.1 is indicative of highly purified RNA. The RNAs were stored at −80° until use.
2.3. Reverse Transcription
Four hundred nanograms of total RNA was reverse-transcribed with 4 μL of buffer 5X, 4.8 μL of MgCl2 25 mM, 2 μL ImpromII (Promega), 1 μL of RNase inhibitor 20 U/L, 0.4 μL random hexamers 250 μM (Promega), 2 μL mix dNTPs 100 mM (Promega), and dd-water in a final volume of 20 μL. The reaction mix was carried out in a GeneAmp PCR system 9700 Thermal Cycle (Applied Biosystems, Foster City, CA, USA) under the following conditions: 5 min at 25 °C, 60 min at 42 °C, and 15 min at 70 °C for the inactivation of the enzyme. The cDNAs were stored at −20 °C until use.
2.4. Transcription Levels of IFNs, ISGs, TRIM28, SETDB1, Pol Genes of HERV-H, -K, and -W, and Env Genes of SYN1 and SYN2 via Real-Time PCR Assays
GAPDH was chosen as the reference gene in all determinations, being one of the most stable among the reference genes and already used in our previous studies [12,36,37,47,69].
Relative expression of transcription levels of IFN-I ISGs (IFI27, IFI44L, ISG15, IFIT1, RSAD2, and SIGLEC) [11], IFN-III (IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4) [13], TRIM28, and SETDB1, as well as of HERV-H-pol, HERV-K-pol, HERV-W-pol, SYN1-env, and SYN2-env was achieved as previously described in detail [12,54], using the primers and probes reported in Table 1. Briefly, 40 ng of cDNA was amplified in a 20 μL total volume reaction, containing 2.5 U goTaQ MaterMix (Promega), 1.25 mmol/L MgCl2, 500 nmol of specific primers, and 200 nmol of specific probes.

Table 1.
Primers and probes used to assess the transcription levels of type I interferon stimulated genes, type III interferon genes, pol genes of HERV-K, -W, and –H, env genes of syncytin 1 and syncytin 2, TRIM28, and SETDB1.
All of the amplifications were run in a 96-well plate at 95 °C for 10 min, followed by 45 cycles at 95 °C for 15 s and at 60 °C for 1 min. Each sample was run in triplicate. Relative quantification of target gene transcripts was performed according to the 2-ΔΔCt method [70]. Briefly, after normalization of the PCR result of each target gene with the housekeeping gene, the method included additional calibration using the expression of the target gene evaluated in a pool of healthy controls. The results, expressed in arbitrary units (called relative expression, RE), show the variations in target gene transcripts relative to the standard set of controls. Since we measured Ct for every target in all samples, we argue that our methods were suitable for HERVs, IFN signatures, TRIM28 and SETDB1 detection, and quantifications.
All analyses were performed in a laboratory of biosafety level 2 (BSL-2), according to the NIH [71] and WHO [72] guidelines.
2.5. Statistical Analysis
A one-way ANOVA test was used to compare the transcriptional levels of IFN-I ISGs, IFN-III, TRIM28, SETDB1, HERV-H-pol, HERV-K-pol, HERV-W-pol, SYN1-env, and SYN2-env, between Group A1 and Group A2 patients and HC. The Mann–Whitney test was used to compare the transcriptional levels of each IFN-I ISG, IFN-III, TRIM28, SETDB1, HERV-H-pol, HERV-K-pol, HERV-W-pol, SYN1-env, and SYN2-env between each group of children with each other. The Spearman correlation test was used to evaluate the correlations between age and mRNA concentrations of each target gene. Statistical analyses were conducted using the Prism software (GraphPad Software, La Jolla, CA, USA). In all analyses, p < 0.05 was taken to be statistically significant.
3. Results
3.1. Study Populations
Group A included 37 children hospitalized for acute RSV bronchiolitis. Their demographics and clinical features are reported in Table 2. Three RSV+ infants with Influenza A coinfection and one with pre-term delivery were excluded from the study.

Table 2.
Demographics and clinical characteristics of children with acute RSV bronchiolitis. n: number; %: percentage; IQR: interquartile range, expressed as 25 and 75 quartile values; yrs: years; SD: standard deviation. Values upper * or below ** normal limit according to age-related cutoffs.
Ten patients had comorbidities: two had asthma, one had metabolic disorders, two had congenital malformations, two had bacterial coinfections, one had neuromuscular disease, one was late pre-term, and one had neuropsychomotor disorder. Three patients had complications: one had bacterial pneumonia, one had cutaneous vasculitis, and one had post-infective anemia. Supplemental oxygen was administered to 31 patients. One patient had been admitted to the ICU. High markers of systemic inflammation, such as C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), and procalcitonin (PCT) levels, were found in 10 (27%) patients.
Patients were subdivided into two subgroups: Group A1 was on steroid treatment, Group A2 was without such therapy (Table 3). No specific criteria were used to select children who underwent steroid treatment; this was the subjective choice of individual pediatricians.

Table 3.
Characteristics of subgroups of RSV+ patients and healthy children. IQR: interquartile range, expressed as 25 and 75 quartile values.
At follow-up, all patients recovered from the acute illness.
As detailed in Table 3, control children encompassed four subgroups of subjects (Group B1, B2, B3, and B4), based on the tests performed.
3.2. Influence of Age on IFN Signature Scores and on Expression Levels of HERVs, TRIM28, and SETDB1
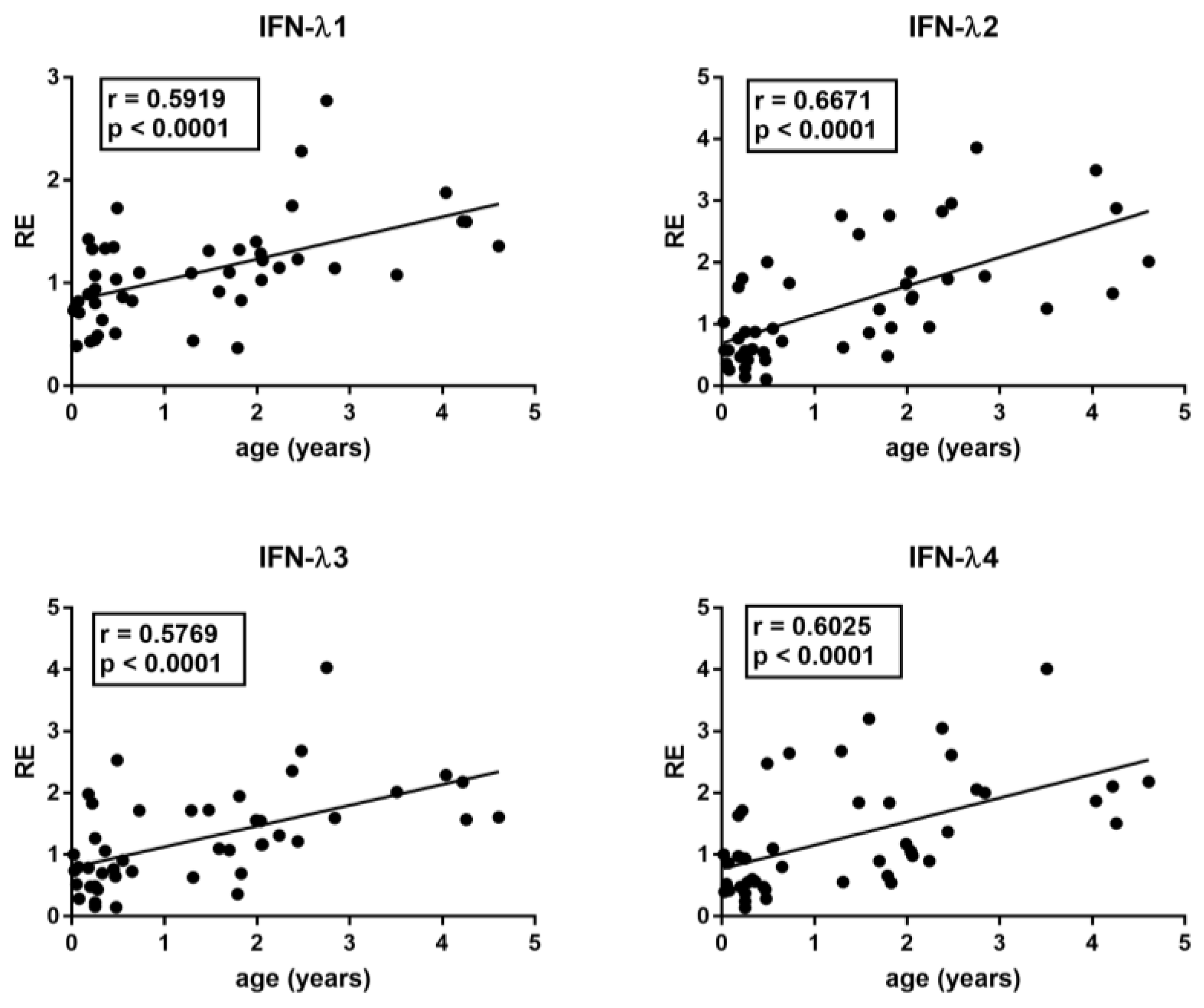
The median age differed between RSV patients and control groups. In particular, Group A children were significantly younger than Group B1, B2, B3, and B4 children (p < 0.0001). However, with the exception of IFN-III, there was no significant correlation between age and the transcriptional levels of each target gene in the control groups, even if there was a trend of negative correlation for TRIM28 and SETDB1. In contrast, there was a significantly positive correlation between the age and the mRNA levels of IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4 in HC (Figure 1).
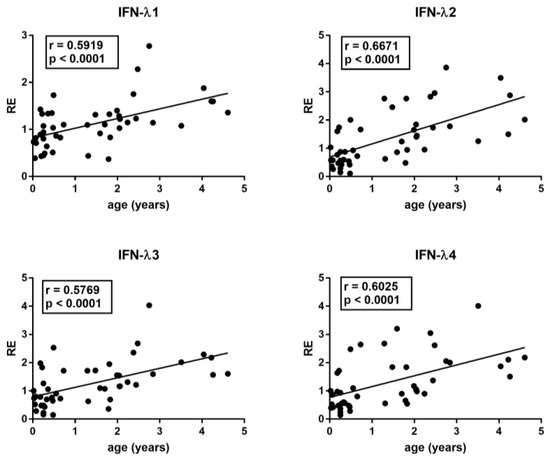
Figure 1.
Correlations between age and transcription levels of IFNλ1, IFNλ2, IFNλ3, and IFN-λ4 in whole blood from 46 healthy children of 0–5 years of age. RE: relative expression. Circles show the mean of three individual measurements. Line: linear regression line. Statistical analysis: Spearman correlation test.
3.3. Type I IFN Signature
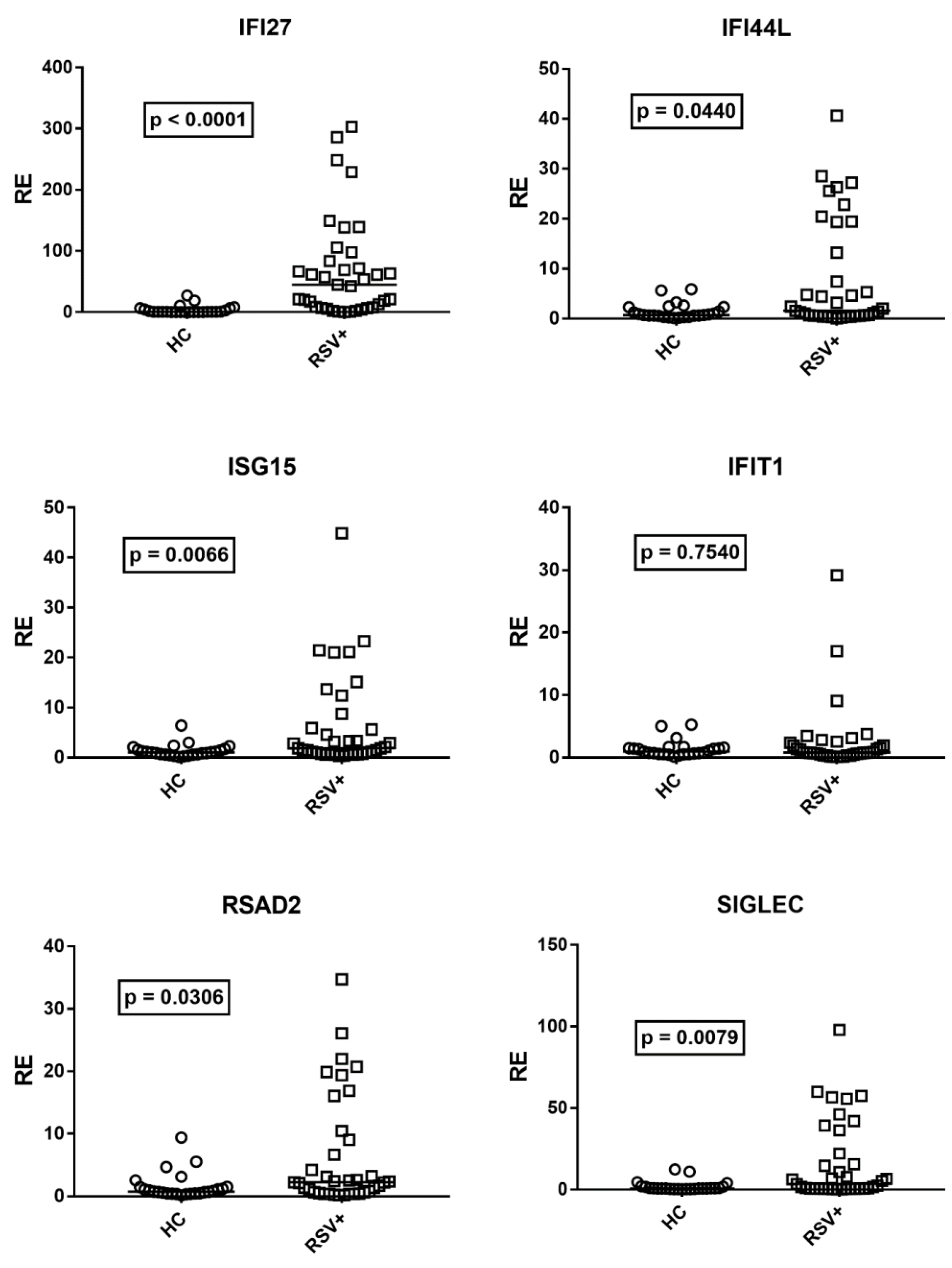
Although there was no correlation between age and expressions of IFN-I ISGs in HC, for analogy with IFN-III, we compared the median transcription levels of IFN-I ISGs in children with RSV bronchiolitis with the subgroup of 25 age-matched HC (median age and 25–75% IQR: RSV+ patients: 0.2, 0.1–0.3; HC: 0.3, 0.2–0.6, p = 0.11). Figure 2 shows that the mRNA levels of IFN-I ISGs were significantly higher in RSV+ patients than in HC for all ISGs, with the exception of IFIT1.
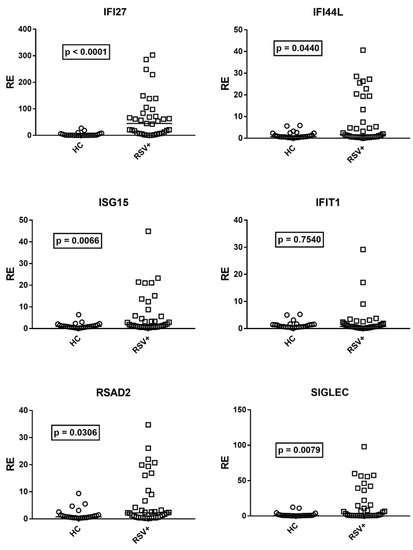
Figure 2.
Expression of type I interferon stimulated genes (ISGs) in whole blood from 37 children with acute RSV bronchiolitis (RSV+) and 25 age-matched healthy children (HC). RE: relative expression. Circles and squares show the mean of three individual measurements; horizontal lines show the median values. Median values and interquartile range 25–75% of ISGs: IFI27: HC 0.55, 0.31–4.60; RSV+ 44.98, 7.90–83; IFI44L: HC 0.72, 0.54–2.36; RSV+ 1.59, 057–13.23; ISG15: HC: 1.02, 0.62–1.50; RSV+ 1.85, 0.78–5.89; IFIT1: HC 0.97, 0.59–1.49; RSV+ 0.79, 0.48–1.91; RSAD2: HC 0.76, 0.46–1.45; RSV+ 2.21, 0.66–9.02; SGLEC: HC 0.73, 0.48–1.63; RSV+ 3.43, 0.68–22.05. Statistical analysis: Mann–Whitney test was used to compare values of each group of children with each other.
No significant differences between males and females were found in the transcription levels of any ISG, both in HC and in RSV+ patients (Supplementary Figures S1 and S2).
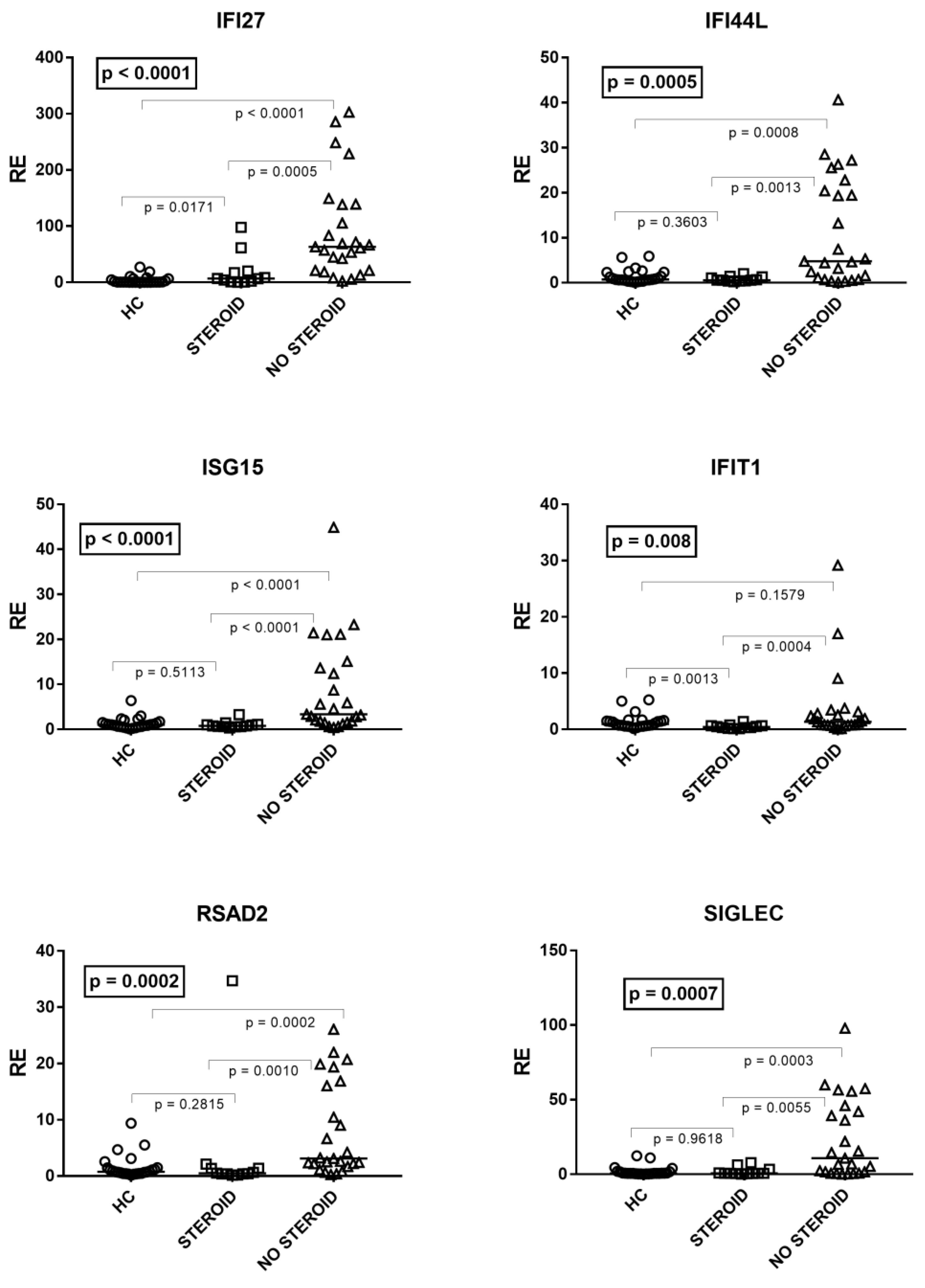
As illustrated in Figure 3, by comparing patients on steroid treatment (Group A1), untreated patients (Group A2), and HC, one-way ANOVA analysis showed significant differences in the mRNA levels of every IFN-I ISG between the three groups of children. In particular, the median transcriptional levels of IFN-I ISGs were significantly higher in untreated patients vs. HC for all genes except IFIT1. The levels of transcripts were significantly lower in treated patients vs. untreated patients for every ISG, whereas in the former vs. HC they remained significantly higher for IFI27, comparable to IFI44L, ISG15, RSAD2, and SIGLE, and reduced for IFIT1 (Figure 3).
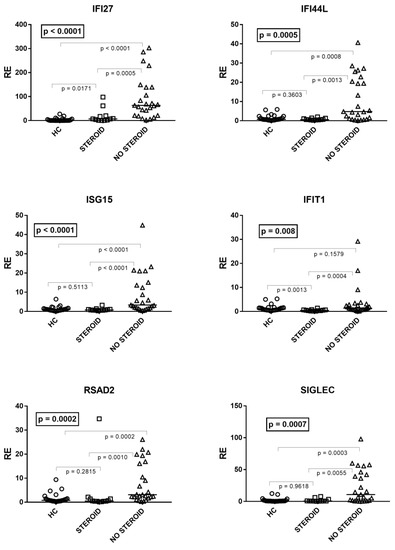
Figure 3.
Expression of type I interferon stimulated genes (ISGs) in whole blood from 25 age-matched healthy children (HC), 12 children with acute RSV bronchiolitis on steroid treatment (Group A1), and 25 untreated patients (Group A2). RE: relative expression. Horizontal lines show the median values; squares, triangles, and circles show the mean of three individual measurements. Median values and interquartile range 25%–75% of six ISGs (values of HC are reported above): IFI27: Group A1 (on steroids) 6.74, 1.90–17.78; Group A2 (untreated) 63.32, 21.13–138.40; IFI44L: Group A1 0.58, 0.43–1.12; Group A2 4.78, 1.12–20.44; ISG15: Group A1 0.77, 0.59–1.05; Group A2 3.35, 1.81–13.66; IFIT1: Group A1 0.41, 0.28–0.66; Group A2 1.36, 0.75–2.82; RSAD2: Group A1 0.47, 0.39-1.36; Group A2 3.11, 2.19–16.04; SIGLEC: Group A1 0.73, 0.59–1.58; Group A2 10.76, 1.67–42.09. Statistical analysis: The transcription levels of each target between the three groups of children were compared using the one-way ANOVA test. The transcription levels of each target between each group of children with each other were compared using the Mann–Whitney test.
3.4. Type III IFNs
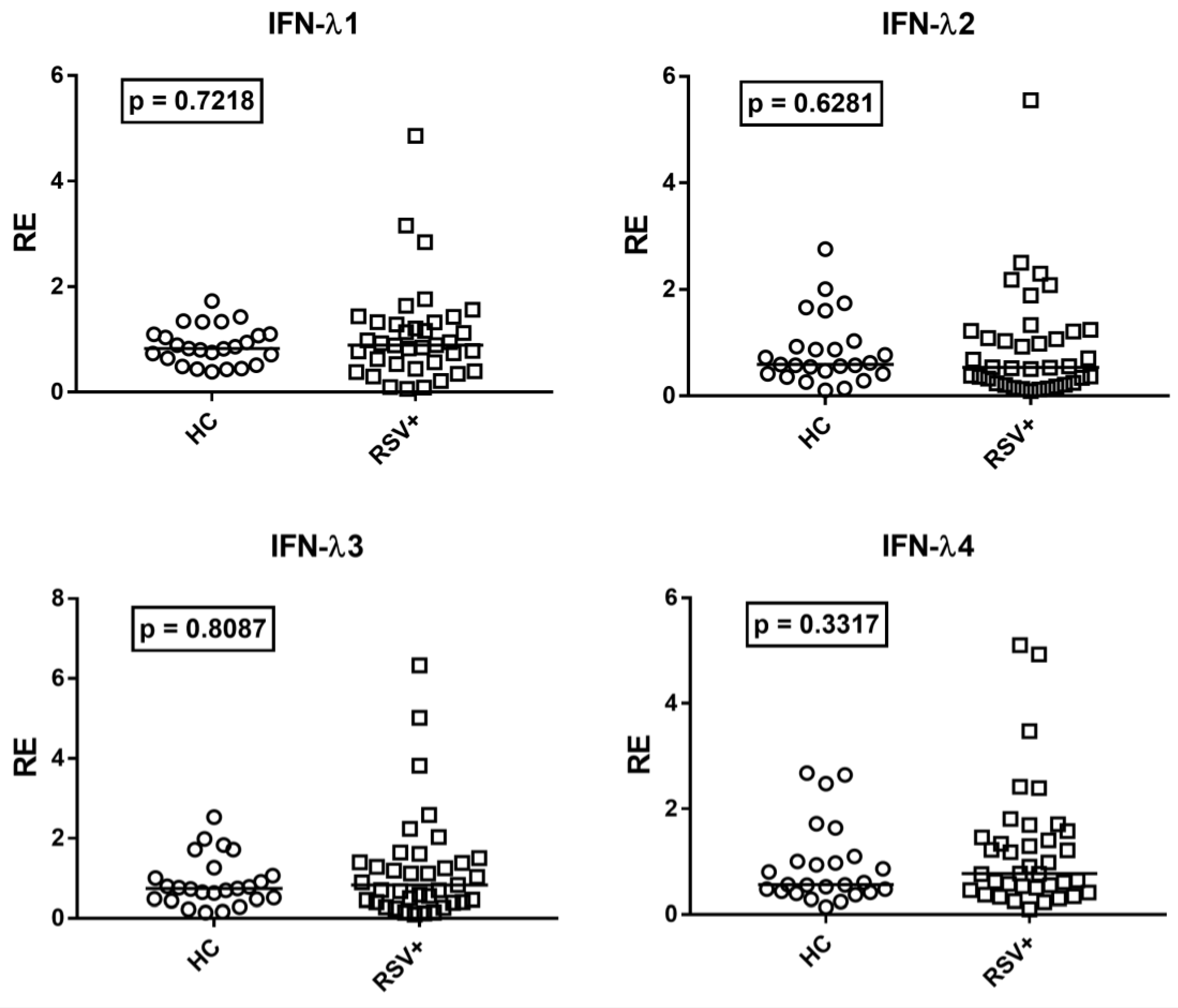
Given the positive correlations between age and IFN-III levels, the transcriptional levels of every IFN-λ in RSV-infected children were compared to the subgroup of the 25 age-matched HC. The transcription levels of all IFN-λs were similar in RSV-infected patients and age-matched HC (Figure 4).
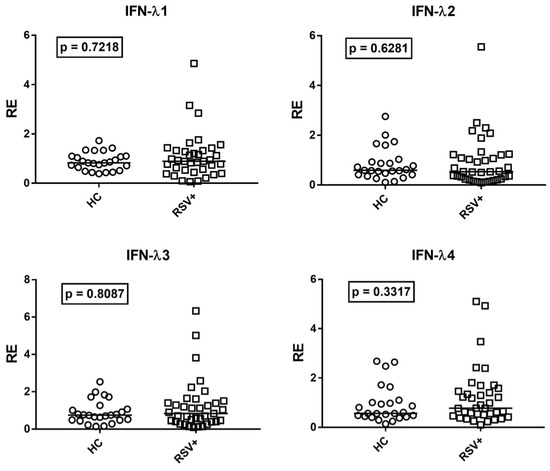
Figure 4.
Expression of type III interferons in whole blood from 37 children with acute RSV bronchiolitis (RSV+) and 25 age-matched healthy children (HC). RE: relative expression. Circles and squares show the mean of three individual measurements; horizontal lines show the median values. Median values and interquartile range 25–75% of IFN-IIIs: IFN-λ1: HC 0.83, 0.64–1.10; RSV+ 0.89, 0.53–1.32; IFN-λ2: HC 0.59, 0.42–0.93; RSV+ 0.54, 0.25–1.21; IFNλ3: HC 0.74, 0.48–1.06; RSV+ 0.83, 0.41–1.39; IFNλ4: HC 0.56, 0.44–1.00; RSV+ 0.77, 0.50–1.46. Statistical analysis: Mann–Whitney test was used to compare values of each group of children with each other.
By comparing Group A1 patients (on steroid therapy) vs. Group A2 patients (untreated), no significant difference emerged in the IFN-III transcript levels. Median values and interquartile range 25%-75% of IFN-IIIs: IFN-λ1: Group A1 0.81, 0.54–1.02; Group A2 0.93, 0.53-1.44; (p = 0.2149); IFN-λ2: Group A1 0.54, 0.23–0.77; Group A2 0.54, 0.34–1.22; (p = 0.4909); IFN-λ3: Group A1: 0.77, IQR 0.45–1.20; Group A2 0.90, 0.41–1.61; (p = 0.6426); IFN-λ4: Group A1 0.83, 0.44–1.27; Group A2 0.77, 0.55–1.46 (p = 0.5322).
No significant differences between genders were observed for IFN-III both in HC and RSV+ patients (Supplementary Figures S3 and S4).
3.5. Expressions of HERV-H-Pol, HERV-K-Pol, HERV-W-Pol, SYN1-Env, and SYN2-Env
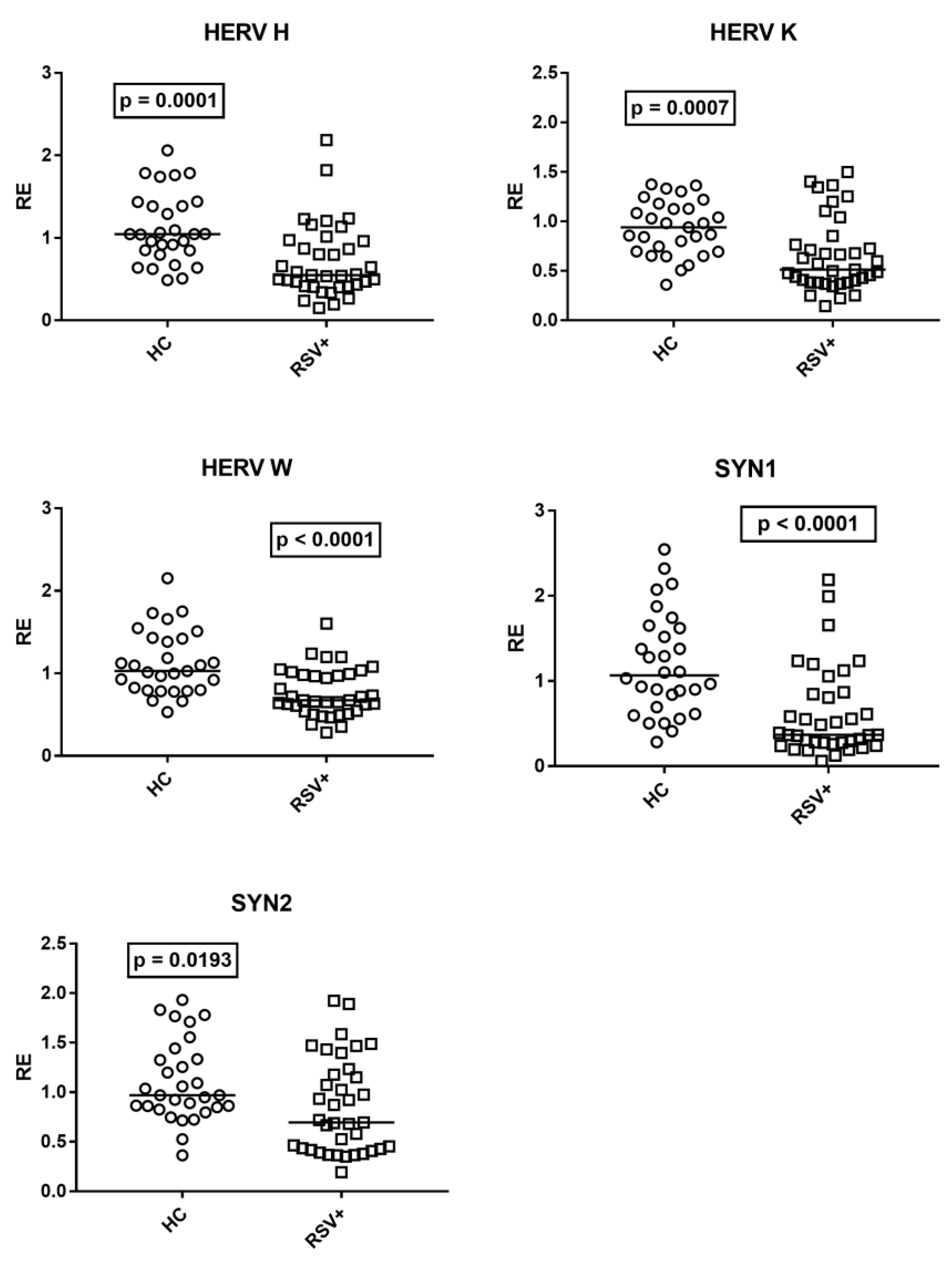
The medians of the transcription levels of HERV-H-pol, HERV-K-pol, and HERV-W-pol, as well as of SYN1-env and SYN2-env, were significantly lower in children with RSV bronchiolitis as compared to HC (Figure 5).
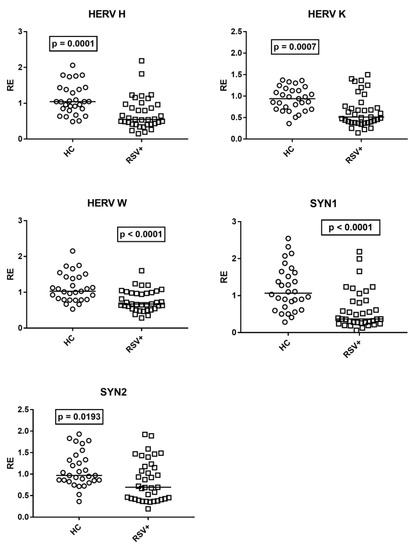
Figure 5.
Transcription levels of pol genes of HERV-H, HERV-K, and HERV-W, and of env genes of syncytin (SYN)1 and syncytin (SYN)2 in whole blood from 37 children with acute RSV bronchiolitis (RSV+), from 29 healthy children (HC) for HERV-pols, and 30 HC for SYN1-env and SYN2-env. RE: relative expression. Circles and squares show the median of three individual measurements; horizontal lines show the median values. Median values and interquartile range 25–75%, HERV-H-pol: HC 1.05, 0.85–1.39; RSV+ 0.55, 0.42–0.96; HERV-K-pol: HC 0.94, 0.70–1.13; RSV+ 0.51, 0.39–0.77; HER-W-pol: HC 1.03, 0.80–1.42; RSV+ 0.67, 0.55–0.98; SYN 1-env: HC 1.07, 0.73–1.60; RSV+ 0.37, 0.28–0.85; SYN2-env: HC 0.97, 0.85–1.33; RSV+ 0.69, 0.43–1.18. Statistical analysis: Mann–Whitney test was used to compare values of each group of children with each other.
No significant differences between genders were observed in the transcription levels of HERVs both in HC and RSV+ patients (Supplementary Figures S5 and S6).
No difference was found between patients with (Group A1) and without (Group A2) steroid treatment. Median values and interquartile range 25–75%, HERV-H-pol: Group A1 0.54, 0.46–0.84; Group A2 0.56, 0.41–0.97; (p = 0.9618); HERV-K-pol: Group A1 0.59, 0.44–0.84; Group A2 0.50, 0.38–0.77; (p = 0.6892); HERV-W-pol: Group A1 0.70, 0.52–1.00; Group A2 0.66, 0.61–0.97; (p > 0.9999); syncytin 1-env: Group A1 0.55, 0.33–1.24; Group A2 0.34, 0.26–0.81; (p = 0.2273); syncytin 2-env: Group A1 0.81, 0.43–1.24; Group A2 0.68, 0.42–1.15; (p = 0.9109).
3.6. Expressions of TRIM28 and SETDB1
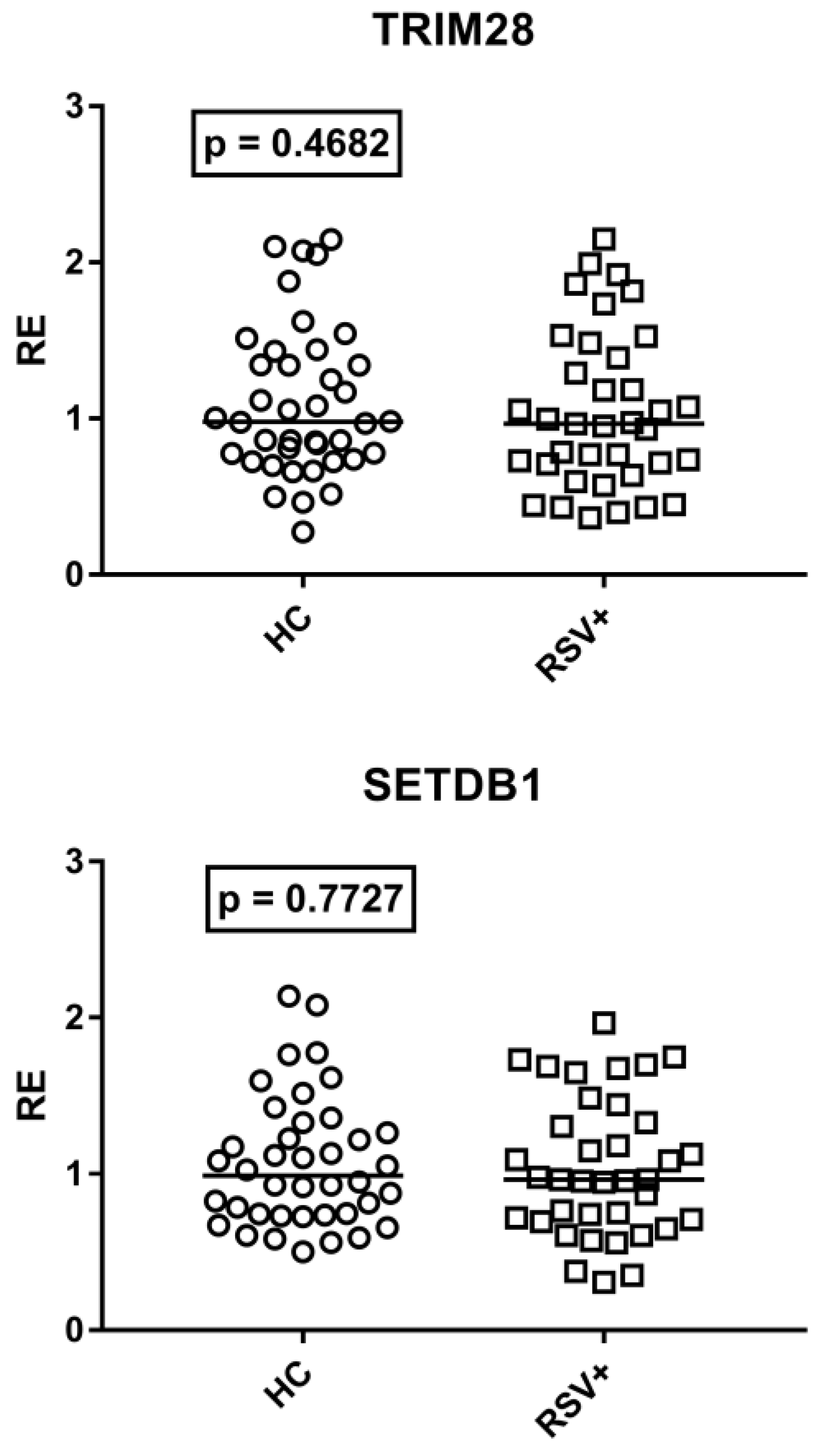
As detailed in Figure 6, TRIM28 and SETDB1 expression levels were comparable between RSV-infected patients and HC.
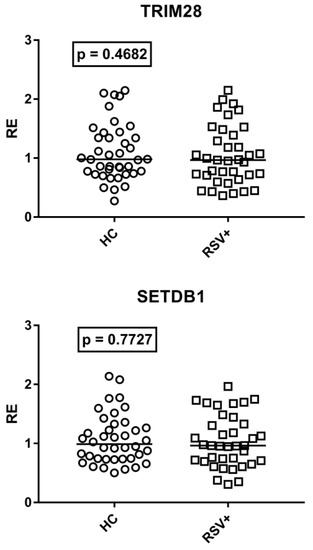
Figure 6.
Expression of TRIM28 and SETDB1 in whole blood from 40 healthy children (HC) and 37 children with acute RSV bronchiolitis. RE: relative expression. Circles and squares show the median of three individual measurements; horizontal lines show the median values. Median values and interquartile range 25–75%: TRIM28: HC 0.98, 0.77–1.36; RSV+ 0.97, 0.71–1.39; SETDB1: HC 0.99, 0.74–1.28; RSV+ 0.96, 0.71–1.33. Statistical analysis: Mann–Whitney test was used to compare values of each group of children with each other.
No difference was found between patients with (Group A1) and without (Group A2) steroid treatment. Median values and interquartile range 25–75%: TRIM28: Group A1 0.87, 0.70–1.53; Group A2 0.97, 0.71–1.18; (p = 0.8604); SETDB1: Group A1 1.03, 0.75–1.50; Group A2 0.96, 0.72–1.30; (p = 0.5753).
TRIM28 and SETDB1 expressions did not show any significant difference between males and females in HC and RSV+ patients (Supplementary Figures S7 and S8).
4. Discussion
The key role of IFNs in the control of viral infections is widely recognized, although their response is qualitatively and quantitatively virus-specific, and their production may vary during the course and the severity of the disease. Type I and type III IFNs are thought to be the main players in controlling the early steps of viral spread. Type I IFN production was found to be impaired in RSV-infected human airway epithelial cells [73]. RSV-nonstructural protein (NS)1, -NS2, and envelope G glycoprotein inhibit IFN-I synthesis [74,75,76,77]. The suppressor of the cytokine signaling (SOCS) family, through a feedback loop, inhibits the IFN-I-dependent antiviral signaling pathway, and NS1 and NS2 proteins upregulate SOCS1 and SOCS3 [78]. All of these findings suggest that RSV relies upon IFN-I downregulation to evade the early antiviral response. Our results, however, highlight that IFN-I signature scores were significantly higher for most ISGs in children hospitalized for acute RSV bronchiolitis without steroid treatment, as compared to age-matched HC. These findings suggest that the RSV infection is actually able to trigger a significant type I IFN synthesis, although the magnitude of this response may be lower than in other acute viral infections. For instance, RSV elicited a type I IFN production in the nasal mucosa of infants, but at significantly lower amounts as compared to the robust influenza virus response [73,79]. The result may be limited to the protection of the spread of RSV to lower airways. The RSV-driven IFN-I production was reduced during the neonatal period in an animal model [80] and in newborns and young children [20,81]. However, we did not observe any significant influence of the age on the expression of IFN-I ISGs in infants and pre-school healthy children. Furthermore, we found a higher expression of IFN-I ISGs by comparing RSV+ patients with age-matched HC.
The pharmacological approach in children with bronchiolitis is still debated. Given their strong anti-inflammatory activity and the lack of side effects in the short term, the administration of steroids is not unusual. However, since there is no evidence that systemic or inhalation steroids provide a significant benefit for bronchiolitis [82,83], their administration is discouraged by several guidelines [84,85]. Nevertheless, these continue to be administered for the management of RSV bronchiolitis in pediatric wards [86,87]. In line with the steroid-induced suppression of IFNs [88,89], our data document that patients on steroid treatment had a significantly impaired transcription of IFN-I ISGs as compared with untreated patients. This provides a biological basis for the more prolonged viral shedding in patients treated with steroids [90], and further supports that these should not be used in RSV bronchiolitis treatment.
The type III IFN machinery seems especially implicated in the protection of mucosal surfaces from viral attacks [14,15]. A first important point emerging from our results is the positive correlation between age and the transcription levels of every IFN-λ in HC. Since IFN-III protects the respiratory and gastrointestinal tract epithelial cells from viral infections [91], the impaired expression of IFN-III in the early period of life may contribute, at least in part, to the typical, high susceptibility of infants and pre-school children to viral infections of airways and the gastrointestinal tract. The second point is the similar concentration of all IFN-λ mRNAs in children with severe RSV bronchiolitis and in age-matched HC. This suggests that the IFN-III pathway remained unstimulated in RSV-infected children with disease progression. TRIM28/SETDB1 and steroids exert relevant regulatory activities in the induction of IFNs, but their expressions remained comparable between RSV-infected patients and HC as well as between patients with and without steroid therapy. IFN-III participates in the first-line defense against RSV via the retinoic-acid-inducible gene-I-(RIG-I)-dependent pathway [92,93]. However, RSV-NS1, -NS2, G proteins, and RSV-driven high concentrations of SOCS-1 and SOCS3 are able to inhibit not only the expression of IFN-I, but also of IFN-III [77,78,94,95]. The suppression of IFN-III was also documented via the RSV-induced elevated levels of prostaglandin 2 [96], and the activation of the epidermal growth factor receptor [97] and of Rab5a [98]. Moreover, the type III IFN response was significantly lower in children with RSV bronchiolitis as compared to children with influenza infection [73]. Therefore, the impaired expression of IFN-III in the first period of life, along with the ability of RSV to downregulate its production, may account for the lack of enhanced transcription levels of IFN-III in our patients and justify the development of severe pulmonary complications. In this context, it must be underlined that blood samples were collected after a long time from symptom onset, when the role-effect of IFN-III could be exhausted. High concentrations of IFN-λ1 were actually found in nasal airway secretions at the onset of acute RSV respiratory illness [99]. Furthermore, we did not assess the RSV strain, which may impact the IFN-I and IFN-III expression profiles and the magnitude of the ISG response [100,101].
Present results highlight, for the first time, that in children hospitalized for acute RSV bronchiolitis, the transcriptional levels of pol genes of HERV-H, -K, and -W, as well as of env genes of SYN1 and SYN2 were significantly lower than in HC. The origin and the clinical significance of such reduced HERV transactivation remain to be elucidated. TRIM28 and SETDB1 are considered to be potent co-repressors of HERV transcription [57,58,59,60]. However, their mRNA levels were similar to those of HC, suggesting that the HERV downregulation was not sustained by the abnormal expression of TRIM28 or SETDB1. Steroids can modulate the activation of retroviral elements [102,103], but we did not find any differences in HERV transcripts between treated and untreated patients. Inflammatory cytokines can stimulate the activation of retroviral sequences [52]. Increased inflammatory markers or alterations in white blood cell count were, however, observed only in a minority of patients. Therefore, the RSV induces strong, local, but poor systemic inflammation in children with severe bronchiolitis. Recently, the nuclear accumulation of RSV NS1 and the matrix (M) protein has been linked to global changes in host gene transcription [104,105]. Whether this also induces similar alterations in retroviral sequences remains to be investigated.
HERVs are involved in the host defense against viral infections and may trigger immune-mediated damages [25,28]. For instance, a HERV-K envelope protein inhibits the release of cytokines [54] and counteracts tetherin-mediated antiviral activity [55]. Since HERV-K elements are highly polymorphic in the human population, interindividual variations among HERV-K-env genes may result in different immune responses against the same viral infection [55,106]. A HERV-W envelope protein exerts potent pathogenic action through CD14/TLR4 stimulation [27,30]. SYN1 antagonizes antiviral responses and increases virus-induced inflammation [32,35,56,107], while SYN2 vigorously suppresses the T cell functions [34]. In general, the lower expression of HERVs in our patients may thus mirror a positive effect for the host deriving from their downregulation. These low HERV transcription levels seem antithetical to their increased activation, usually elicited by exogenous viral infections. An enhanced HERV expression was noticed, however, in chronic infections, such as those due to herpesviruses [38,39,40,41,42,43], HIV [44,45], and hepatitis viruses [46,47], or in vitro due to influenza viruses [48]. Upon an acute infection, such as SARS-CoV-2 infection, children with mild/moderate symptoms showed enhanced HERV transcription, whereas those with severe clinical pictures and a long duration of disease had significant declines in their mRNA concentrations, even below the normal values [12]. The present patients were all hospitalized for severe bronchiolitis and blood samples were collected after a long time from the beginning of the infection. Taken together, these findings suggest that reduced HERV expressions might characterize a severe, prolonged course of acute viral infections. This could be due to an exhaustion of the virus-driven stimulatory mechanisms and/or the upregulation of specific inhibitory checkpoints. For instance, the sterile alpha motif and HD-domain-containing protein 1 (SAMHD1) are able to block inflammatory responses and the activation of retroviruses during viral infections [108]. Based on this, children with mild forms of RSV infection and/or tested in the first days of illness could exhibit normal or increased levels of HERV mRNAs, with low concentrations of transcripts being a feature of a progressive acute disease.
The final biologic effects of HERVs on cell homeostasis are expected to be mediated by proteins. We assessed their transcriptional profiles, not their protein coding capacities. This is a limitation of our study, which, however, does not undermine the potential impact of HERVs on the evolution of RSV infection. As non-coding regulatory elements, HERVs can act as promoters and enhancers of cellular genes [25,26]. Their RNAs may be reintegrated everywhere into the DNA or recognized as non-self by viral RNA receptors triggering innate and adaptive immune responses [26,27,28,29].
RSV infection may induce epigenetic changes with possible clinical consequences [109,110]. Many members of the large family of TRIM proteins contribute to blocking viral replication [111] and growing data shed light on the pivotal roles of TRIM28/SETDB1 in epigenetic control of the immune response, including antiviral effects and T cell differentiation and functions [60,61,62,63,64,65,66]. The normal mRNA concentrations of TRIM28/SETDB1 that emerged in our patients, however, suggest that they are not key players in RSV-driven epigenetic variations. On the other hand, the activation of TRIM28/SETDB1 is a highly dynamic process that is removed rapidly by specific deconjugating proteases [112,113]. The prolonged time interval between symptom onset and blood collection could thus have influenced their transcription levels.
5. Conclusions
Our study shows the enhanced expression of many IFN-I ISGs in children hospitalized for severe RSV bronchiolitis without steroid treatment. The significant suppression of ISGs following the administration of steroids further points out that these should not be used in RSV bronchiolitis. The reduced expression of IFN-λs in the first period of life in HC and the lack of their activation in patients with disease progression may mirror an inadequate IFN-III protection of the airway mucosa surface that exposes RSV-infected infants to a high risk of pulmonary involvement. Type III IFN is of therapeutic interest due to its low proinflammatory profile. Our data support its potential use in RSV-infected at-risk infants to prevent the development of respiratory complications. The positive correlation between age and the expression of IFN-III may account for the high frequency of viral infections of the respiratory and gastrointestinal tracts in infants and pre-school children. The significantly impaired HERV transcription in children with RSV bronchiolitis was an unexpected feature. Its underlying biochemical mechanisms and clinical consequences remain to be elucidated. It does not seem imputable to inhibitory effects of corticosteroids or to alterations in TRIM28/SETDB1 repressors. The potential negative impact of HERV activation on the immune system is increasingly recognized. Its downregulation in children with severe RSV bronchiolitis may be a biomarker of disease progression, representing an attempt to limit its negative effects on the host defensive mechanisms. In contrast, given the essential role of TRIM28/SETDB1 in innate and adaptive immune responses, its preserved expression may contribute to maintaining effective immune protection against infection.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cimb45020079/s1: Supporting information on the differences between males and females in the transcription levels of all variables studied can be downloaded in the Supplementary Figures S1–S8 at https://www.mdpi.com/. Figure S1. Expression of type I interferon stimulated genes (ISGs) in whole blood from 25 healthy children: 16 males and 9 females. Figure S2. Expression of type I interferon stimulated genes (ISGs) in whole blood from 37 children with acute RSV bronchiolitis: 15 males and 22 females. Figure S3. Expression of type III interferons in whole blood from 25 healthy children: 16 males and 9 females. Figure S4. Expression of type III interferons in whole blood from 37 children with acute RSV bronchiolitis: 15 males and 22 females. Figure S5. Transcription levels of pol genes of HERV-H, HERV-K, and HERV-W in whole blood from 29 healthy children (HC), 17 males and 12 females, and of env genes of syncytin (SYN)1 and syncytin (SYN)2 from 30 HC, 17 males and 13 females. Figure S6. Transcription levels of pol genes of HERV-H, HERV-K, and HERV-W, and of env genes of syncytin (SYN)1 and syncytin (SYN)2 in whole blood from 37 children with acute RSV bronchiolitis: 15 males and 22 females. Figure S7. Expression of TRIM28 and SETDB1 in whole blood from 40 healthy children (HC): 23 males and 17 females. Figure S8. Expression of TRIM28 and SETDB1 in whole blood from 37 children with acute RSV bronchiolitis (RSV+): 15 males and 22 females.
Author Contributions
P.-A.T., substantial contributions to the conception of the work, drafting the work, and revising it critically for important intellectual contents and its final approval; S.G., F.S., G.P., G.F. and E.F., enrollment and follow-up of patients, revising the work critically; V.D., M.D. and I.G., performed laboratory tests, substantial contributions to analysis and interpretation of data; M.B., substantial contributions to the conception and design of the work, analysis, and interpretation of data, and drafting of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Clinical data were treated in accordance with the principles of the Helsinki Declaration. The study protocol was approved by the ethics committee of the Azienda Ospedaliera-Universitaria Città della Salute e della Scienza, Turin (protocol code 289571, approved 20.04.2020).
Informed Consent Statement
Blood samples were collected from leftovers of laboratory samples after parents gave informed consent.
Data Availability Statement
No additional data were created.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| HERVs | human endogenous retroviruses |
| IFN | interferon |
| ISGs | interferon-stimulated genes |
| KRAB-ZFPs | Kruppel-associated box domain zinc finger proteins |
| NS1 | RSV-nonstructural protein 1 |
| NS2 | RSV-nonstructural protein 2 |
| PICU | pediatric intensive care unit |
| PBMCs | peripheral blood mononuclear cells |
| PRR | pattern recognition receptor |
| RSV | respiratory syncytial virus |
| SETDB1 | domain bifurcated histone lysine methyltrasferase 1 |
| SOCS | suppressor of cytokine signaling |
| SYN1 | syncytin 1 |
| SYN2 | syncytin 2 |
| TRIM28 | tripartite motif containing 28 |
| TLR | Toll-like receptor |
References
- Hasegawa, K.; Tsugawa, Y.; Brown, D.F.; Mansbach, J.M.; Camargo, C.A., Jr. Trends in bronchiolitis hospitalizations in the United States, 2000–2009. Pediatrics 2013, 132, 28–36. [Google Scholar] [CrossRef]
- Ghazaly, M.; Nadel, S. Characteristics of children admitted to intensive care with acute bronchiolitis. Eur. J. Pediatr. 2018, 177, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Checchia, P.A.; Paes, B.; Bont, L.; Manzoni, P.; Simoes, E.A.; Fauroux, B.; Figueras-Aloy, J.; Carbonell-Estrany, X. Defining the risk and associated morbidity and mortality of severe respiratory syncytial virus infection among infants with congenital heart disease. Infect. Dis. Ther. 2017, 6, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.T.; Bont, L.J.; Zar, H.; Polack, F.P.; Park, C.; Claxton, A.; Borok, G.; Butylkova, Y.; Wegzyn, C. Respiratory syncytial virus hospitalization and mortality: Systematic review and meta-analysis. Pediatr. Pulmonol. 2017, 52, 556–569. [Google Scholar] [CrossRef]
- Lu, S.; Hartert, T.V.; Everard, M.L.; Giezek, H.; Nelsen, L.; Mehta, A.; Patel, H.; Knorr, B.; Reiss, T.F. Predictors of asthma following severe respiratory syncytial virus (RSV) bronchiolitis in early childhood. Pediatr. Pulmonol. 2016, 51, 1382–1392. [Google Scholar] [CrossRef] [PubMed]
- Dupont, W.; Wu, P.; Gebretsadik, T.; Hartert, T. RSV prevention in infancy and asthma in later life. Lancet Respir. Med. 2018, 6, e32. [Google Scholar] [CrossRef]
- Falsey, A.R.; Hennessey, P.A.; Formica, M.A.; Cox, C.; Walsh, E.E. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005, 352, 1749–1759. [Google Scholar] [CrossRef]
- Carvajal, J.J.; Avellaneda, A.M.; Salazar-Ardiles, C.; Maya, J.E.; Kalergis, A.M.; Lay, M.K. Host components contributing to respiratory syncytial virus pathogenesis. Front. Immunol. 2019, 10, 2152. [Google Scholar] [CrossRef]
- Hardy, M.P.; Owczarek, C.M.; Jermiin, L.S.; Ejdebäck, M.; Hertzog, P.J. Characterization of the type I interferon locus and identification of novel genes. Genomics 2004, 84, 331–345. [Google Scholar] [CrossRef]
- Schreiber, G. The molecular basis for differential type I interferon signaling. J. Biol. Chem. 2017, 292, 7285–7294. [Google Scholar] [CrossRef]
- Lamot, L.; Niemietz, I.; Brown, K.L. Comparable type I interferon score determination from PAXgene and Tempus whole blood RNA collection and isolation systems. BMC Res. Notes. 2019, 12, 511. [Google Scholar] [CrossRef] [PubMed]
- Tovo, P.A.; Garazzino, S.; Daprà, V.; Pruccoli, G.; Calvi, C.; Mignone, F.; Alliaudi, C.; Denina, M.; Scolfaro, C.; Zoppo, M.; et al. COVID-19 in children: Expressions of type I/II/III interferons, TRIM28, SETDB1, and endogenous retroviruses in mild and severe cases. Int. J. Mol. Sci. 2021, 22, 7481. [Google Scholar] [CrossRef] [PubMed]
- Andreakos, E.; Zanoni, I.; Galani, I.E. Lambda interferons come to light: Dual function cytokines mediating antiviral immunity and damage control. Curr. Opin. Immunol. 2019, 56, 67–75. [Google Scholar] [CrossRef]
- Syedbasha, M.; Egli, A. Interferon lambda: Modulating immunity in infectious diseases. Front. Immunol. 2017, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Kotenko, S.V.; Rivera, A.; Parker, D.; Durbin, J.E. Type III IFNs: Beyond antiviral protection. Semin. Immunol. 2019, 43, 101303. [Google Scholar] [CrossRef]
- Ye, L.; Schnepf, D.; Becker, J.; Ebert, K.; Tanriver, Y.; Bernasconi, V.; Gad, H.H.; Hartmann, R.; Lycke, N.; Staeheli, P. Interferon-λ enhances adaptive mucosal immunity by boosting release of thymic stromal lymphopoietin. Nat. Immunol. 2019, 20, 593–601. [Google Scholar] [CrossRef]
- Xia, C.; Vijayan, M.; Pritzl, C.J.; Fuchs, S.Y.; McDermott, A.B.; Hahm, B. Hemagglutinin of influenza A virus antagonizes type I interferon (IFN) responses by inducing degradation of type I IFN receptor 1. J. Virol. 2015, 90, 2403–2417. [Google Scholar] [CrossRef]
- Konno, Y.; Kimura, I.; Uriu, K.; Fukushi, M.; Irie, T.; Koyanagi, Y.; Sauter, D.; Gifford, R.J.; Nakagawa, S.; Sato, K. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020, 32, 108185. [Google Scholar] [CrossRef]
- Graham, B.S.; Anderson, L.J. Challenges and opportunities for respiratory syncytial virus vaccines. Curr. Top. Microbiol. Immunol. 2013, 372, 391–404. [Google Scholar]
- Hijano, D.R.; Vu, L.D.; Kauvar, L.M.; Tripp, R.A.; Polack, F.P.; Cormier, S.A. Role of type I interferon (IFN) in the respiratory syncytial virus (RSV) immune response and disease severity. Front. Immunol. 2019, 10, 566. [Google Scholar] [CrossRef]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef]
- Blond, J.L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.L. An envelope glycoprotein of the human endogenous retrovirus herv-w is expressed in the human placenta and fuses cells expressing the type d mammalian retrovirus receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef] [PubMed]
- Blaise, S.; de Parseval, N.; Benit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef] [PubMed]
- Lokossou, A.G.; Toudic, C.; Barbeau, B. Implication of human endogenous retrovirus envelope proteins in placental functions. Viruses 2014, 6, 4609–4627. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Tramontano, E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front. Immunol. 2018, 9, 2039. [Google Scholar]
- Isbel, L.; Whitelaw, E. Endogenous retroviruses in mammals: An emerging picture of how ERVs modify expression of adjacent genes. BioEssays 2012, 34, 734–738. [Google Scholar] [CrossRef]
- Rolland, A.; Jouvin-Marche, E.; Viret, C.; Faure, M.; Perron, H.; Marche, P.N. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J. Immunol. 2006, 76, 7636–7644. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Mu, X.; Ahmad, S.; Hur, S. Endogenous retroelements and the host innate immune sensors. Adv. Immunol. 2016, 132, 47–69. [Google Scholar]
- Madeira, A.; Burgelin, I.; Perron, H.; Curtin, F.; Lang, A.B.; Faucard, R. MSRV envelope protein is a potent, endogenous and pathogenic agonist of human toll-like receptor 4: Relevance of GNbAC1 in multiple sclerosis treatment. J. Neuroimmunol. 2016, 291, 29–38. [Google Scholar] [CrossRef]
- Yu, P. The potential role of retroviruses in autoimmunity. Immunol. Rev. 2016, 269, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Holder, B.S.; Tower, C.L.; Forbes, K.; Mulla, M.J.; Aplin, J.D.; Abrahams, V.M. Immune cell activation by trophoblast-derived microvesicles is mediated by syncytin 1. Immunology 2012, 136, 184–191. [Google Scholar] [PubMed]
- Hummel, J.; Kämmerer, U.; Müller, N.; Avota, E.; Schneider-Schaulies, S. Human endogenous retrovirus envelope proteins target dendritic cells to suppress T-cell activation. Eur. J. Immunol. 2015, 45, 1748–1759. [Google Scholar] [CrossRef]
- Lokossou, A.G.; Toudic, C.; Nguyen, P.T.; Elisseeff, X.; Vargas, A.; Rassart, É.; Lafond, J.; LeDuc, L.; Bourgault, S.; Gilbert, C.; et al. Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells. Biol. Reprod. 2020, 102, 185–198. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Rodriguez-Martin, E.; Ramos-Mozo, P.; Ortega-Madueño, I.; Dominguez-Mozo, M.I.; Arias-Leal, A.; García-Martínez, M.Á.; Casanova, I.; Galan, V.; Arroyo, R.; et al. Syncytin-1/HERV-W envelope is an early activation marker of leukocytes and is upregulated in multiple sclerosis patients. Eur. J. Immunol. 2020, 50, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Tovo, P.A.; Rabbone, I.; Tinti, D.; Galliano, I.; Trada, M.; Daprà, V.; Cerutti, F.; Bergallo, M. Enhanced expression of human endogenous retroviruses in new-onset type 1 diabetes: Pathogenetic and therapeutic implications. Autoimmunity 2020, 53, 283–288. [Google Scholar]
- Tovo, P.A.; Monti, G.; Daprà, V.; Montanari, P.; Calvi, C.; Alliaudi, C.; Sardo, A.; Galliano, I.; Bergallo, M. Enhanced expression of endogenous retroviruses and of TRIM28 and SETDB1 in children with food allergy. Clin. Transl. Allergy 2022, 12, e12124. [Google Scholar]
- Ruprecht, K.; Obojes, K.; Wengel, V.; Gronen, F.; Kim, K.S.; Perron, H.; Schneider-Schaulies, J.; Rieckmann, P. Regulation of human endogenous retrovirus W protein expression by herpes simplex virus type 1: Implications for multiple sclerosis. J. Neurovirol. 2006, 12, 65–71. [Google Scholar] [CrossRef]
- Brudek, T.; Lühdorf, P.; Christensen, T.; Hansen, H.J.; Møller-Larsen, A. Activation of endogenous retrovirus reverse transcriptase in multiple sclerosis patient lymphocytes by inactivated HSV-1, HHV-6 and VZV. J. Neuroimmunol. 2007, 187, 147–155. [Google Scholar]
- Tai, A.K.; Luka, J.; Ablashi, D.; Huber, B.T. HHV-6A Infection induces expression of HERV-K18-encoded superantigen. J. Clin. Virol. 2009, 46, 47–48. [Google Scholar] [CrossRef]
- Hsiao, F.C.; Tai, A.K.; Deglon, A.; Sutkowski, N.; Longnecker, R.; Huber, B.T. EBV LMP-2A employs a novel mechanism to transactivate the HERV-K18 superantigen through its ITAM. Virology 2009, 385, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Mameli, G.; Poddighe, L.; Mei, A.; Uleri, E.; Sotgiu, S.; Serra, C.; Manetti, R.; Dolei, A. Expression and activation by Epstein Barr virus of human endogenous retroviruses-W in blood cells and astrocytes: Inference for multiple sclerosis. PLoS ONE 2012, 7, e44991. [Google Scholar] [CrossRef] [PubMed]
- Assinger, A.; Yaiw, K.C.; Göttesdorfer, I.; Leib-Mösch, C.; Söderberg-Nauclér, C. Human cytomegalovirus (HCMV) induces human endogenous retrovirus (HERV) transcription. Retrovirology 2013, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Van der Kuyl, A.C. HIV infection and HERV expression: A review. Retrovirology 2012, 9, 6. [Google Scholar] [CrossRef]
- Vincendeau, M.; Göttesdorfer, I.; Schreml, J.M.; Wetie, A.G.; Mayer, J.; Greenwood, A.D.; Helfer, M.; Kramer, S.; Seifarth, W.; Hadian, K.; et al. Modulation of human endogenous retrovirus (HERV) transcription during persistent and de novo HIV-1 infection. Retrovirology 2015, 12, 27. [Google Scholar] [CrossRef]
- Liu, C.; Liu, L.; Wang, X.; Liu, Y.; Wang, M.; Zhu, F. HBV X Protein induces overexpression of HERV-W env through NF-kappaB in HepG2 cells. Virus Genes 2017, 53, 797–806. [Google Scholar] [CrossRef]
- Tovo, P.A.; Garazzino, S.; Daprà, V.; Alliaudi, C.; Silvestro, E.; Calvi, C.; Montanari, P.; Galliano, I.; Bergallo, M. Chronic HCV infection is associated with overexpression of human endogenous retroviruses that persists after drug-induced viral clearance. Int. J. Mol. Sci. 2020, 21, 3980. [Google Scholar] [CrossRef]
- Schmidt, N.; Domingues, P.; Golebiowski, F.; Patzina, C.; Tatham, M.H.; Hay, R.T.; Hale, B.G. An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17399–17408. [Google Scholar] [CrossRef]
- Balestrieri, E.; Minutolo, A.; Petrone, V.; Fanelli, M.; Iannetta, M.; Malagnino, V.; Zordan, M.; Vitale, P.; Charvet, B.; Horvat, B.; et al. Evidence of the pathogenic HERV-W envelope expression in T lymphocytes in association with the respiratory outcome of COVID-19 patients. EBioMedicine 2021, 66, 103341. [Google Scholar] [CrossRef]
- Marston, J.L.; Greenig, M.; Singh, M.; Bendall, M.L.; Duarte, R.R.R.; Feschotte, C.; Iñiguez, L.P.; Nixon, D.F. SARS-CoV-2 infection mediates differential expression of human endogenous retroviruses and long interspersed nuclear elements. JCI Insight 2021, 6, e147170. [Google Scholar] [CrossRef]
- Temerozo, J.R.; Fintelman-Rodrigues, N.; Dos Santos, M.C.; Hottz, E.D.; Sacramento, C.Q.; de Paula Dias da Silva, A.; Mandacaru, S.C.; Dos Santos Moraes, E.C.; Trugilho, M.R.O.; Gesto, J.S.M.; et al. Human endogenous retrovirus K in the respiratory tract is associated with COVID-19 physiopathology. Microbiome 2022, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Manghera, M.; Ferguson-Parry, J.; Lin, R.; Douville, R.N. NF-κB and IRF1 induce endogenous retrovirus K expression via interferon-stimulated response elements in its 5′ long terminal repeat. J. Virol. 2016, 90, 9338–9349. [Google Scholar] [CrossRef] [PubMed]
- Srinivasachar Badarinarayan, S.; Sauter, D. Switching sides: How endogenous retroviruses protect us from viral infections. J. Virol. 2021, 95, e02299-20. [Google Scholar] [CrossRef]
- Morozov, V.A.; Dao Thi, V.L.; Denner, J. The transmembrane protein of the human endogenous retrovirus—K (HERV-K) modulates cytokine release and gene expression. PLoS ONE 2013, 8, e70399. [Google Scholar] [CrossRef] [PubMed]
- Lemaître, C.; Harper, F.; Pierron, G.; Heidmann, T.; Dewannieux, M. The HERV-K human endogenous retrovirus envelope protein antagonizes Tetherin antiviral activity. J. Virol. 2014, 88, 13626–13637. [Google Scholar] [CrossRef]
- Tolosa, J.M.; Parsons, K.S.; Hansbro, P.M.; Smith, R.; Wark, P.A. The placental protein syncytin-1 impairs antiviral responses and exaggerates inflammatory responses to influenza. PLoS ONE 2015, 10, e0118629. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Fredericks, W.J.; Jensen, D.E.; Speicher, D.W.; Huang, X.P.; Neilson, E.G.; Rauscher, F.J., 3rd. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996, 10, 2067–2078. [Google Scholar] [CrossRef]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J., III. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Nisole, S.; Stoye, J.P.; Saïb, A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 2005, 3, 799–808. [Google Scholar] [CrossRef]
- Adoue, V.; Binet, B.; Malbec, A.; Fourquet, J.; Romagnoli, P.; van Meerwijk, J.P.M.; Amigorena, S.; Joffre, O.P. The histone methyltransferase SETDB1 controls T helper cell lineage integrity by repressing endogenous retroviruses. Immunity 2019, 50, 629–644.e8. [Google Scholar] [CrossRef]
- Zhou, X.F.; Yu, J.; Chang, M.; Zhang, M.; Zhou, D.; Cammas, F.; Sun, S.C. TRIM28 mediates chromatin modifications at the TCRα enhancer and regulates the development of T and natural killer T cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20083–20088. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, U.; Burbage, M.; Zueva, E.; Goudot, C.; Esnault, C.; Ye, M.; Carpier, J.M.; Burgdorf, N.; Hoyler, T.; Suarez, G.; et al. Critical role for TRIM28 and HP1β/γ in the epigenetic control of T cell metabolic reprograming and effector differentiation. Proc. Natl. Acad. Sci. USA 2019, 116, 25839–25849. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, P.; Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.A. Melanoma stem cell-like phenotype and significant suppression of immune response within a tumor are regulated by TRIM28 protein. Cancers 2020, 12, 2998. [Google Scholar] [CrossRef]
- Kamitani, S.; Ohbayashi, N.; Ikeda, O.; Togi, S.; Muromoto, R.; Sekine, Y.; Ohta, K.; Ishiyama, H.; Matsuda, T. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem. Biophys. Res. Commun. 2008, 370, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Deng, H.; Li, X.; Wu, X.; Tang, Q.; Chang, T.H.; Peng, H.; Rauscher, F.J., 3rd; Ozato, K.; Zhu, F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 2011, 187, 4754–4763. [Google Scholar] [CrossRef]
- Cuellar, T.L.; Herzner, A.M.; Zhang, X.; Goyal, Y.; Watanabe, C.; Friedman, B.A.; Janakiraman, V.; Durinck, S.; Stinson, J.; Arnott, D. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell Biol. 2017, 216, 3535–3549. [Google Scholar] [CrossRef]
- Bergallo, M.; Galliano, I.; Pirra, A.; Daprà, V.; Licciardi, F.; Montanari, P.; Coscia, A.; Bertino, E.; Tovo, P.-A. Transcriptional activity of human endogenous retriviruses is higher at birth in inversed correlation with gestationale age. Infect. Genet. Evol. 2019, 68, 273–279. [Google Scholar] [CrossRef]
- Balestrieri, E.; Cipriani, C.; Matteucci, C.; Benvenuto, A.; Coniglio, A.; Argaw-Denboba, A.; Toschi, N.; Bucci, I.; Miele, M.T.; Grelli, S.; et al. Children with autism spectrum disorder and their mothers share abnormal expression of selected endogenous retroviruses families and cytokines. Front. Immunol. 2019, 10, 2244. [Google Scholar] [CrossRef]
- Tovo, P.A.; Davico, C.; Marcotulli, D.; Vitiello, B.; Daprà, V.; Calvi, C.; Montanari, P.; Carpino, A.; Galliano, I.; Bergallo, M. Enhanced expression of human endogenous retroviruses, TRIM28 and SETDB1 in autism spectrum disorder. Int. J. Mol. Sci. 2022, 23, 5964. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgenb, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- NIH. NIH Guidelines for Research Involving Recombinant or Synthetic Nucleic Acid Molecules. 2019. Available online: https://osp.od.nih.gov/wp-content/uploads/2019_NIH_Guidelines.htm (accessed on 25 April 2019).
- WHO. Laboratory Biosafety Guidance Related to Coronavirus Disease (COVID-19): Interim Guidance. 2020. Available online: https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-(covid-19) (accessed on 13 May 2020).
- Ioannidis, I.; McNally, B.; Willette, M.; Peeples, M.E.; Chaussabel, D.; Durbin, J.E.; Ramilo, O.; Mejias, A.; Flaño. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J. Virol. 2012, 86, 5422–5436. [Google Scholar] [CrossRef] [PubMed]
- Ban, J.; Lee, N.R.; Lee, N.J.; Lee, J.K.; Quan, F.S.; Inn, K.S. Human respiratory syncytial virus NS 1 targets TRIM25 to suppress RIG-I ubiquitination and subsequent RIG-I-mediated antiviral signaling. Viruses 2018, 10, 10. [Google Scholar] [CrossRef]
- Ling, Z.; Tran, K.C.; Teng, M.N. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 2009, 83, 3734–3742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, L.; Zan, Y.; Du, N.; Yang, Y.; Tien, P. Human respiratory syncytial virus infection is inhibited by IFN-induced transmembrane proteins. J. Gen. Virol. 2015, 96, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Spann, K.M.; Tran, K.C.; Chi, B.; Rabin, R.L.; Collins, P.L. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 2004, 78, 4363.e9. [Google Scholar] [CrossRef]
- Zheng, J.; Yang, P.; Tang, Y.; Pan, Z.; Zhao, D. Respiratory syncytial virus nonstructural proteins upregulate SOCS1 and SOCS3 in the different manner from endogenous IFN signaling. J. Immunol. Res. 2015, 2015, 738547. [Google Scholar] [CrossRef]
- Melendi, G.A.; Coviello, S.; Bhat, N.; Zea-Hernandez, J.; Ferolla, F.M.; Polack, F.P. Breastfeeding is associated with the production of type I interferon in infants infected with influenza virus. Acta Paediatr. 2010, 99, 1517–1521. [Google Scholar] [CrossRef]
- Cormier, S.A.; Shrestha, B.; Saravia, J.; Lee, G.I.; Shen, L.; DeVincenzo, J.P.; Kim, Y.I.; You, D. Limited type I interferons and plasmacytoid dendritic cells during neonatal respiratory syncytial virus infection permit immunopathogenesis upon reinfection. J. Virol. 2014, 88, 9350–9360. [Google Scholar] [CrossRef]
- Marr, N.; Wang, T.I.; Kam, S.H.; Hu, Y.S.; Sharma, A.A.; Lam, A.; Markowski, J.; Solimano, A.; Lavoie, P.M.; Turvey, S.E. Attenuation of respiratory syncytial virus-induced and RIG-I-dependent type I IFN responses in human neonates and very young children. J. Immunol. 2014, 192, 948–957. [Google Scholar] [CrossRef]
- Corneli, H.M.; Zorc, J.J.; Mahajan, P.; Shaw, K.N.; Holubkov, R.; Reeves, S.D.; Ruddy, R.M.; Malik, B.; Nelson, K.A.; Bregstein, J.S.; et al. Bronchiolitis study group of the pediatric emergency care applied research network (pecarn). A multicenter, randomized, controlled trial of dexamethasone for bronchiolitis. N. Engl. J. Med. 2007, 357, 331–339. [Google Scholar] [CrossRef]
- Fernandes, R.M.; Bialy, L.M.; Vandermeer, B.; Tjosvold, L.; Plint, A.C.; Patel, H.; Johnson, D.; Klassen, T.P.; Hartling, L. Glucocorticoids for acute viral bronchiolitis in infants and young children. Cochrane Database Syst. Rev. 2013, 6, CD004878. [Google Scholar] [CrossRef] [PubMed]
- Ralston, S.L.; Lieberthal, A.S.; Meissner, H.C.; Alverson, B.K.; Baley, J.E.; Gadomski, A.M.; Johnson, D.W.; Light, M.J.; Maraqa, N.F.; Mendonca, E.A.; et al. American Academy of Pediatrics. Clinical practice guideline: The diagnosis, management, and prevention of bronchiolitis. Pediatrics 2014, 134, e1474–e1502. [Google Scholar] [PubMed]
- National Institute for Health and Care Excellence. Bronchiolitis in Children: Diagnosis and Management. In NICE Guidelines; 2015. London, United Kingdom. Available online: https://www.nice.org.uk/guidance/ng9 (accessed on 21 July 2022).
- Cutrera, R.; Baraldi, E.; Indinnimeo, L.; Miraglia Del Giudice, M.; Piacentini, G.; Scaglione, F.; Ullmann, N.; Moschino, L.; Galdo, F.; Duse, M. Management of acute respiratory diseases in the pediatric population: The role of oral corticosteroids. Ital. J. Pediatr. 2017, 43, 31. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Lera, E.; Peremiquel-Trillas, P.; Martínez, L.; Barceló, I.; Andrés, C.; Rodrigo-Pendás, J.Á.; Antón, A.; Rodrigo, C. Management of hospitalized respiratory syncytial virus bronchiolitis in the pediatric ward in Spain: Assessing the impact of a new clinical practice protocol. Paediatr. Drugs 2022, 24, 63–71. [Google Scholar] [CrossRef]
- Shimba, A.; Ikuta, K. Control of immunity by glucocorticoids in health and disease. Semin. Immunopathol. 2020, 42, 669–680. [Google Scholar] [CrossRef]
- Finney, L.J.; Glanville, N.; Farne, H.; Aniscenko, J.; Fenwick, P.; Kemp, S.V.; Trujillo-Torralbo, M.B.; Loo, S.L.; Calderazzo, M.A.; Wedzicha, J.A.; et al. Inhaled corticosteroids downregulate the SARS-CoV-2 receptor ACE2 in COPD through suppression of type I interferon. J. Allergy Clin. Immunol. 2021, 147, 510–519.e5. [Google Scholar] [CrossRef]
- Hall, C.B.; Powell, K.R.; MacDonald, N.E.; Gala, C.; Menegus, M.E.; Suffin, S.C.; Cohen, H.J. Respiratory syncytial viral infection in children with compromised immune function. N. Engl. J. Med. 1986, 315, 77–81. [Google Scholar] [CrossRef]
- Mordstein, M.; Neugebauer, E.; Ditt, V.; Jessen, B.; Rieger, T.; Falcone, V.; Sorgeloos, F.; Ehl, S.; Mayer, D.; Kochs, G.; et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010, 84, 5670–5677. [Google Scholar] [CrossRef]
- Liu, P.; Jamaluddin, M.; Li, K.; Garofalo, R.P.; Casola, A.; Brasier, A.R. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 2007, 81, 1401–1411. [Google Scholar] [CrossRef]
- Okabayashi, T.; Kojima, T.; Masaki, T.; Yokota, S.; Imaizumi, T.; Tsutsumi, H.; Himi, T.; Fujii, N.; Sawada, N. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011, 160, 360–366. [Google Scholar] [CrossRef]
- Bakre, A.A.; Harcourt, J.L.; Haynes, L.M.; Anderson, L.J.; Tripp, R.A. The central conserved region (CCR) of respiratory syncytial virus (RSV) G protein modulates host miRNA expression and alters the cellular response to infection. Vaccines 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, L.; Benveniste, E. Viral exploitation of host SOCS protein functions. J. Virol. 2010, 85, 1912–1921. [Google Scholar] [CrossRef] [PubMed]
- Werder, R.B.; Lynch, J.P.; Simpson, J.C.; Zhang, V.; Hodge, N.H.; Poh, M.; Forbes-Blom, E.; Kulis, C.; Smythe, M.L.; Upham, J.W.; et al. PGD2/DP2 receptor activation promotes severe viral bronchiolitis by suppressing IFN-λ production. Sci. Transl. Med. 2018, 10, eaao0052. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, A.; Galen, B.T.; Ueki, I.F.; Sun, Y.; Mulenos, A.; Osafo-Addo, A.; Clark, B.; Joerns, J.; Liu, W.; Nadel, J.A.; et al. Respiratory syncytial virus activates epidermal growth factor receptor to suppress interferon regulatory factor 1-dependent interferon-lambda and antiviral defense in airway epithelium. Mucosal Immunol. 2018, 11, 958–967. [Google Scholar] [CrossRef]
- Mo, S.; Tang, W.; Xie, J.; Chen, S.; Ren, L.; Zang, N.; Xie, X.; Deng, Y.; Gao, L.; Liu, E. Respiratory syncytial virus activates Rab5a to suppress IRF1-dependent IFN-λ production, subverting the antiviral defense of airway epithelial cells. J. Virol. 2021, 95, e02333-20. [Google Scholar] [CrossRef]
- Salka, K.; Arroyo, M.; Chorvinsky, E.; Abutaleb, K.; Perez, G.F.; Wolf, S.; Xuchen, X.; Weinstock, J.; Gutierrez, M.J.; Pérez-Losada, M.; et al. Innate IFN-lambda responses to dsRNA in the human infant airway epithelium and clinical regulatory factors during viral respiratory infections in early life. Clin. Exp. Allergy 2020, 50, 1044–1054. [Google Scholar] [CrossRef]
- Hillyer, P.; Mane, V.P.; Chen, A.; Dos Santos, M.B.; Schramm, L.M.; Shepard, R.E.; Luongo, C.; Le Nouen, C.; Huang, L.; Yan, L.; et al. Respiratory syncytial virus infection induces a subset of types I and III interferons in human dendritic cells. Virology 2017, 504, 63–72. [Google Scholar] [CrossRef]
- Pierangeli, A.; Viscido, A.; Bitossi, C.; Frasca, F.; Gentile, M.; Oliveto, G.; Frassanito, A.; Nenna, R.; Midulla, F.; Scagnolari, C. Differential interferon gene expression in bronchiolitis caused by respiratory syncytial virus-A genotype ON1. Med. Microbiol. Immunol. 2020, 209, 23–28. [Google Scholar] [CrossRef]
- Fiegl, M.; Strasser-Wozak, E.; Geley, S.; Gsur, A.; Drach, J.; Kofler, R. Glucocorticoid-mediated immunomodulation: Hydrocortisone enhances immunosuppressive endogenous retroviral protein (p15E) expression in mouse immune cells. Clin. Exp. Immunol. 1995, 101, 259–264. [Google Scholar]
- Hsu, K.; Lee, Y.K.; Chew, A.; Chiu, S.; Lim, D.; Greenhalgh, D.G.; Cho, K. Inherently variable responses to glucocorticoid stress among endogenous retroviruses isolated from 23 mouse strains. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2594–2600. [Google Scholar]
- Pei, J.; Beri, N.R.; Zou, A.J.; Hubel, P.; Dorando, H.K.; Bergant, V.; Andrews, R.D.; Pan, J.; Andrews, J.M.; Sheehan, K.C.F.; et al. Nuclear-localized human respiratory syncytial virus NS1 protein modulates host gene transcription. Cell Rep. 2021, 37, 109803. [Google Scholar] [CrossRef] [PubMed]
- Li, H.M.; Ghildyal, R.; Hu, M.; Tran, K.C.; Starrs, L.M.; Mills, J.; Teng, M.N.; Jans, D.A. Respiratory syncytial virus matrix protein-chromatin association is key to transcriptional inhibition in infected cells. Cells 2021, 10, 2786. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Sechi, L.A.; Kelvin, D.J. Human endogenous retrovirus K (HML-2) in health and disease. Front. Microbiol. 2020, 11, 1690. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Z.; Wang, P.; Li, S.; Zeng, J.; Tu, X.; Yan, Q.; Xiao, Z.; Pan, M.; Zhu, F. Syncytin-1, an endogenous retroviral protein, triggers the activation of CRP via TLR3 signal cascade in glial cells. Brain Behav. Immun. 2018, 67, 324–334. [Google Scholar] [CrossRef]
- Chen, S.; Bonifati, S.; Qin, Z.; St Gelais, C.; Kodigepalli, K.M.; Barrett, B.S.; Kim, S.H.; Antonucci, J.M.; Ladner, K.J.; Buzovetsky, O.; et al. SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kB. and interferon pathways. Proc. Natl. Acad. Sci. USA 2018, 115, E3798–E3807. [Google Scholar] [PubMed]
- Ptaschinski, C.; Mukherjee, S.; Moore, M.L.; Albert, M.; Helin, K.; Kunkel, S.L.; Lukacs, N.W. RSV-Induced H3K4 demethylase KDM5B leads to regulation of dendritic cell-derived innate cytokines and exacerbates pathogenesis in vivo. PLoS Pathog. 2015, 11, e1004978. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.J.; Scheltema, N.M.; Qi, C.; Vedder, R.; Klein, L.B.C.; Nibbelke, E.E.; van der Ent, C.K.; Bont, L.J.; Koppelman, G.H. Infant RSV immunoprophylaxis changes nasal epithelial DNA methylation at 6 years of age. Pediatr. Pulmonol. 2021, 56, 3822–3831. [Google Scholar] [CrossRef]
- Van Tol, S.; Hage, A.; Giraldo, M.I.; Bharaj, P.; Rajsbaum, R. The TRIMendous role of TRIMs in virus-host interactions. Vaccines 2017, 5, 23. [Google Scholar] [CrossRef]
- Mukhopadhyay, D.; Dasso, M. Modification in reverse: The SUMO proteases. Trends Biochem. Sci. 2007, 32, 286–295. [Google Scholar] [CrossRef]
- Zheng, Q.; Cao, Y.; Chen, Y.; Wang, J.; Fan, Q.; Huang, X.; Wang, Y.; Wang, T.; Wang, X.; Ma, J.; et al. Senp2 regulates adipose lipid storage by de-SUMOylation of Setdb1. J. Mol. Cell Biol. 2018, 10, 258–266. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).