Transcriptomic and Hormone Analyses Provide Insight into the Regulation of Axillary Bud Outgrowth of Eucommia ulmoides Oliver
Abstract
:1. Introduction
2. Materials and Methods
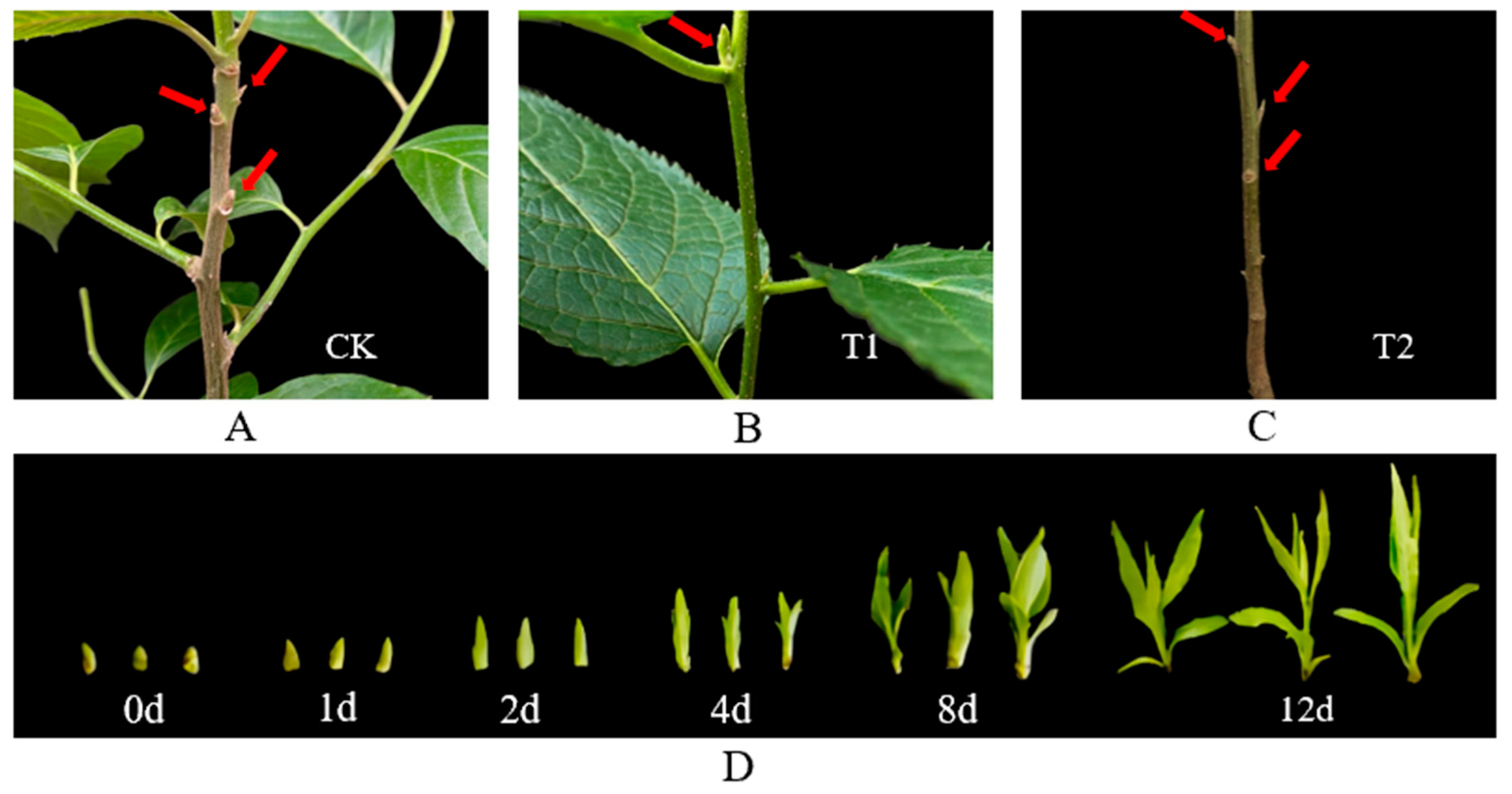
2.1. Plant Materials and Growth Conditions
2.2. RNA Extraction, Library Preparation, and Transcriptome Sequencing
2.3. De Novo Assembly and Functional Annotation Analysis of Illumina Sequencing
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. Measurements of Relevant Hormone Contents
2.6. Quantitative Real-Time PCR Validation
3. Results
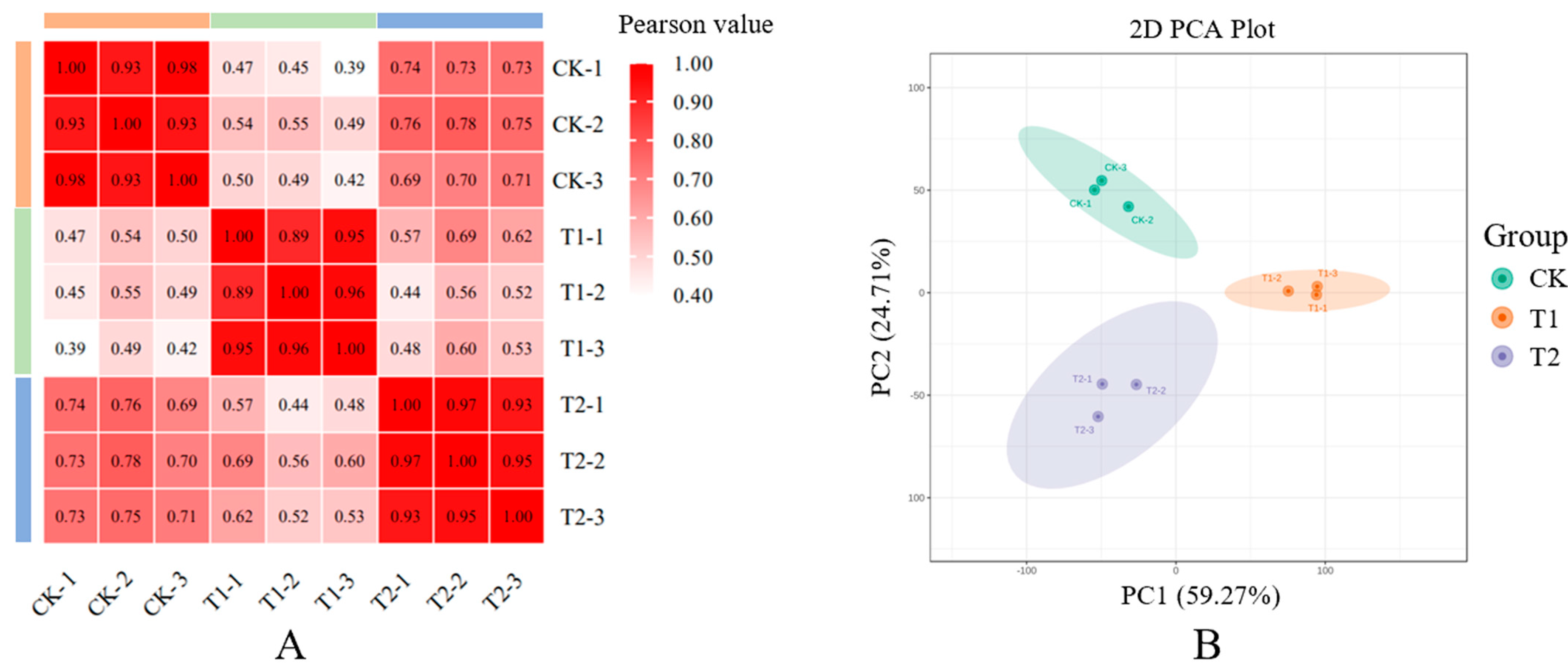
3.1. Quality Assessment and Repeat Correlation Analysis of RNA-seq Data
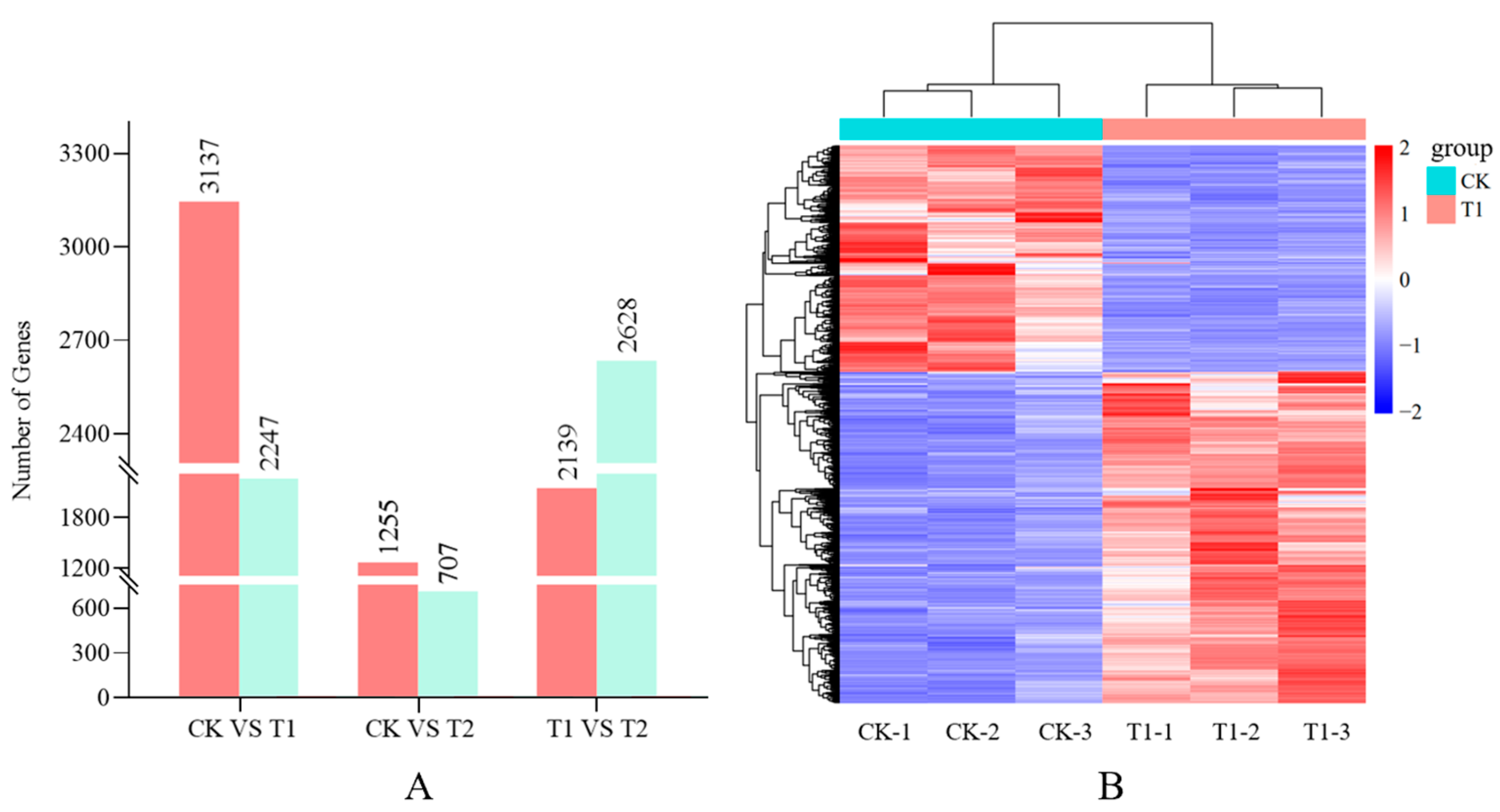
3.2. Analysis of DEGs in Different Comparison Groups
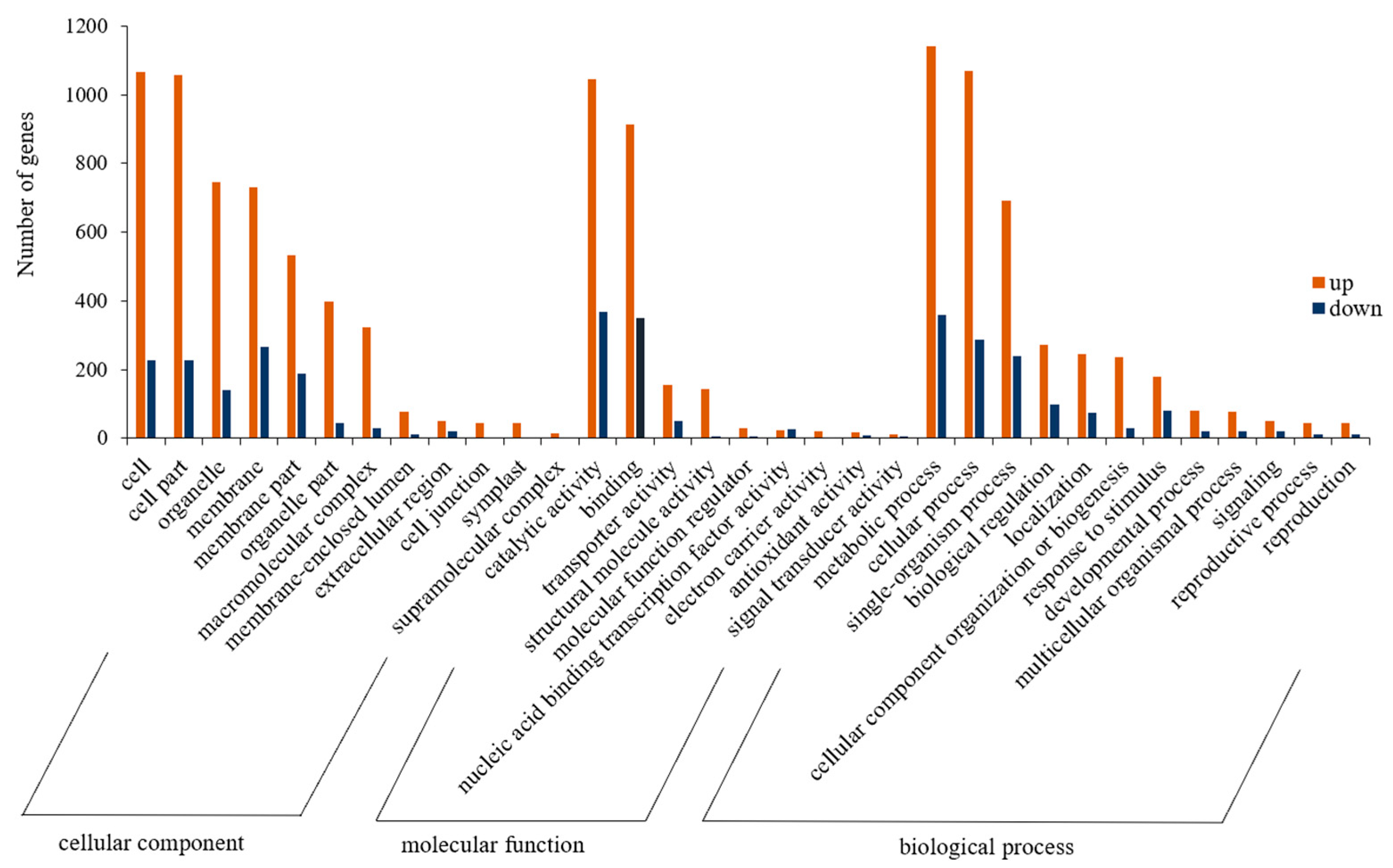
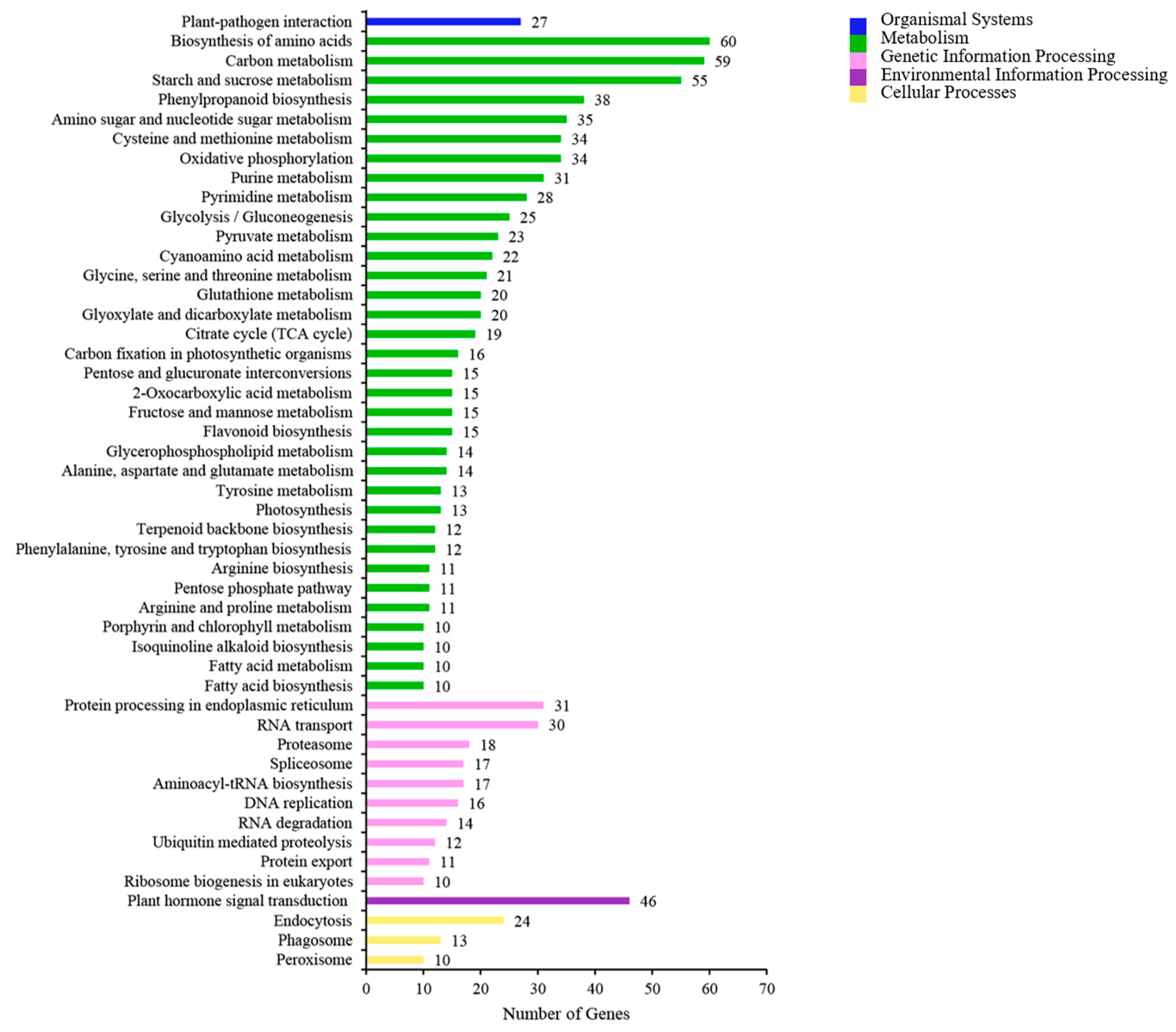
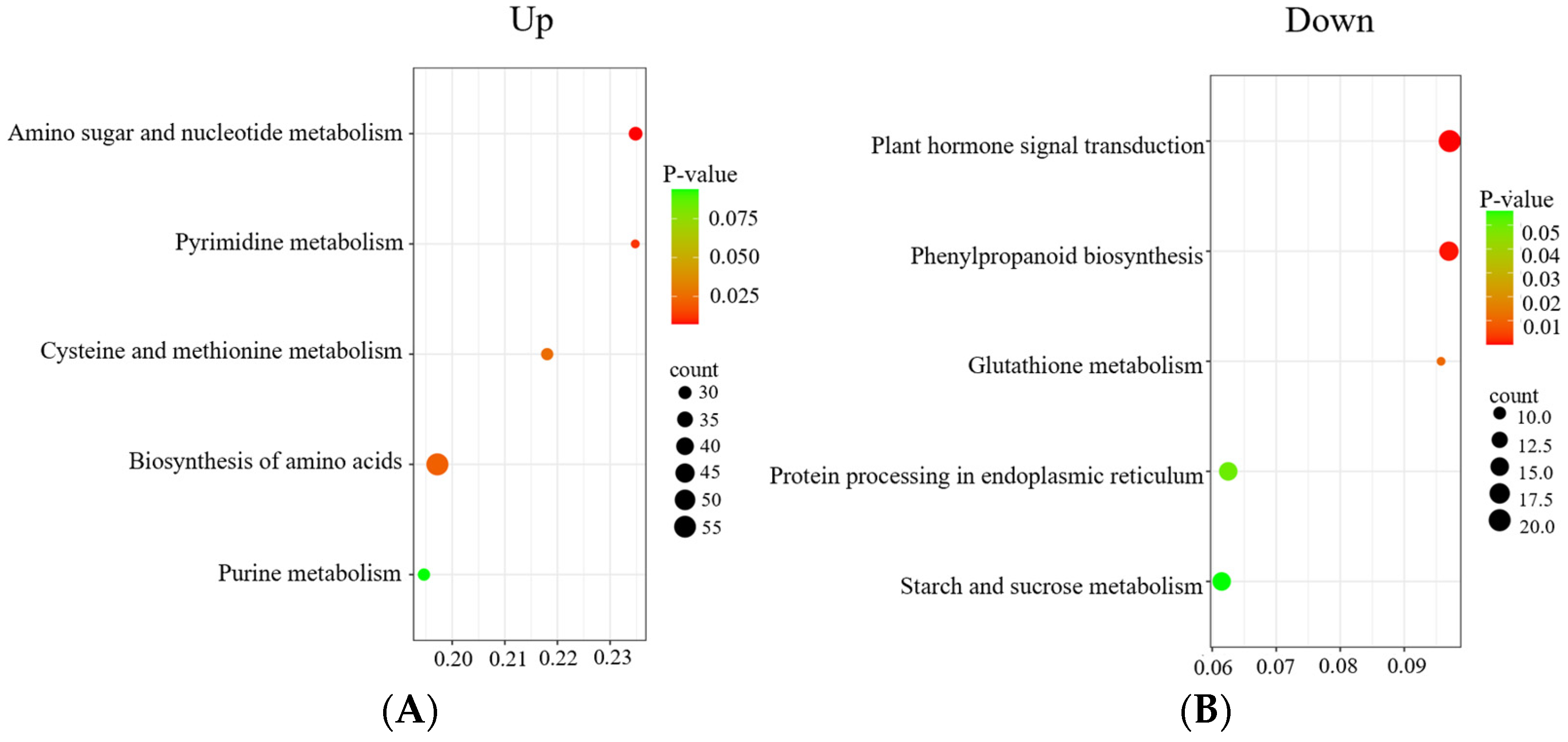
3.3. GO and KEGG Enrichment Analysis of DEGs
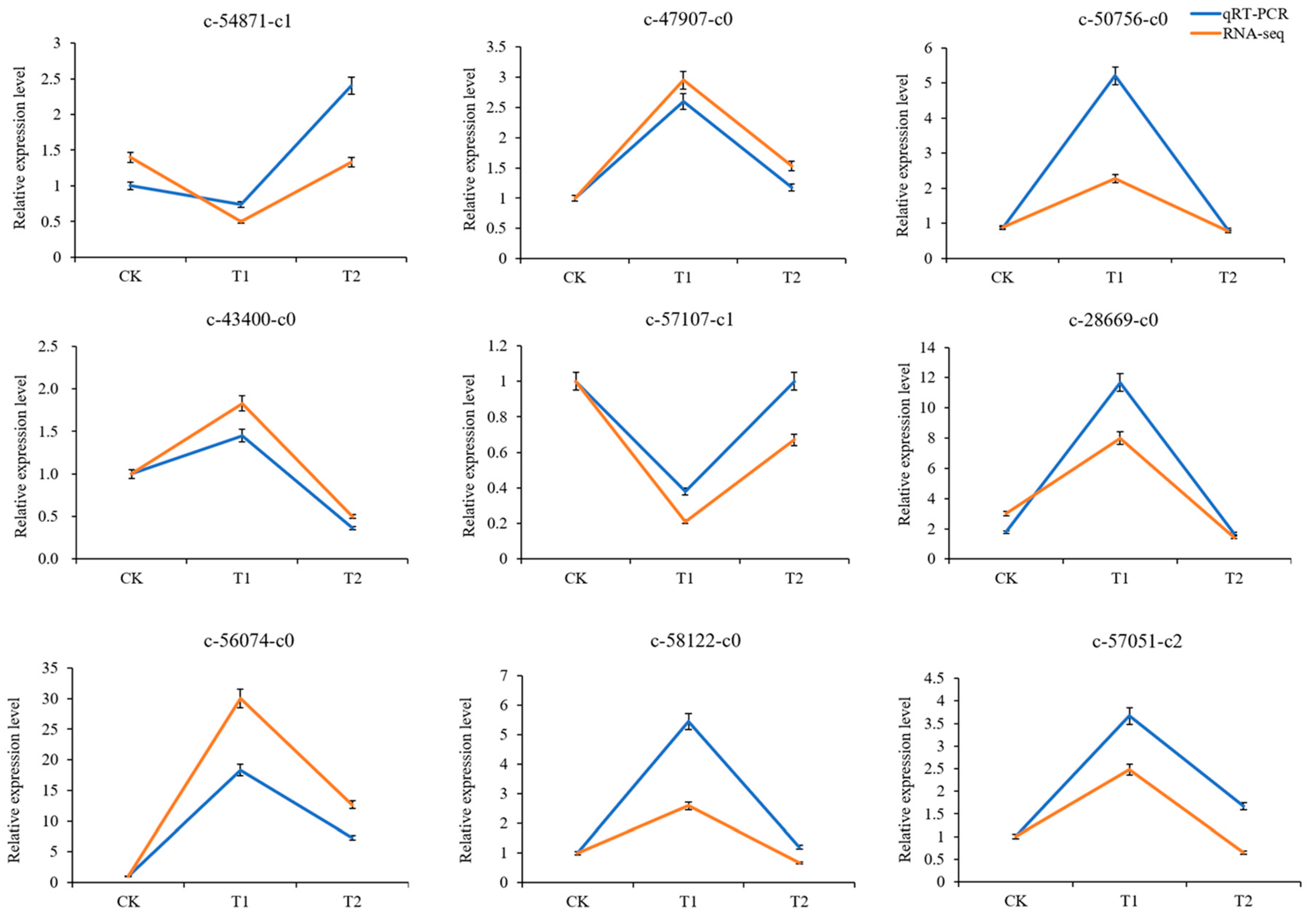
3.4. Validation of RNA-seq Data by RT‒qPCR
3.5. Quantification of Changes in Endogenous Plant Hormone Contents during Bud Development in E. ulmoides
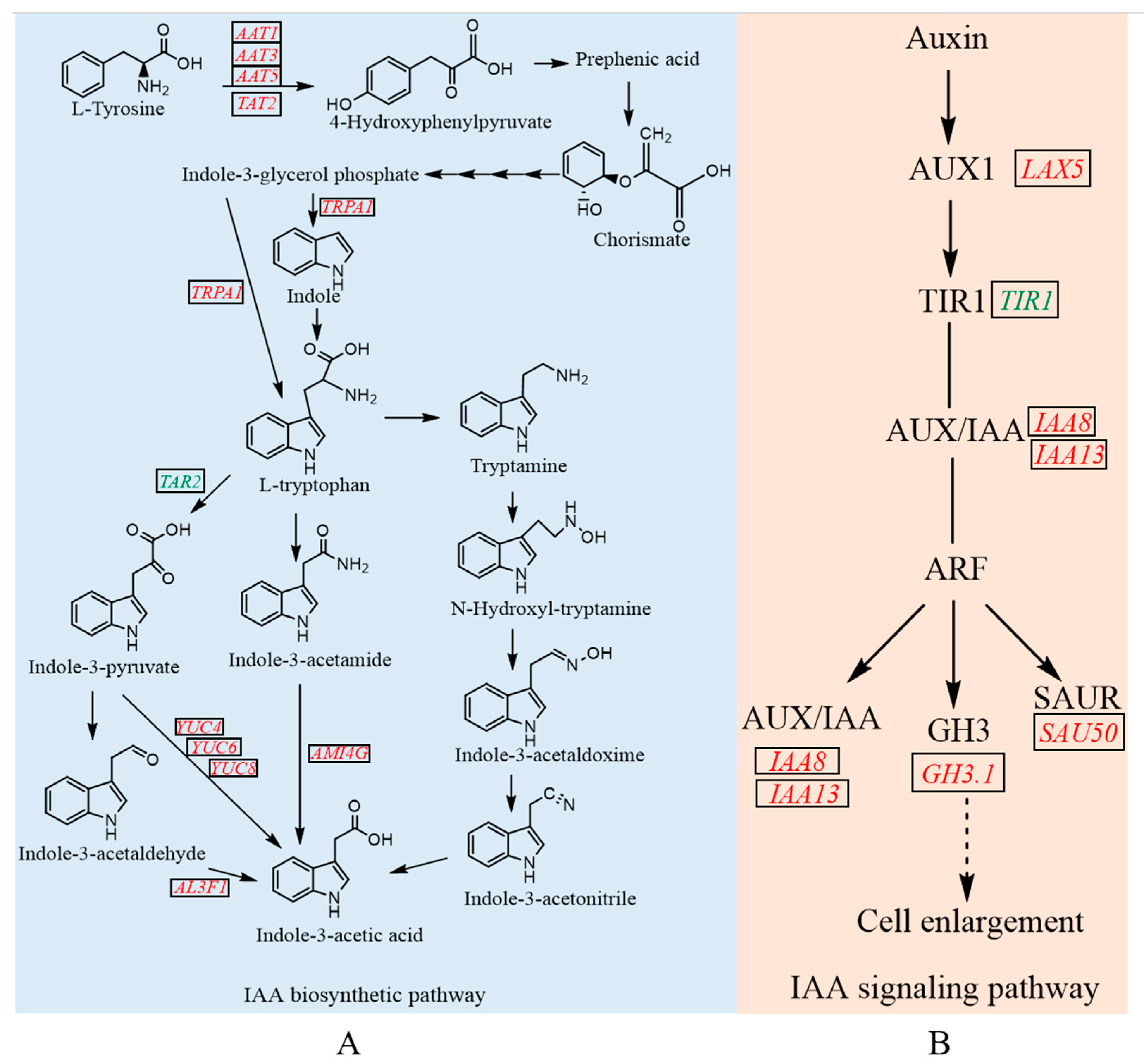
3.6. The Integrated Analysis of DEGs Related to IAA Biosynthesis and Signaling Pathway in Tip Bud
3.7. Validation and Expression Analysis of Key Enzyme Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Tang, L.; Li, L.; Zhou, Q.; Li, Y.; Li, J.; Wang, Y. Geographic authentication of Eucommia ulmoides leaves using multivariate analysis and preliminary study on the compositional response to environment. Front. Plant Sci. 2020, 11, 79. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, K.; Mo, L.; Ahmad, A.; Wang, D.; Rong, Z.; Peng, H.; Cai, H.; Liu, G. Eucommia ulmoides polysaccharides attenuate rabbit osteoarthritis by regulating the function of macrophages. Front. Pharmacol. 2021, 12, 730557. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, J.; Li, M.; Hao, D.; Yang, Y.; Zhang, C.; He, R.; Tao, R. Eucommia ulmoides Oliv.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2014, 151, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lyu, Q.; Zheng, W.; Yang, Q.; Cao, G. Traditional application and modern pharmacological research of Eucommia ulmoides Oliv. Chin. Med. 2021, 16, 73. [Google Scholar] [CrossRef] [PubMed]
- Do, M.; Hur, J.; Choi, J.; Kim, M.; Kim, M.; Kim, Y.; Ha, S. Eucommia ulmoides ameliorates glucotoxicity by suppressing advanced glycation end-products in diabetic mice kidney. Nutrients 2018, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 741. [Google Scholar] [CrossRef]
- Ward, S.P.; Leyser, O. Shoot branching. Curr. Opin. Plant Biol. 2004, 7, 73–78. [Google Scholar] [CrossRef]
- Tarancón, C.; González-Grandío, E.; Oliveros, J.C.; Nicolas, M.; Cubas, P. A conserved carbon starvation response underlies bud dormancy in woody and herbaceous species. Front. Plant Sci. 2017, 8, 788. [Google Scholar] [CrossRef]
- Seale, M.; Bennett, T.; Leyser, O. BRC1 expression regulates bud activation potential, but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 2017, 144, 161–1673. [Google Scholar]
- Porcher, A.; Guérin, V.; Leduc, N.; Lebrec, A.; Lothier, J.; Vian, A. Ascorbate–glutathione pathways mediated by cytokinin regulate H2O2 levels in light-controlled rose bud burst. Plant Physiol. 2021, 186, 910–928. [Google Scholar] [CrossRef]
- Beveridge, C.A. Axillary bud outgrowth: Sending a message. Curr. Opin. Plant Biol. 2006, 9, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Kudo, T.; Kojima, M.; Sakakibara, H. Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 2011, 62, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Barraza, A.; Cabrera-Ponce, J.L.; Gamboa-Becerra, R.; Luna-Martínez, F.; Winkler, R.; Álvarez-Venegas, R. The phaseolus vulgaris PvTRX1h gene regulates plant hormone biosynthesis in embryogenic callus from common bean. Front. Plant Sci. 2015, 6, 577. [Google Scholar] [CrossRef]
- Skalický, V.; Vojtková, T.; Pěnčík, A.; Vrána, J.; Juzoń, K.; Koláčková, V.; Sedlářová, M.; Kubeš, M.F.; Novák, O. Auxin metabolite profiling in isolated and intact plant nuclei. Int. J. Mol. Sci. 2021, 22, 12369. [Google Scholar] [CrossRef]
- Salinas-Grenet, H.; Herrera-Vásquez, A.; Parra, S.; Cortez, A.; Gutiérrez, L.; Pollmann, S.; León, G.; Blanco-Herrera, F. Modulation of auxin levels in pollen grains affects stamen development and anther dehiscence in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2480. [Google Scholar] [CrossRef]
- Waqas, M.; Khan, A.L.; Shahzad, R.; Ullah, I.; Khan, A.R.; Lee, I.J. Mutualistic fungal endophytes produce phytohormones and organic acids that promote japonica rice plant growth under prolonged heat stress. J. Zhejiang Univ. Sci. B 2015, 16, 1011–1018. [Google Scholar] [CrossRef]
- Parmar, S.; Sharma, V.K.; Li, T.; Tang, W.; Li, H. Fungal seed endophyte FZT214 improves dysphania ambrosioides cd tolerance throughout different developmental stages. Front. Microbiol. 2022, 12, 783475. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Jain, M.; Garg, R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 2014, 5, 789. [Google Scholar] [CrossRef]
- Cackett, L.; Cannistraci, C.V.; Meier, S.; Ferrandi, P.; Pěnčík, A.; Gehring, C.; Novák, O.; Ingle, R.A.; Donaldson, L. Salt-specific gene expression reveals elevated auxin levels in Arabidopsis thaliana plants grown under saline conditions. Front. Plant Sci. 2022, 13, 804716. [Google Scholar] [CrossRef]
- Asselbergh, B.; De Vleesschauwer, D.; Höfte, M. Global switches and fine-tuning-aba modulates plant pathogen defense. Mol. Plant Microbe Interact. 2008, 21, 709–719. [Google Scholar] [CrossRef]
- Hamayun, M.; Hussain, A.; Khan, S.A.; Irshad, M.; Khan, A.L.; Waqas, M.; Shahzad, R.; Iqbal, A.; Ullah, N.; Rehman, G.; et al. Kinetin modulates physio-hormonal attributes and isoflavone contents of Soybean grown under salinity stress. Front. Plant Sci. 2015, 6, 377. [Google Scholar] [CrossRef] [PubMed]
- Ling, A.P.; Tan, K.P.; Hussein, S. Comparative effects of plant growth regulators on leaf and stem explants of Labisia pumila var. Alata. J. Zhejiang Univ. Sci. B 2013, 14, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Chopra, R.; Burow, G.; Burke, J.J.; Gladman, N.; Xin, Z. Genome-wide association analysis of seedling traits in diverse sorghum germplasm under thermal stress. BMC Plant Biol. 2017, 17, 12. [Google Scholar] [CrossRef]
- Zhang, W.; Xia, L.; Peng, F.; Song, C.; Manzoor, M.A.; Cai, Y.; Jin, Q. Transcriptomics and metabolomics changes triggered by exogenous 6-benzylaminopurine in relieving epicotyl dormancy of Polygonatum cyrtonema Hua seeds. Front. Plant Sci. 2022, 13, 961899. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Khan, T.A.; Shahzad, S.; Ullah, S.; Shahzadi, I.; Ali, A.; Akram, W.; Yasin, N.A.; Yusuf, M. Bioclay nanosheets infused with GA3 ameliorate the combined stress of hexachlorobenzene and temperature extremes in Brassica alboglabra plants. Front. Plant Sci. 2022, 13, 964041. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, C.; Hou, L.; Wu, X.; Wang, D.; Zhang, L.; Liu, P. Exogenous sa affects rice seed germination under salt stress by regulating Na+/K+ balance and endogenous gas and aba homeostasis. Int. J. Mol. Sci. 2022, 23, 3293. [Google Scholar] [CrossRef]
- Balzan, S.; Johal, G.S.; Carraro, N. The role of auxin transporters in monocots development. Front. Plant Sci. 2014, 5, 393. [Google Scholar] [CrossRef]
- Wei, W.; Inaba, J.; Zhao, Y.; Mowery, J.D.; Hammond, R. Phytoplasma infection blocks starch breakdown and triggers chloroplast degradation, leading to premature leaf senescence, sucrose reallocation, and spatiotemporal redistribution of phytohormones. Int. J. Mol. Sci. 2022, 23, 1810. [Google Scholar] [CrossRef]
- Rubio-Moraga, A.; Ahrazem, O.; Pérez-Clemente, R.M.; Gómez-Cadenas, A.; Yoneyama, K.; López-Ráez, J.A.; Molina, R.V.; Gómez-Gómez, L. Apical dominance in saffron and the involvement of the branching enzymes CCD7 and CCD8 in the control of bud sprouting. BMC Plant Biol. 2014, 14, 171. [Google Scholar] [CrossRef]
- Wild, M.; Davière, J.; Cheminant, S.; Regnault, T.; Baumberger, N.; Heintz, D.; Baltz, R.; Genschik, P.; Achard, P. The Arabidopsis DELLARGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 2012, 24, 3307–3319. [Google Scholar] [CrossRef]
- Mader, J.C.; Emery, R.J.N.; Turnbull, C.G.N. Spatial and temporal changes in multiple hormone groups during lateral bud release shortly following apex decapitation of chickpea (Cicer arietinum) seedlings. Physiol. Plant. 2003, 119, 295–308. [Google Scholar] [CrossRef]
- Tamas, I.A.; Ozbun, J.L.; Wallace, D.H. Effect of fruits on dormancy and abscisic acid concentration in the axillary buds of Phaseolus vulgaris L. Plant Physiol. 1979, 64, 615–619. [Google Scholar] [CrossRef]
- Bowler, C.S.Z.N.; Chua, N.H. Emerging themes of plant signal transduction. Plant Cell 1994, 6, 1529–1541. [Google Scholar]
- Chai, L.; Chai, P.; Chen, S.; Flaishman, M.A.; Ma, H. Transcriptome analysis unravels spatiotemporal modulation of phytohormone-pathway expression underlying gibberellin-induced parthenocarpic fruit set in San Pedro-type fig (Ficus carica L.). BMC Plant Biol. 2018, 18, 100. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.P.G.E.; Theologis, A. Early genes and auxin action. Plant Physiol. 1996, 111, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Tente, E.; Ereful, N.; Rodriguez, A.C.; Grant, P.; O Sullivan, D.M.; Boyd, L.A.; Gordon, A. Reprogramming of the wheat transcriptome in response to infection with Claviceps purpurea, the causal agent of ergot. BMC Plant Biol. 2021, 21, 316. [Google Scholar] [CrossRef]
- Peng, Z.; Li, W.; Gan, X.; Zhao, C.; Paudel, D.; Su, W.; Lv, J.; Lin, S.; Liu, Z.; Yang, X. Genome-wide analysis of saur gene family identifies a candidate associated with fruit size in loquat (Eriobotrya japonica Lindl.). Int. J. Mol. Sci. 2022, 23, 13271. [Google Scholar] [CrossRef]
- Luo, W.; Xiao, N.; Wu, F.; Mo, B.; Kong, W.; Yu, Y. Genome-wide identification and characterization of YUCCA gene family in mikania micrantha. Int. J. Mol. Sci. 2022, 23, 13037. [Google Scholar] [CrossRef]
- Liu, H.; Fu, J.; Du, H.; Hu, J.; Wuyun, T. De novo sequencing of Eucommia ulmoides flower bud transcriptomes for identification of genes related to floral development. Genom. Data 2016, 9, 105–110. [Google Scholar] [CrossRef]
- Li, L.; Liu, M.; Shi, K.; Yu, Z.; Zhou, Y.; Fan, R.; Shi, Q. Dynamic changes in metabolite accumulation and the transcriptome during leaf growth and development in Eucommia ulmoides. Int. J. Mol. Sci. 2019, 20, 4030. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, L.; Fu, J.; Li, F.; Wang, L.; Tan, X.; Mo, W.; Cao, H. Characterization of glycolytic pathway genes using RNA-Seq in developing kernels of Eucommia ulmoides. J. Agric. Food Chem. 2016, 64, 3712–3731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, K.; Zeng, S.; Teixeira Da Silva, J.A.; Zhao, X.; Tian, C.; Xia, H.; Duan, J. Transcriptome analysis of Cymbidium sinense and its application to the identification of genes associated with floral devel-opment. BMC Genom. 2013, 14, 279. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Mori, H. Control of outgrowth and dormancy in axillary buds. Plant Physiol. 2001, 127, 1405–1413. [Google Scholar] [CrossRef]
- Muntha, S.T.; Zhang, L.; Zhou, Y.; Zhao, X.; Hu, Z.; Yang, J.; Zhang, M. Phytochrome a signal transduction 1 and CONSTANS-LIKE 13 coordinately orchestrate shoot branching and flowering in leafy Brassica juncea. Plant Biotechnol. J. 2019, 17, 1333–1343. [Google Scholar] [CrossRef]
- Ni, J.; Gao, C.; Chen, M.; Pan, B.; Ye, K.; Xu, Z. Gibberellin promotes shoot branching in the perennial woody plant Jatroph Curcas. Plant Cell Physiol. 2015, 56, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Raya-González, J.; Ortiz-Castro, R.; Ruíz-Herrera, L.F.; Kazan, K.; López-Bucio, J. Phytochrome and flowering TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in Arabidopsis. Plant Physiol. 2014, 165, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant. J. 2001, 28, 465–474. [Google Scholar] [CrossRef]
- Ongaro, V.; Leyser, O. Hormonal control of shoot branching. J. Exp. Bot. 2007, 59, 67–74. [Google Scholar] [CrossRef]
- Balla, J.; Kalousek, P.; Reinöhl, V.; Friml, J.; Procházka, S. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J. 2011, 65, 571–577. [Google Scholar] [CrossRef]
- Gu, J.; Zeng, Z.; Wang, Y.; Lyu, Y. Transcriptome analysis of carbohydrate metabolism genes and molecular regulation of sucrose transport gene LoSUT on the flowering process of developing oriental hybrid lily ‘Sorbonne’ bulb. Int. J. Mol. Sci. 2020, 21, 3092. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, Y.; Li, J.; Ding, X.; Xiao, S.; Fan, S.; Song, Z.; Chen, W.; Li, X. Exogenous 2,4-epibrassinolide treatment maintains the quality of carambola fruit associated with enhanced antioxidant capacity and alternative respiratory metabolism. Front. Plant Sci. 2021, 12, 678295. [Google Scholar] [CrossRef]
- Zhang, X.; Luo, G.; Wang, R.; Wang, J.; Himelrick, D.G. Growth and developmental responses of seeded and seedless grape berries to shoot girdling. J. Am. Soc. Hortic. Sci. 2003, 128, 316–323. [Google Scholar] [CrossRef]
- Cai, T.; Meng, X.; Liu, X.; Liu, T.; Wang, H.; Jia, Z.; Yang, D.; Ren, X. Exogenous hormonal application regulates the occurrence of wheat tillers by changing endogenous hormones. Front. Plant Sci. 2018, 9, 1886. [Google Scholar] [CrossRef] [PubMed]
- Khaksar, G.; Sirikantaramas, S. Auxin response factor 2A is part of the regulatory network mediating fruit ripening through auxin-ethylene crosstalk in durian. Front. Plant Sci. 2020, 11, 543747. [Google Scholar] [CrossRef] [PubMed]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an auxin response factor in auxin signaling. Plant Cell 2005, 17, 1387–1396. [Google Scholar] [CrossRef]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef]
- Bottcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase gh3-1 in grape berry (Vitis vinifera L.) And the proposed role of auxin conjugation during ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar] [CrossRef] [PubMed]
- Mellidou, I.; Ainalidou, A.; Papadopoulou, A.; Leontidou, K.; Genitsaris, S.; Karagiannis, E.; Van de Poel, B.; Karamanoli, K. Comparative transcriptomics and metabolomics reveal an intricate priming mechanism involved in PGPR-mediated salt tolerance in tomato. Front. Plant Sci. 2021, 12, 713984. [Google Scholar] [CrossRef]
- Aoi, Y.; Tanaka, K.; Cook, S.D.; Hayashi, K.; Kasahara, H. Gh3 auxin-amido synthetases alter the ratio of indole-3-acetic acid and phenylacetic acid in Arabidopsis. Plant Cell Physiol. 2020, 61, 596–605. [Google Scholar] [CrossRef]
- Yadav, S.; Yugandhar, P.; Alavilli, H.; Raliya, R.; Singh, A.; Sahi, S.V.; Sarkar, A.K.; Jain, A. Potassium chloroaurate-mediated in vitro synthesis of gold nanoparticles improved root growth by crosstalk with sucrose and nutrient-dependent auxin homeostasis in Arabidopsis thaliana. Nanomaterials 2022, 12, 2099. [Google Scholar] [CrossRef]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Reads | Mapped Reads | Q30 (%) | GC Content (%) | Total Reads (%) | Mapped Reads (%) | Uniquely Mapped Reads (%) | Multiple Mapped Reads (%) |
|---|---|---|---|---|---|---|---|---|
| CK-1 | 21,090,187 | 16,622,007 | 93.13% | 46.44% | 100% | 78.81% | 31.39% | 68.61% |
| CK-2 | 42,863,152 | 34,186,250 | 92.71% | 46.39% | 100% | 79.76% | 31.41% | 68.59% |
| CK-3 | 19,543,667 | 15,452,823 | 93.02% | 46.60% | 100% | 79.07% | 31.73% | 68.27% |
| T1-1 | 21,905,904 | 17,739,521 | 91.11% | 46.46% | 100% | 80.98% | 31.56% | 68.44% |
| T1-2 | 29,965,914 | 24,595,359 | 93.04% | 47.39% | 100% | 82.08% | 31.96% | 68.04% |
| T1-3 | 37,593,894 | 30,876,786 | 92.97% | 47.12% | 100% | 82.13% | 32.25% | 67.75% |
| T2-1 | 21,323,173 | 16,947,499 | 91.49% | 47.17% | 100% | 79.48% | 30.62% | 69.38% |
| T2-2 | 21,976,083 | 17,881,395 | 92.68% | 48.21% | 100% | 81.37% | 30.14% | 69.86% |
| T2-3 | 21,443,839 | 17,003,703 | 91.71% | 47.87% | 100% | 79.29% | 30.40% | 69.60% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Du, D.; Wei, H.; Xie, S.; Tian, X.; Yang, J.; Xiao, S.; Tang, Z.; Li, D.; Liu, Y. Transcriptomic and Hormone Analyses Provide Insight into the Regulation of Axillary Bud Outgrowth of Eucommia ulmoides Oliver. Curr. Issues Mol. Biol. 2023, 45, 7304-7318. https://doi.org/10.3390/cimb45090462
Zhang Y, Du D, Wei H, Xie S, Tian X, Yang J, Xiao S, Tang Z, Li D, Liu Y. Transcriptomic and Hormone Analyses Provide Insight into the Regulation of Axillary Bud Outgrowth of Eucommia ulmoides Oliver. Current Issues in Molecular Biology. 2023; 45(9):7304-7318. https://doi.org/10.3390/cimb45090462
Chicago/Turabian StyleZhang, Ying, Dandan Du, Hongling Wei, Shengnan Xie, Xuchen Tian, Jing Yang, Siqiu Xiao, Zhonghua Tang, Dewen Li, and Ying Liu. 2023. "Transcriptomic and Hormone Analyses Provide Insight into the Regulation of Axillary Bud Outgrowth of Eucommia ulmoides Oliver" Current Issues in Molecular Biology 45, no. 9: 7304-7318. https://doi.org/10.3390/cimb45090462
APA StyleZhang, Y., Du, D., Wei, H., Xie, S., Tian, X., Yang, J., Xiao, S., Tang, Z., Li, D., & Liu, Y. (2023). Transcriptomic and Hormone Analyses Provide Insight into the Regulation of Axillary Bud Outgrowth of Eucommia ulmoides Oliver. Current Issues in Molecular Biology, 45(9), 7304-7318. https://doi.org/10.3390/cimb45090462







