PCID2 Subunit of the Drosophila TREX-2 Complex Has Two RNA-Binding Regions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Radiolabeled RNA Fragments
2.2. Electrophoretic Mobility Shift Assay (EMSA)
2.3. Drosophila Cell Culture Extracts
2.4. Antibodies
2.5. Statistical Analysis
2.6. Biomolecular Resources
3. Results
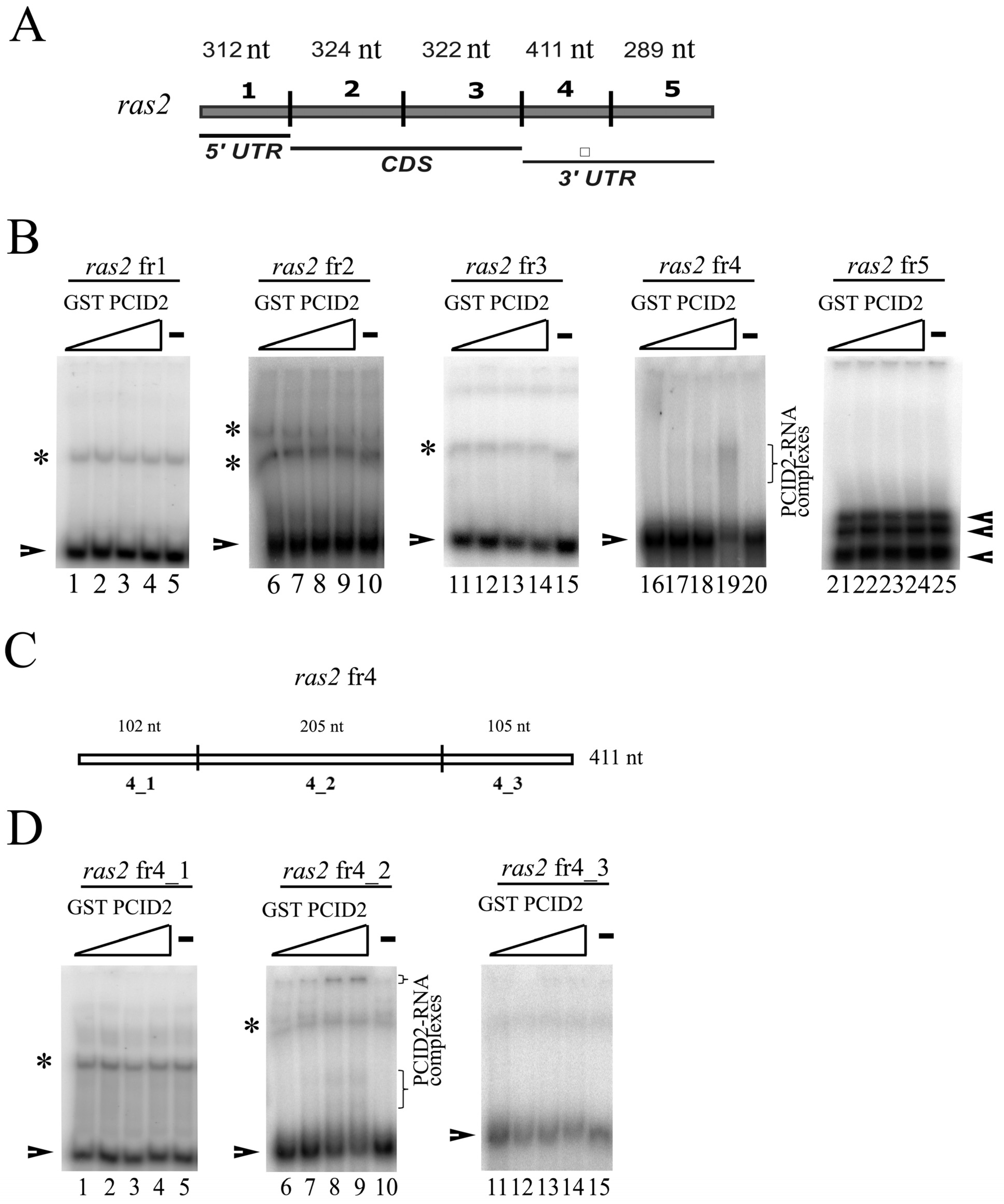
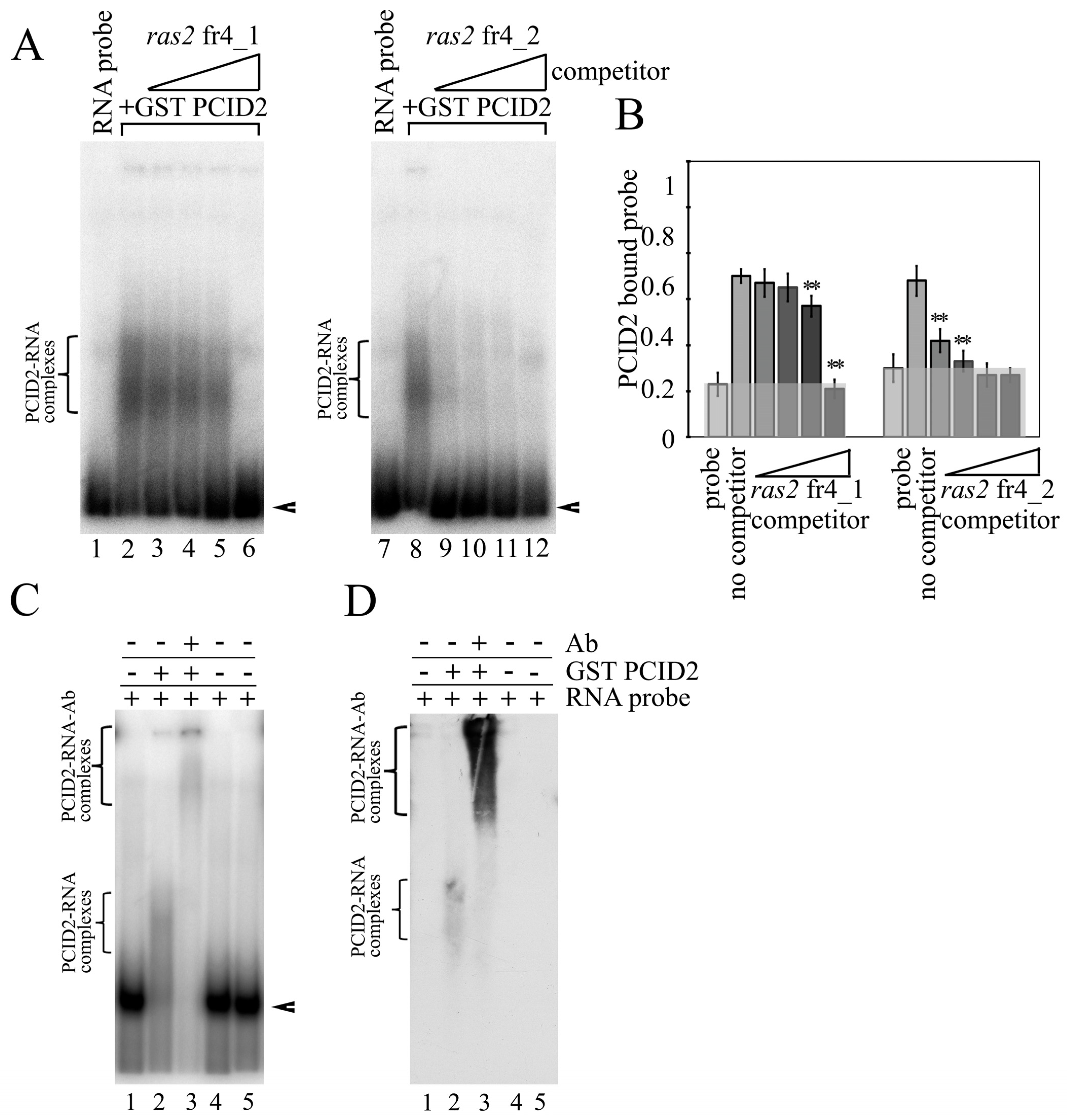
3.1. PCID2 Specifically Interacts with a Fragment of the 3′-Noncoding Region of the ras2 mRNA Independently of Xmas-2
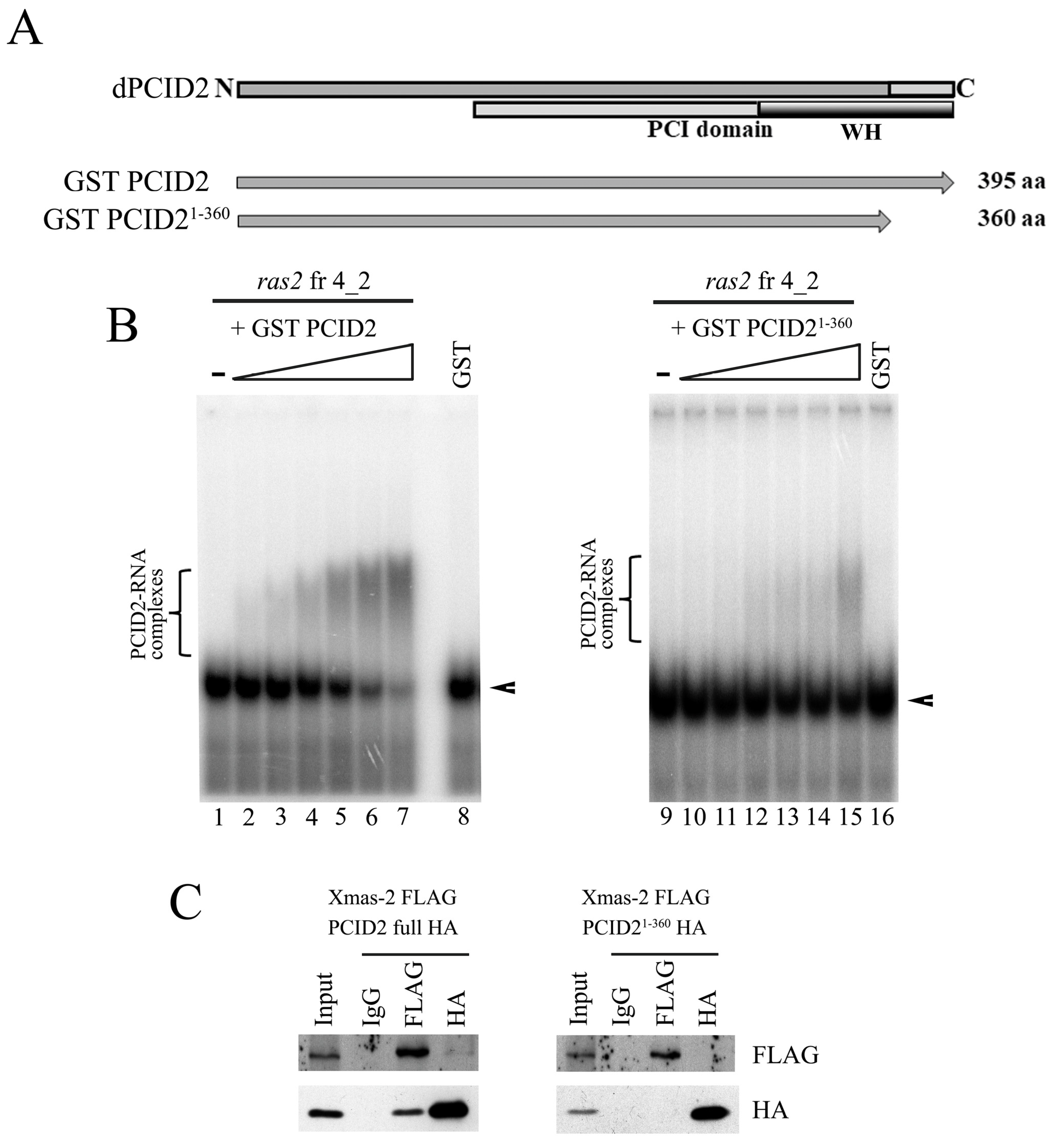
3.2. The C-Terminal Fragment of the PCI domain Interacts with the 3′-Noncoding Region of the ras2 mRNA
3.3. The C-Terminal Fragment of the PCI domain Interacts with the 3′-Noncoding Region of the ras2 mRNA
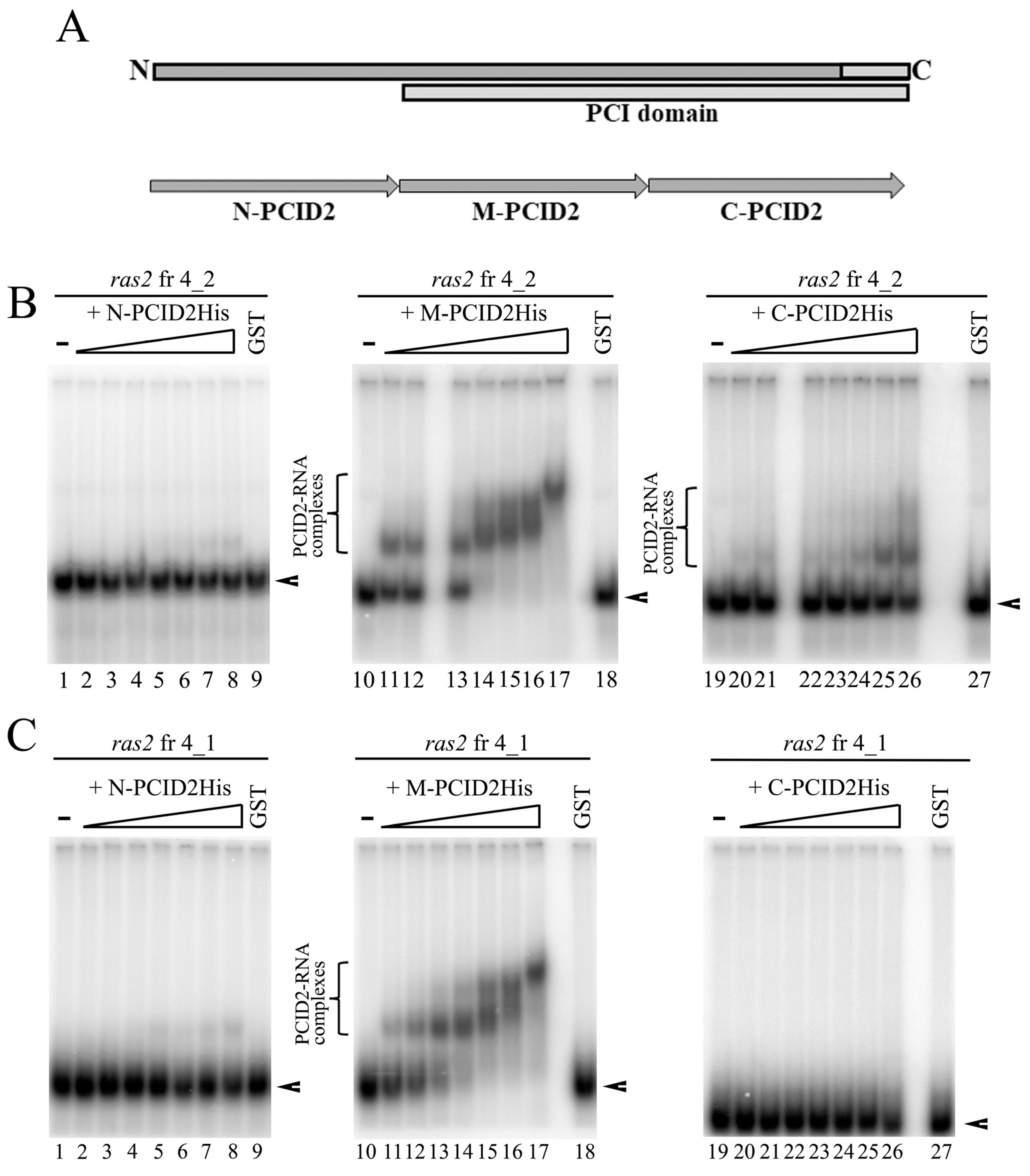
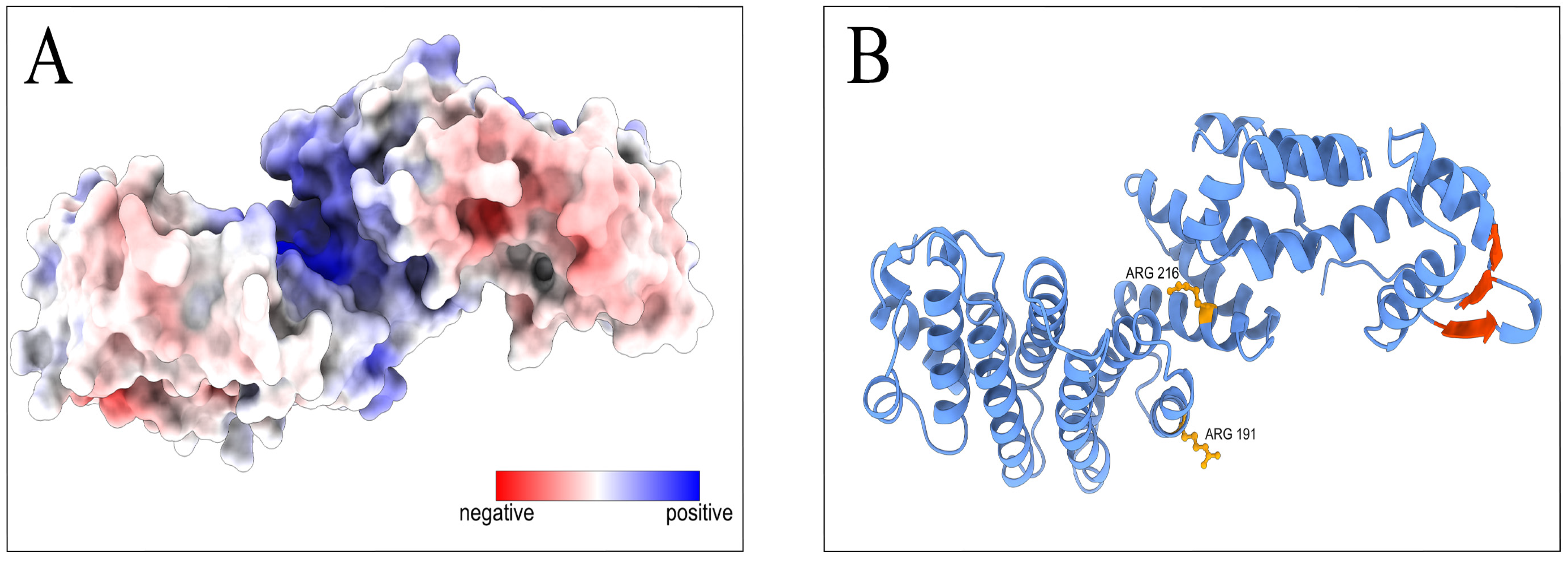
3.4. There Is an Additional RNA Interaction Region in PCID2
3.5. Point Mutations of the M Domain Abolish PCID2 Binding with RNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fischer, T.; Strässer, K.; Rácz, A.; Rodriguez-Navarro, S.; Oppizzi, M.; Ihrig, P.; Lechner, J.; Hurt, E. The mRNA Export Machinery Requires the Novel Sac3p-Thp1p Complex to Dock at the Nucleoplasmic Entrance of the Nuclear Pores. EMBO J. 2002, 21, 5843–5852. [Google Scholar] [CrossRef] [Green Version]
- Luna, R.; González-Aguilera, C.; Aguilera, A. Transcription at the Proximity of the Nuclear Pore: A Role for the THP1-SAC3-SUS1-CDC31 (THSC) Complex. RNA Biol. 2009, 6, 145–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, T.; Rodríguez-Navarro, S.; Pereira, G.; Rácz, A.; Schiebel, E.; Hurt, E. Yeast Centrin Cdc31 Is Linked to the Nuclear mRNA Export Machinery. Nat. Cell Biol. 2004, 6, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Kurshakova, M.M.; Krasnov, A.N.; Kopytova, D.V.; Shidlovskii, Y.V.; Nikolenko, J.V.; Nabirochkina, E.N.; Spehner, D.; Schultz, P.; Tora, L.; Georgieva, S.G. SAGA and a Novel Drosophila Export Complex Anchor Efficient Transcription and mRNA Export to NPC. EMBO J. 2007, 26, 4956–4965. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, V.O.; Stewart, M.; Laskey, R.A. GANP Enhances the Efficiency of mRNA Nuclear Export in Mammalian Cells. Nucleus 2010, 1, 393–396. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Tang, X.; Tian, G.; Wang, F.; Liu, K.; Nguyen, V.; Kohalmi, S.E.; Keller, W.A.; Tsang, E.W.T.; Harada, J.J.; et al. Arabidopsis Homolog of the Yeast TREX-2 mRNA Export Complex: Components and Anchoring Nucleoporin. Plant J. 2010, 61, 259–270. [Google Scholar] [CrossRef]
- Ellisdon, A.M.; Dimitrova, L.; Hurt, E.; Stewart, M. Structural Basis for the Assembly and Nucleic Acid Binding of the TREX-2 Transcription-Export Complex. Nat. Struct. Mol. Biol. 2012, 19, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Jani, D.; Lutz, S.; Hurt, E.; Laskey, R.A.; Stewart, M.; Wickramasinghe, V.O. Functional and Structural Characterization of the Mammalian TREX-2 Complex That Links Transcription with Nuclear Messenger RNA Export. Nucleic Acids Res. 2012, 40, 4562–4573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Navarro, S.; Fischer, T.; Luo, M.-J.; Antúnez, O.; Brettschneider, S.; Lechner, J.; Pérez-Ortín, J.E.; Reed, R.; Hurt, E. Sus1, a Functional Component of the SAGA Histone Acetylase Complex and the Nuclear Pore-Associated mRNA Export Machinery. Cell 2004, 116, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Pascual-García, P.; Govind, C.K.; Queralt, E.; Cuenca-Bono, B.; Llopis, A.; Chavez, S.; Hinnebusch, A.G.; Rodríguez-Navarro, S. Sus1 Is Recruited to Coding Regions and Functions during Transcription Elongation in Association with SAGA and TREX2. Genes. Dev. 2008, 22, 2811–2822. [Google Scholar] [CrossRef] [Green Version]
- González-Aguilera, C.; Tous, C.; Gómez-González, B.; Huertas, P.; Luna, R.; Aguilera, A. The THP1-SAC3-SUS1-CDC31 Complex Works in Transcription Elongation-mRNA Export Preventing RNA-Mediated Genome Instability. Mol. Biol. Cell 2008, 19, 4310–4318. [Google Scholar] [CrossRef] [Green Version]
- Evangelista, F.M.; Maglott-Roth, A.; Stierle, M.; Brino, L.; Soutoglou, E.; Tora, L. Transcription and mRNA Export Machineries SAGA and TREX-2 Maintain Monoubiquitinated H2B Balance Required for DNA Repair. J. Cell Biol. 2018, 217, 3382–3397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faza, M.B.; Kemmler, S.; Jimeno, S.; González-Aguilera, C.; Aguilera, A.; Hurt, E.; Panse, V.G. Sem1 Is a Functional Component of the Nuclear Pore Complex-Associated Messenger RNA Export Machinery. J. Cell Biol. 2009, 184, 833–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glukhova, A.A.; Kurshakova, M.M.; Nabirochkina, E.N.; Georgieva, S.G.; Kopytova, D.V. PCID2, a Subunit of the Drosophila TREX-2 Nuclear Export Complex, Is Essential for Both mRNA Nuclear Export and Its Subsequent Cytoplasmic Trafficking. RNA Biol. 2021, 18, 1969–1980. [Google Scholar] [CrossRef]
- Wilmes, G.M.; Bergkessel, M.; Bandyopadhyay, S.; Shales, M.; Braberg, H.; Cagney, G.; Collins, S.R.; Whitworth, G.B.; Kress, T.L.; Weissman, J.S.; et al. A Genetic Interaction Map of RNA-Processing Factors Reveals Links between Sem1/Dss1-Containing Complexes and mRNA Export and Splicing. Mol. Cell 2008, 32, 735–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jani, D.; Lutz, S.; Marshall, N.J.; Fischer, T.; Köhler, A.; Ellisdon, A.M.; Hurt, E.; Stewart, M. Sus1, Cdc31, and the Sac3 CID Region Form a Conserved Interaction Platform That Promotes Nuclear Pore Association and mRNA Export. Mol. Cell 2009, 33, 727–737. [Google Scholar] [CrossRef]
- Umlauf, D.; Bonnet, J.; Waharte, F.; Fournier, M.; Stierle, M.; Fischer, B.; Brino, L.; Devys, D.; Tora, L. The Human TREX-2 Complex Is Stably Associated with the Nuclear Pore Basket. J. Cell Sci. 2013, 126, 2656–2667. [Google Scholar] [CrossRef] [Green Version]
- Aksenova, V.; Smith, A.; Lee, H.; Bhat, P.; Esnault, C.; Chen, S.; Iben, J.; Kaufhold, R.; Yau, K.C.; Echeverria, C.; et al. Nucleoporin TPR Is an Integral Component of the TREX-2 mRNA Export Pathway. Nat. Commun. 2020, 11, 4577. [Google Scholar] [CrossRef] [PubMed]
- Kopytova, D.; Popova, V.; Kurshakova, M.; Shidlovskii, Y.; Nabirochkina, E.; Brechalov, A.; Georgiev, G.; Georgieva, S. ORC Interacts with THSC/TREX-2 and Its Subunits Promote Nxf1 Association with mRNP and mRNA Export in Drosophila. Nucleic Acids Res. 2016, 44, 4920–4933. [Google Scholar] [CrossRef] [Green Version]
- Dimitrova, L.; Valkov, E.; Aibara, S.; Flemming, D.; McLaughlin, S.H.; Hurt, E.; Stewart, M. Structural Characterization of the Chaetomium Thermophilum TREX-2 Complex and Its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2. Structure 2015, 23, 1246–1257. [Google Scholar] [CrossRef] [Green Version]
- Kopytova, D.V.; Orlova, A.V.; Krasnov, A.N.; Gurskiy, D.Y.; Nikolenko, J.V.; Nabirochkina, E.N.; Shidlovskii, Y.V.; Georgieva, S.G. Multifunctional Factor ENY2 Is Associated with the THO Complex and Promotes Its Recruitment onto Nascent mRNA. Genes. Dev. 2010, 24, 86–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, M.; Hellerschmied, D.; Schubert, T.; Amlacher, S.; Vinayachandran, V.; Reja, R.; Pugh, B.F.; Clausen, T.; Köhler, A. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell 2015, 162, 1016–1028. [Google Scholar] [CrossRef] [Green Version]
- García-Oliver, E.; García-Molinero, V.; Rodríguez-Navarro, S. MRNA Export and Gene Expression: The SAGA-TREX-2 Connection. Biochim. Biophys. Acta 2012, 1819, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Aibara, S.; Bai, X.-C.; Stewart, M. The Sac3 TPR-like Region in the Saccharomyces Cerevisiae TREX-2 Complex Is More Extensive but Independent of the CID Region. J. Struct. Biol. 2016, 195, 316–324. [Google Scholar] [CrossRef] [Green Version]
- Stewart, M. Structure and Function of the TREX-2 Complex. Subcell. Biochem. 2019, 93, 461–470. [Google Scholar] [CrossRef]
- Ellisdon, A.M.; Stewart, M. Structural Biology of the PCI-Protein Fold. Bioarchitecture 2012, 2, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Gajiwala, K.S.; Burley, S.K. Winged Helix Proteins. Curr. Opin. Struct. Biol. 2000, 10, 110–116. [Google Scholar] [CrossRef]
- Pathare, G.R.; Nagy, I.; Bohn, S.; Unverdorben, P.; Hubert, A.; Körner, R.; Nickell, S.; Lasker, K.; Sali, A.; Tamura, T.; et al. The Proteasomal Subunit Rpn6 Is a Molecular Clamp Holding the Core and Regulatory Subcomplexes Together. Proc. Natl. Acad. Sci. USA 2012, 109, 149–154. [Google Scholar] [CrossRef]
- Gallardo, M.; Luna, R.; Erdjument-Bromage, H.; Tempst, P.; Aguilera, A. Nab2p and the Thp1p-Sac3p Complex Functionally Interact at the Interface between Transcription and mRNA Metabolism. J. Biol. Chem. 2003, 278, 24225–24232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryder, S.P.; Recht, M.I.; Williamson, J.R. Quantitative Analysis of Protein-RNA Interactions by Gel Mobility Shift. Methods Mol. Biol. 2008, 488, 99–115. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, J.A.; Kugel, J.F. Studying the Affinity, Kinetic Stability, and Specificity of RNA/Protein Interactions: SINE NcRNA/Pol II Complexes as a Model System. Methods Mol. Biol. 2015, 1206, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Popova, V.V.; Orlova, A.V.; Kurshakova, M.M.; Nikolenko, J.V.; Nabirochkina, E.N.; Georgieva, S.G.; Kopytova, D.V. The Role of SAGA Coactivator Complex in SnRNA Transcription. Cell Cycle 2018, 17, 1859–1870. [Google Scholar] [CrossRef] [Green Version]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [Green Version]
- Tuvshinjargal, N.; Lee, W.; Park, B.; Han, K. PRIdictor: Protein-RNA Interaction Predictor. Biosystems 2016, 139, 17–22. [Google Scholar] [CrossRef]
- Li, S.; Yamashita, K.; Amada, K.M.; Standley, D.M. Quantifying Sequence and Structural Features of Protein-RNA Interactions. Nucleic Acids Res. 2014, 42, 10086–10098. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vdovina, Y.A.; Kurshakova, M.M.; Georgieva, S.G.; Kopytova, D.V. PCID2 Subunit of the Drosophila TREX-2 Complex Has Two RNA-Binding Regions. Curr. Issues Mol. Biol. 2023, 45, 5662-5676. https://doi.org/10.3390/cimb45070357
Vdovina YA, Kurshakova MM, Georgieva SG, Kopytova DV. PCID2 Subunit of the Drosophila TREX-2 Complex Has Two RNA-Binding Regions. Current Issues in Molecular Biology. 2023; 45(7):5662-5676. https://doi.org/10.3390/cimb45070357
Chicago/Turabian StyleVdovina, Yulia A., Maria M. Kurshakova, Sofia G. Georgieva, and Daria V. Kopytova. 2023. "PCID2 Subunit of the Drosophila TREX-2 Complex Has Two RNA-Binding Regions" Current Issues in Molecular Biology 45, no. 7: 5662-5676. https://doi.org/10.3390/cimb45070357
APA StyleVdovina, Y. A., Kurshakova, M. M., Georgieva, S. G., & Kopytova, D. V. (2023). PCID2 Subunit of the Drosophila TREX-2 Complex Has Two RNA-Binding Regions. Current Issues in Molecular Biology, 45(7), 5662-5676. https://doi.org/10.3390/cimb45070357





