Unveiling Nilaparvata lugens Stål Genes Defining Compatible and Incompatible Interactions with Rice through Transcriptome Analysis and Gene Silencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Insect Materials
2.2. BPH Bioassays
2.3. Transcriptome Sequencing and Analysis
2.4. Expression Analysis via RT-qPCR
2.5. Analysis of Glucose and Trehalose Contents in BPH
2.6. Double-Stranded RNA (dsRNA) Synthesis and RNAi Assays in BPH
2.7. Statistical Analysis
3. Results
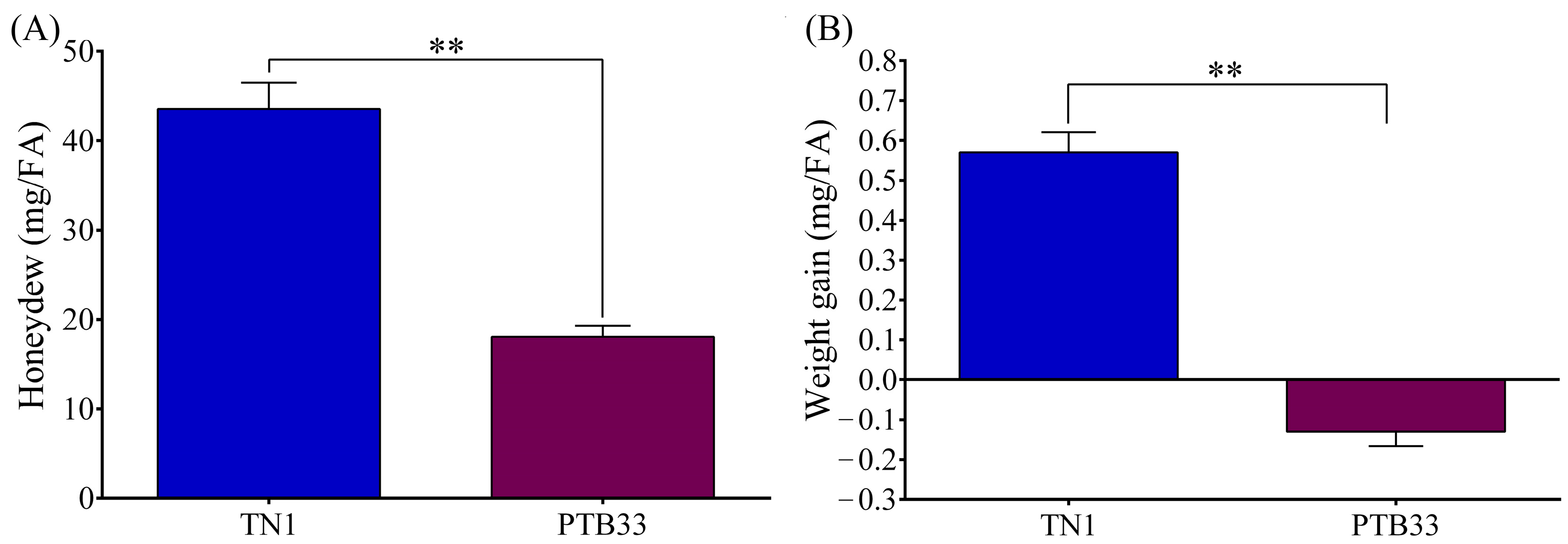
3.1. BPH Performance on TN1 and PTB33 Rice
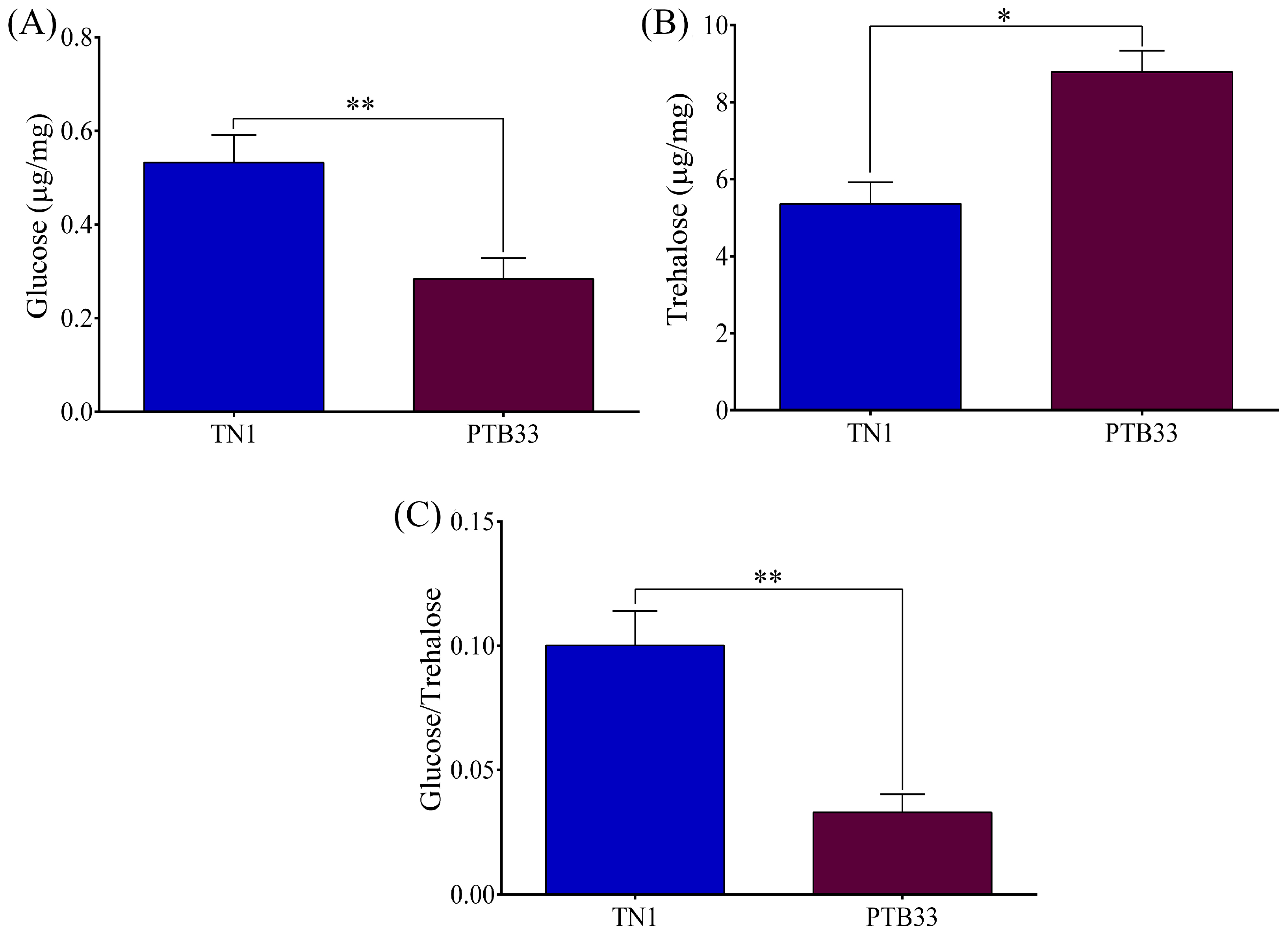
3.2. Analysis of Glucose and Trehalose Content in the BPH Infested on TN1 and PTB33 Rice
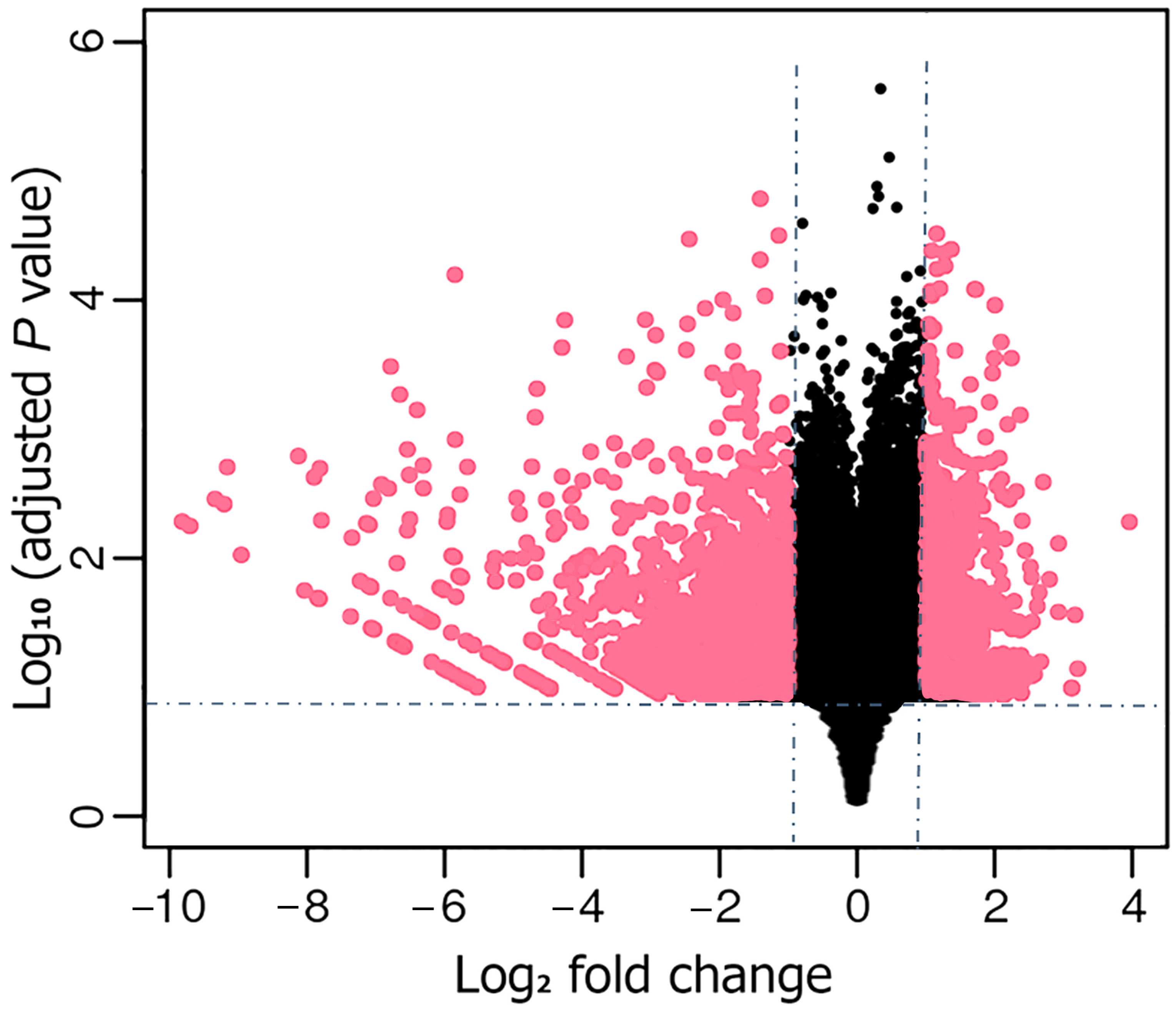
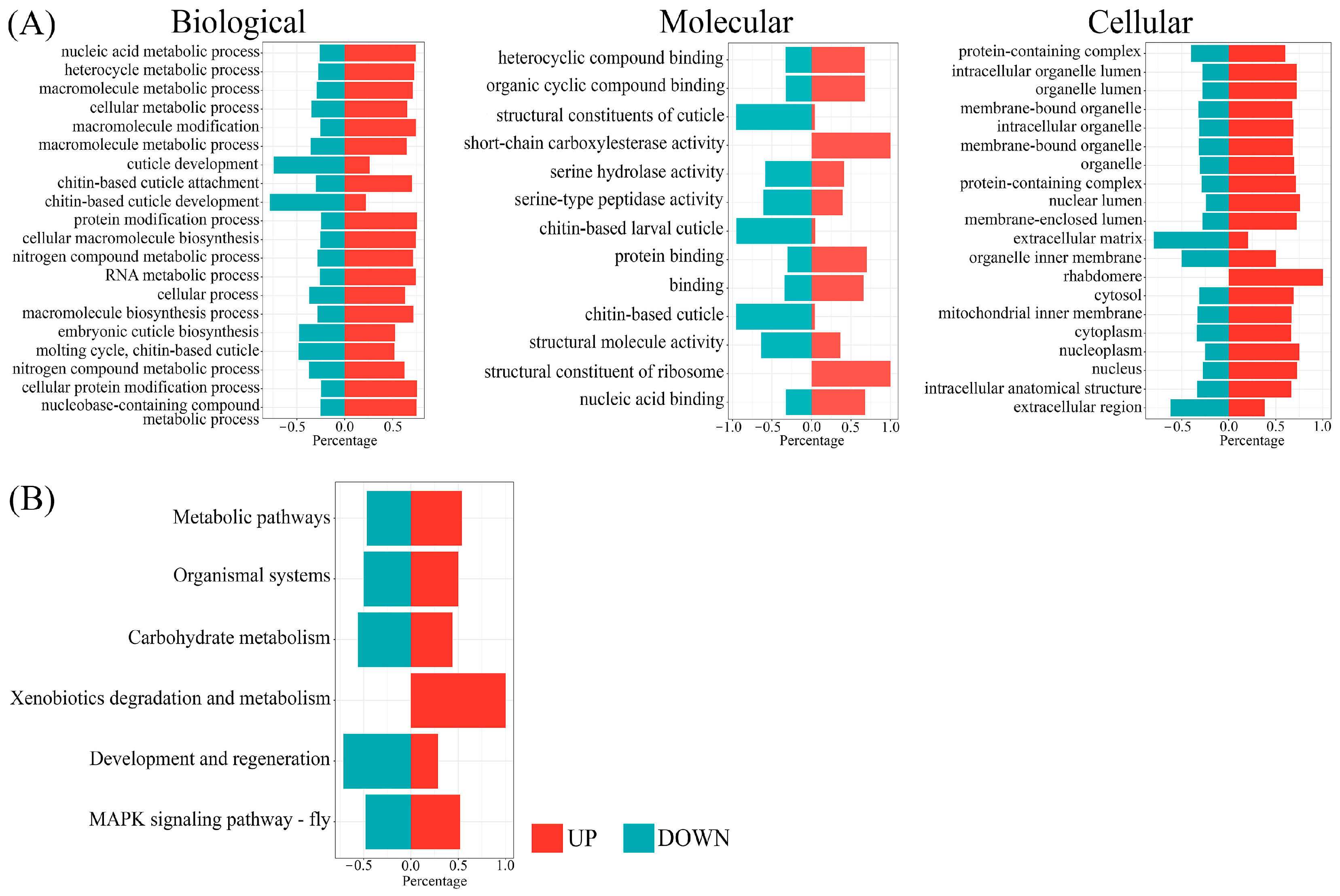
3.3. Identification and Analysis of DEGs
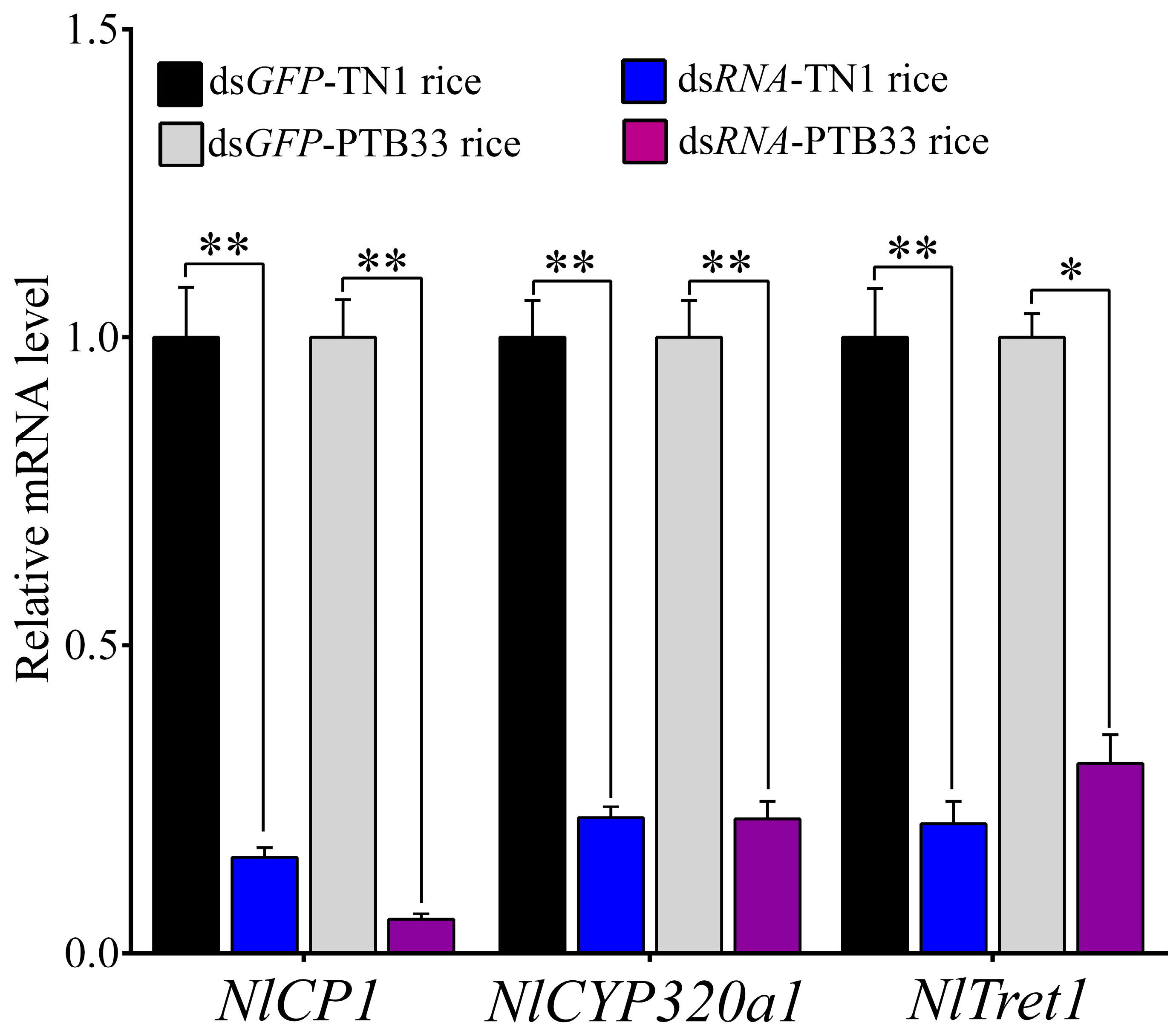
3.4. Silencing of Selected DEGs and Effect Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nanda, S.; Wan, P.J.; Yuan, S.-Y.; Lai, F.-X.; Wang, W.-X.; Fu, Q. Differential responses of OsMPKs in IR56 rice to two BPH populations of different virulence levels. Int. J. Mol. Sci. 2018, 19, 4030. [Google Scholar] [CrossRef] [PubMed]
- Cabauatan, P.Q.; Cabunagan, R.C.; Choi, I.R. Rice viruses transmitted by the brown planthopper Nilaparvata lugens Stål. In Planthoppers: New Threats to the Sustainability to of Intensive Rice Production Systems in Asia; Heong, K.L., Hardy, B., Eds.; International Rice Research Institute: Los Baños, Philippines, 2010; pp. 357–368. [Google Scholar]
- Sani Haliru, B.; Rafii, M.Y.; Mazlan, N.; Ramlee, S.I.; Muhammad, I.; Silas Akos, I.; Halidu, J.; Swaray, S.; Rini Bashir, Y. Recent strategies for detection and improvement of brown planthopper resistance genes in rice: A review. Plants 2020, 9, 1202. [Google Scholar] [CrossRef] [PubMed]
- Athwal, D.S.; Pathak, M.D.; Bacalangco, E.H.; Pura, C.D. Genetics of resistance to brown planthoppers and green leafhoppers in Oryza sativa L. Crop Sci. 1971, 11, 747–750. [Google Scholar] [CrossRef]
- Li, C.-P.; Wu, D.-H.; Huang, S.-H.; Meng, M.; Shih, H.-T.; Lai, M.-H.; Chen, L.-J.; Jena, K.K.; Hechanova, S.L.; Ke, T.-J.; et al. The Bph45 gene confers resistance against brown planthopper in rice by reducing the production of limonene. Int. J. Mol. Sci. 2023, 24, 1798. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhu, L.; He, G. Towards understanding of molecular interactions between rice and the brown planthopper. Mol. Plant. 2013, 6, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Sogawa, K. Planthopper outbreaks in different paddy ecosystems in Asia: Man-made hopper plagues that threatened the green revolution in rice. In Rice Planthoppers: Ecology, Management, Socio Economics and Policy; Heong, K.L., Cheng, J., Escalada, M., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 33–63. [Google Scholar]
- Zheng, Y.; He, J.; Wan, P.; Lai, F.; Sun, Y.; Lin, J.; Fu, Q. Virulence characteristics of Nilaparvata lugens (Stål) reared on resistant rice variety IR56. Chin. J. Rice Sci. 2016, 30, 552–558. [Google Scholar]
- Mogilicherla, K.; Roy, A. Epigenetic regulations as drivers of insecticide resistance and resilience to climate change in arthropod pests. Front. Genet. 2023, 13, 1044980. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.M. Epigenetic responses to environmental change and their evolutionary implications. Phil. Trans. R. Soc. B 2009, 364, 3403–3418. [Google Scholar] [CrossRef]
- Zhang, J.; Xing, Y.; Li, Y.; Yin, C.; Ge, C.; Li, F. DNA methyltransferases have an essential role in female fecundity in Brown planthopper, Nilaparvata lugens. Biochem. Biophys. Res. Commun. 2015, 464, 83–88. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, J.L.; Liang, S.K.; Li, G.H.; Wang, F.H.; Ahmad, I. Differentially methylated genomic fragments related with sexual dimorphism of rice pests, Sogatella furcifera. Insect Sci. 2015, 22, 731–738. [Google Scholar] [CrossRef]
- Gupta, A.; Nair, S. Heritable epigenomic modifications influence stress resilience and rapid adaptations in the brown planthopper (Nilaparvata lugens). Int. J. Mol. Sci. 2022, 23, 8728. [Google Scholar] [CrossRef]
- Jing, S.; Zhang, L.; Ma, Y.; Liu, B.; Zhao, Y.; Yu, H.; Zhou, X.; Qin, R.; Zhu, L.; He, G. Genome-wide mapping of virulence in brown planthopper identifies loci that break down host plant resistance. PLoS ONE 2014, 9, e98911. [Google Scholar] [CrossRef]
- Kobayashi, T.; Yamamoto, K.; Suetsugu, Y.; Kuwazaki, S.; Hattori, M.; Jairin, J.; Sanada-Morimura, S.; Matsumura, M. Genetic mapping of the rice resistance-breaking gene of the brown planthopper Nilaparvata lugens. Proc. R. Soc. B Biol. Sci. 2014, 281, 20140726. [Google Scholar] [CrossRef]
- Barr, K.L.; Hearne, L.B.; Briesacher, S.; Clark, T.L.; Davis, G.E. Microbial symbionts in insects influence down-regulation of defense genes in maize. PLoS ONE 2010, 5, e11339. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Du, B.; Hao, F.; Lei, H.; Wan, Q.; He, G.; Wang, Y.; Tang, H. Dynamic metabolic responses of brown planthoppers towards susceptible and resistant rice plants. Plant Biotechnol. J. 2017, 15, 1346–1357. [Google Scholar] [CrossRef] [PubMed]
- Wan, P.-J.; Zhou, R.-N.; Nanda, S.; He, J.-C.; Yuan, S.-Y.; Wang, W.-X.; Lai, F.-X.; Fu, Q. Phenotypic and transcriptomic responses of two Nilaparvata lugens Populations to the mudgo rice containing Bph1. Sci. Rep. 2019, 9, 14049. [Google Scholar] [CrossRef]
- AB Ghaffar, M.B.; Pritchard, J.; Ford-Lloyd, B. Brown planthopper (N. lugens Stal) feeding behaviour on rice germplasm as an indicator of resistance. PLoS ONE 2011, 6, e22137. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, M.; Feng, F.; He, R. Differentially regulated genes in the salivary glands of brown planthopper after feeding in resistant versus susceptible rice varieties. Arch. Insect Biochem. Physiol. 2015, 89, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Noda, H.; Kawai, S.; Koizumi, Y.; Matsui, K.; Zhang, Q.; Furukawa, S.; Shimomura, M.; Mita, K. Annotated ESTs from various tissues of the brown planthopper Nilaparvata lugens: A genomic resource for studying agricultural pests. BMC Genom. 2008, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Yu, H.; Fu, Q.; Chen, H.; Ye, W.; Li, S.; Lou, Y. Comparative transcriptome analysis of salivary glands of two populations of rice Brown planthopper, Nilaparvata lugens, that differ in virulence. PLoS ONE 2013, 8, e79612. [Google Scholar] [CrossRef]
- Rao, W.; Zheng, X.; Liu, B.; Guo, Q.; Guo, J.; Wu, Y.; Shangguan, X.; Wang, H.; Wu, D.; Wang, Z.; et al. Secretome analysis and in planta expression of salivary proteins identify candidate effectors from the brown planthopper Nilaparvata lugens. Mol. Plant Microbe Interact. 2019, 32, 227–239. [Google Scholar] [CrossRef]
- Nanda, S.; Yuan, S.-Y.; Lai, F.-X.; Wang, W.-X.; Fu, Q.; Wan, P.-J. Identification and analysis of MiRNAs in IR56 rice in response to BPH infestations different virulence levels. Sci. Rep. 2020, 10, 19093. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, D.; Gao, D.; Zhao, W.; Du, H.; Qiu, Z.; Huang, J.; Wen, P.; Wang, Y.; Li, Q.; et al. Cytokinin confers brown planthopper resistance by elevating jasmonic acid pathway in rice. Int. J. Mol. Sci. 2022, 23, 5946. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Yuan, L.-Y.; Li, Y.-F.; Xiao, H.-X.; Li, Y.-F.; Zhang, Y.; Wu, W.-J.; Zhang, Z.-F. Salivary protein 7 of the brown planthopper functions as an effector for mediating tricin metabolism in rice plants. Sci. Rep. 2022, 12, 3205. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Shi, Y.; Wang, L.; Tian, T.; Li, J.; Gong, L.; Zheng, Z.; Jing, M.; Fang, J.; Ji, R. Planthopper-secreted salivary calmodulin acts as an effector for defense responses in rice. Front. Plant. Sci. 2022, 13, 841378. [Google Scholar] [CrossRef]
- Guo, J.; Wang, H.; Guan, W.; Guo, Q.; Wang, J.; Yang, J.; Peng, Y.; Shan, J.; Gao, M.; Shi, S.; et al. A tripartite rheostat controls self-regulated host plant resistance to insects. Nature 2023, 618, 799–807. [Google Scholar] [CrossRef]
- Ten Broeke, C.J.M.; Dicke, M.; van Loon, J.J.A. Feeding behaviour and performance of different populations of the black currant-tettuce aphid, Nasonovia ribisnigri, on resistant and susceptible Lettuce. Entomol. Exp. Appl. 2013, 148, 130–141. [Google Scholar] [CrossRef]
- Gu, J.; Shao, Y.; Zhang, C.; Liu, Z.; Zhang, Y. Characterization of putative soluble and membrane-bound trehalases in a hemipteran Insect, Nilaparvata lugens. J. Insect Physiol. 2009, 55, 997–1002. [Google Scholar] [CrossRef]
- Peng, L.; Zhao, Y.; Wang, H.; Zhang, J.; Song, C.; Shangguan, X.; Zhu, L.; He, G. comparative metabolomics of the interaction between rice and the brown planthopper. Metabolomics 2016, 12, 132. [Google Scholar] [CrossRef]
- Hao, P.; Liu, C.; Wang, Y.; Chen, R.; Tang, M.; Du, B.; Zhu, L.; He, G. Herbivore-induced callose deposition on the sieve plates of rice: An important mechanism for host resistance. Plant Physiol. 2008, 146, 1810–1820. [Google Scholar] [CrossRef]
- Li, C.; Xiong, Z.; Fang, C.; Liu, K. Transcriptome and metabolome analyses reveal the responses of brown planthoppers to RH resistant rice cultivar. Front. Physiol. 2022, 13, 1018470. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Jia, H.; Li, Z.; Liu, Y.; Hou, M. Transcriptomic and expression analysis of the salivary glands in brown planthoppers, Nilaparvata lugens (Hemiptera: Delphacidae). J. Econ. Entomol. 2018, 111, 2884–2893. [Google Scholar] [CrossRef] [PubMed]
- Kikuta, S.; Nakamura, Y.; Hattori, M.; Sato, R.; Kikawada, T.; Noda, H. Herbivory-induced glucose transporter gene expression in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 2015, 64, 60–67. [Google Scholar] [CrossRef]
- Després, L.; David, J.-P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef]
- Ross, M.K.; Streit, T.M.; Herring, K.L. Carboxylesterases: Dual roles in lipid and pesticide metabolism. J. Pestic. Sci. 2010, 35, 257–264. [Google Scholar] [CrossRef]
- Bao, Y.-Y.; Wang, Y.; Wu, W.-J.; Zhao, D.; Xue, J.; Zhang, B.-Q.; Shen, Z.-C.; Zhang, C.-X. De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics 2012, 99, 256–264. [Google Scholar] [CrossRef]
- Kehr, J. Phloem sap proteins: Their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 2006, 57, 767–774. [Google Scholar] [CrossRef]
- Ye, W.; Yu, H.; Jian, Y.; Zeng, J.; Ji, R.; Chen, H.; Lou, Y. A salivary EF-Hand calcium-binding protein of the brown planthopper Nilaparvata lugens functions as an effector for defense responses in Rice. Sci. Rep. 2017, 7, 40498. [Google Scholar] [CrossRef]
- Chen, S.; Nanda, S.; Guo, M.; Kong, L.; Yang, C.; Liu, Z.; Gao, R.; Qiu, B.; Zhang, Y.; Zhou, X.; et al. Tyrosine hydroxylase involved in cuticle tanning and reproduction in the 28-Spotted Potato Ladybeetle, Henosepilachna vigintioctopunctata. Pest. Manag. Sci. 2022, 78, 3859–3870. [Google Scholar] [CrossRef]
- Zhang, W.; Liang, G.; Ma, L.; Jiang, T.; Xiao, H. Dissecting the role of juvenile hormone binding protein in response to hormone and starvation in the cotton bollworm, Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae). J. Econ. Entomol. 2019, 112, 1411–1417. [Google Scholar] [CrossRef]
- Kim, I.H.; Castillo, J.C.; Aryan, A.; Martin-Martin, I.; Nouzova, M.; Noriega, F.G.; Barletta, A.B.F.; Calvo, E.; Adelman, Z.N.; Ribeiro, J.M.C.; et al. A mosquito juvenile hormone binding protein (MJHBP) regulates the activation of innate immune defenses and hemocyte development. PLoS Pathog. 2020, 16, e1008288. [Google Scholar] [CrossRef]
- Muduli, L.; Pradhan, S.K.; Mishra, A.; Bastia, D.N.; Samal, K.C.; Agrawal, P.K.; Dash, M. Understanding brown planthopper resistance in rice: Genetics, biochemical and molecular breeding approaches. Rice Sci. 2021, 28, 532–546. [Google Scholar] [CrossRef]
- Balabanidou, V.; Grigoraki, L.; Vontas, J. Insect cuticle: A critical determinant of insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 68–74. [Google Scholar] [CrossRef]
- Nauen, R.; Bass, C.; Feyereisen, R.; Vontas, J. The role of cytochrome P450s in insect toxicology and resistance. Annu. Rev. Entomol. 2022, 67, 105–124. [Google Scholar] [CrossRef] [PubMed]


| Sample Name | Raw Sequence | Trimmed Sequence | % GC | Average Length | % Aligned |
|---|---|---|---|---|---|
| BPH_PTB33 | 34.2 million | 34.18 million | 35 | 141 | 92 |
| BPH_TN1 | 35.08 million | 34.97 million | 35 | 140 | 91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rout, P.; Ravindranath, N.; Gaikwad, D.; Nanda, S. Unveiling Nilaparvata lugens Stål Genes Defining Compatible and Incompatible Interactions with Rice through Transcriptome Analysis and Gene Silencing. Curr. Issues Mol. Biol. 2023, 45, 6790-6803. https://doi.org/10.3390/cimb45080429
Rout P, Ravindranath N, Gaikwad D, Nanda S. Unveiling Nilaparvata lugens Stål Genes Defining Compatible and Incompatible Interactions with Rice through Transcriptome Analysis and Gene Silencing. Current Issues in Molecular Biology. 2023; 45(8):6790-6803. https://doi.org/10.3390/cimb45080429
Chicago/Turabian StyleRout, Priyadarshini, Nihal Ravindranath, Dinkar Gaikwad, and Satyabrata Nanda. 2023. "Unveiling Nilaparvata lugens Stål Genes Defining Compatible and Incompatible Interactions with Rice through Transcriptome Analysis and Gene Silencing" Current Issues in Molecular Biology 45, no. 8: 6790-6803. https://doi.org/10.3390/cimb45080429
APA StyleRout, P., Ravindranath, N., Gaikwad, D., & Nanda, S. (2023). Unveiling Nilaparvata lugens Stål Genes Defining Compatible and Incompatible Interactions with Rice through Transcriptome Analysis and Gene Silencing. Current Issues in Molecular Biology, 45(8), 6790-6803. https://doi.org/10.3390/cimb45080429






