Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Whole Cell Protein Extract and Western Blot
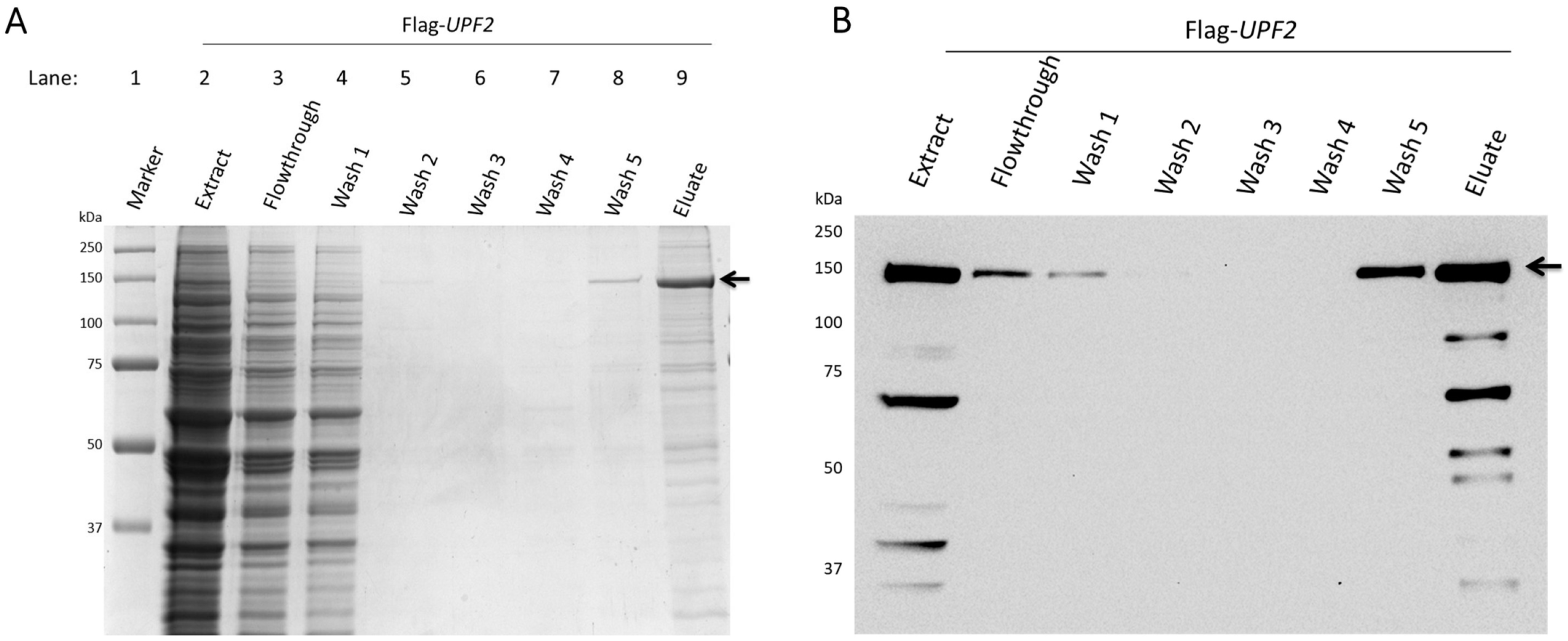
2.3. Upf2 Protein Purification
2.4. Tandem Mass Spectrometry Analysis
2.5. Mass Spectrometry Data Analysis
2.6. RNA Isolation and Northern Blot
2.7. can1-100 Nonsense Suppression Assay
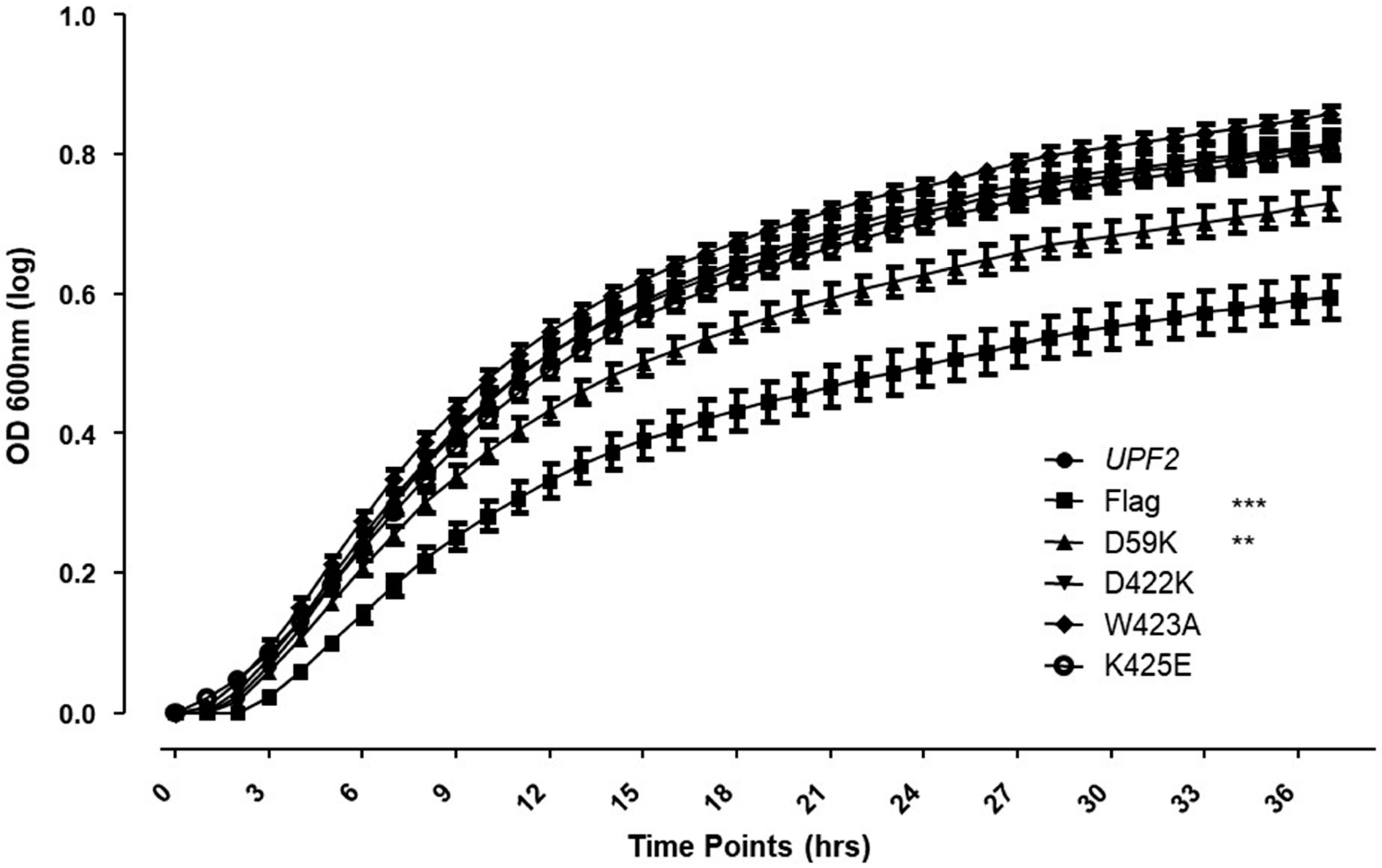
2.8. can1-100 Nonsense Suppression Assay Growth Curves
3. Results
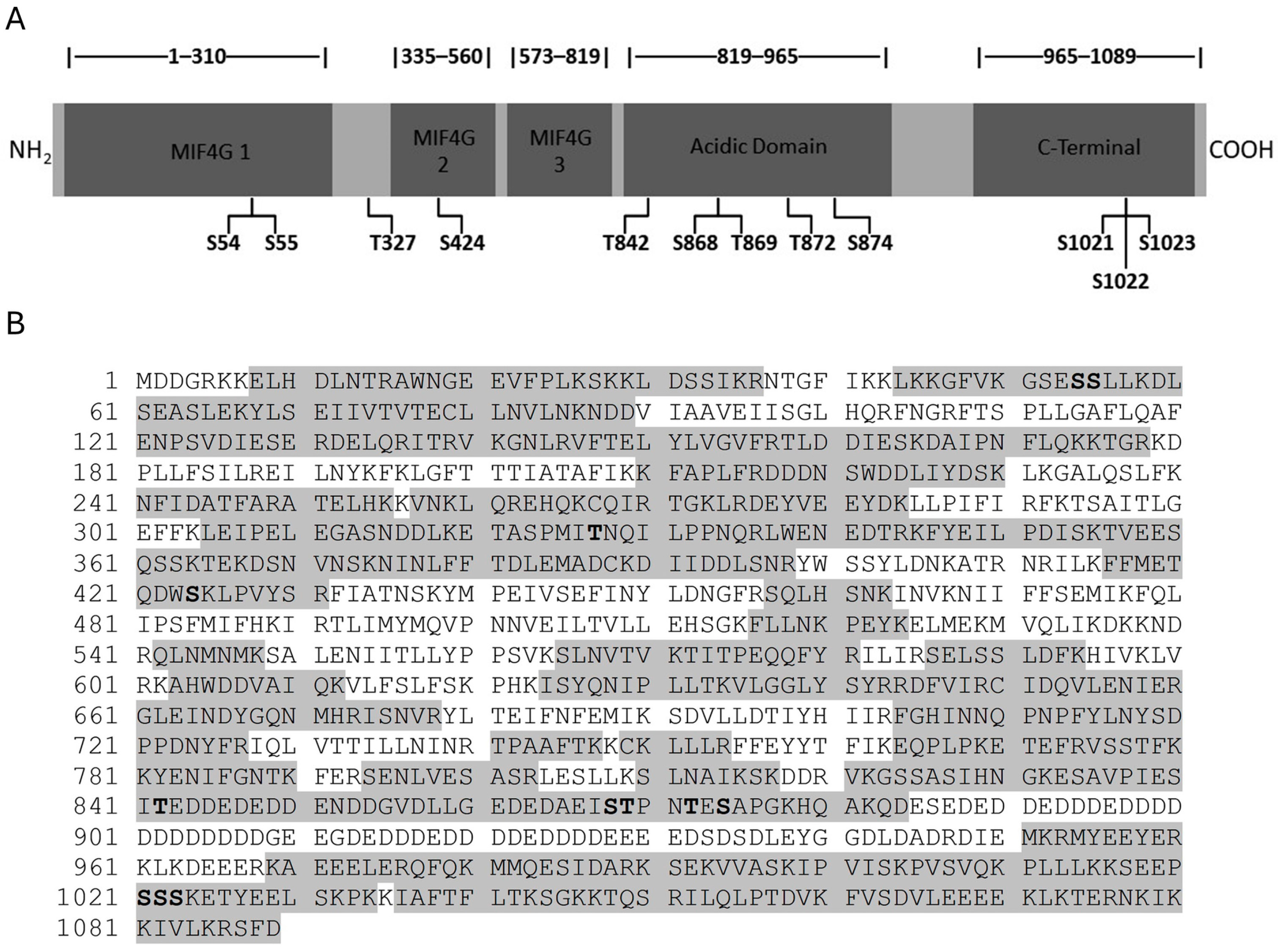
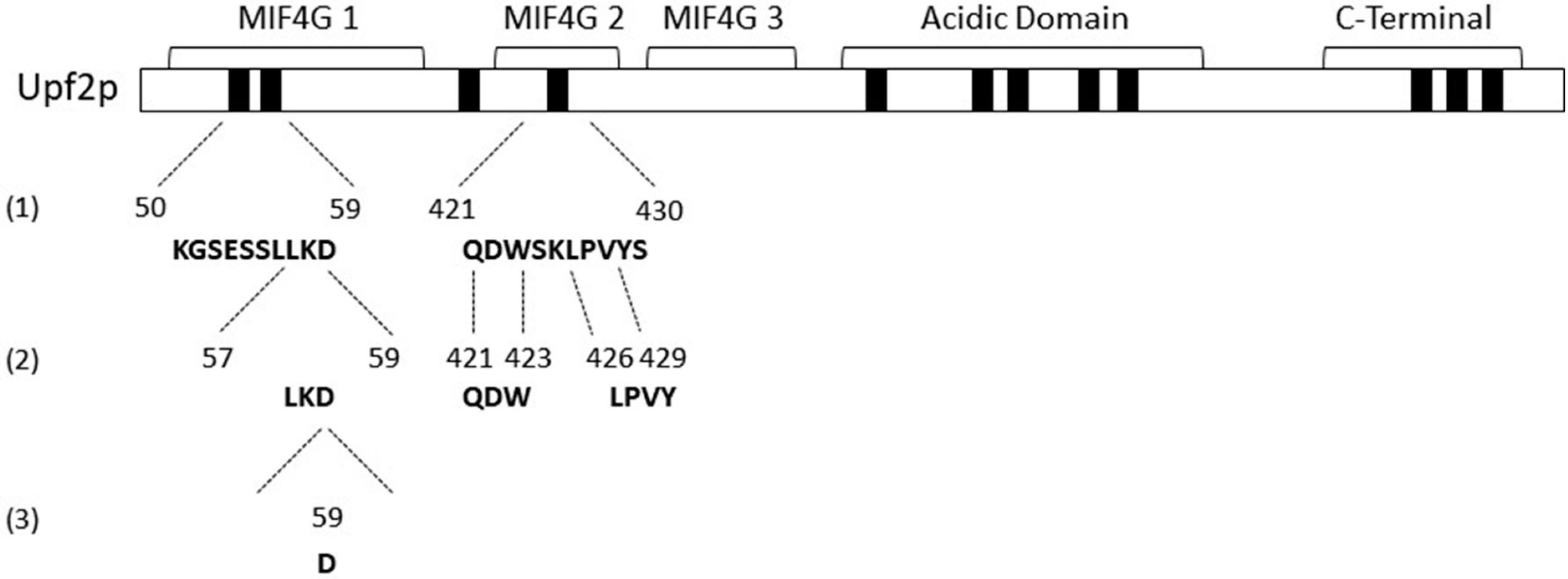
3.1. Identification of Novel Phosphorylation Sites in Yeast Upf2p mIF4G Domains
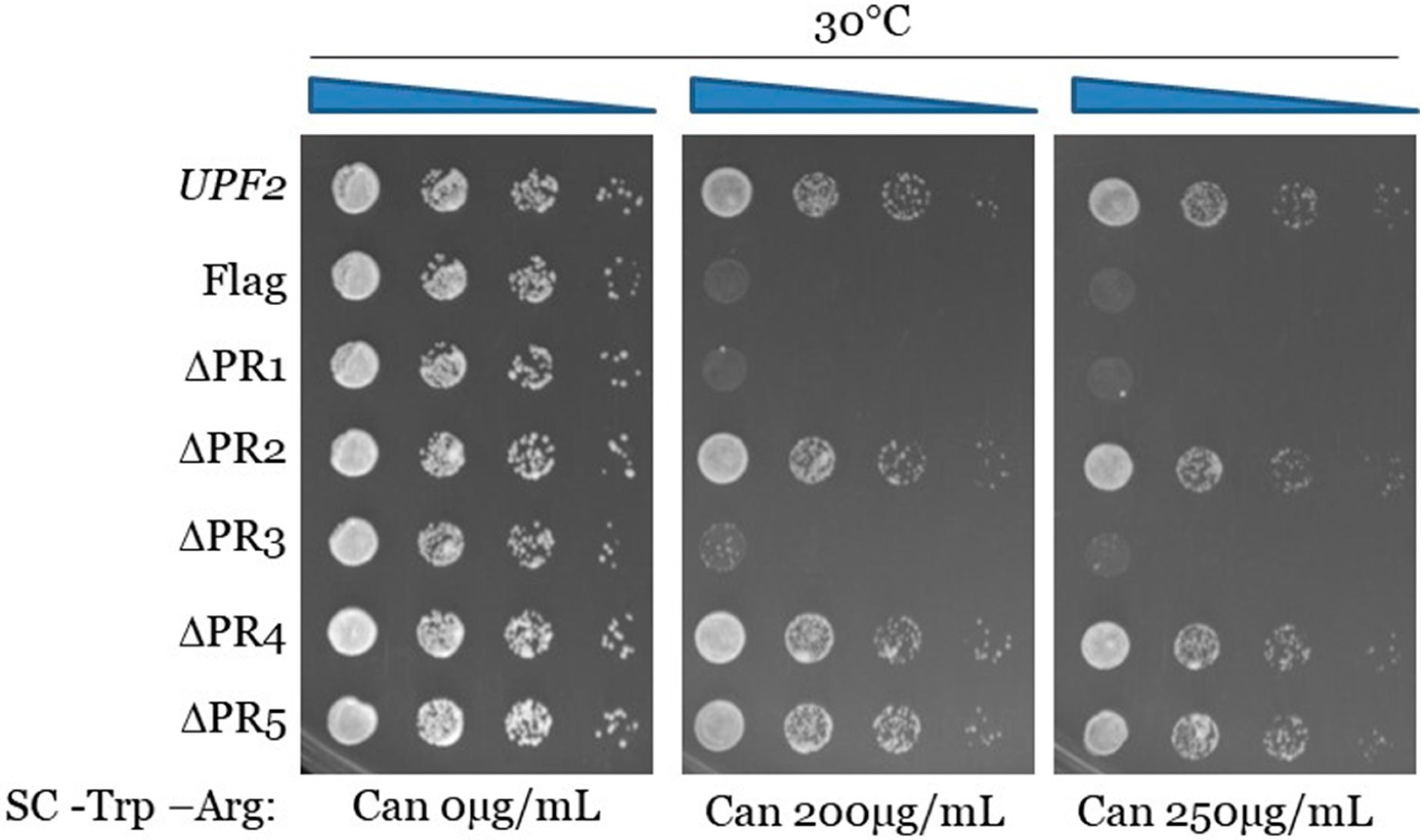
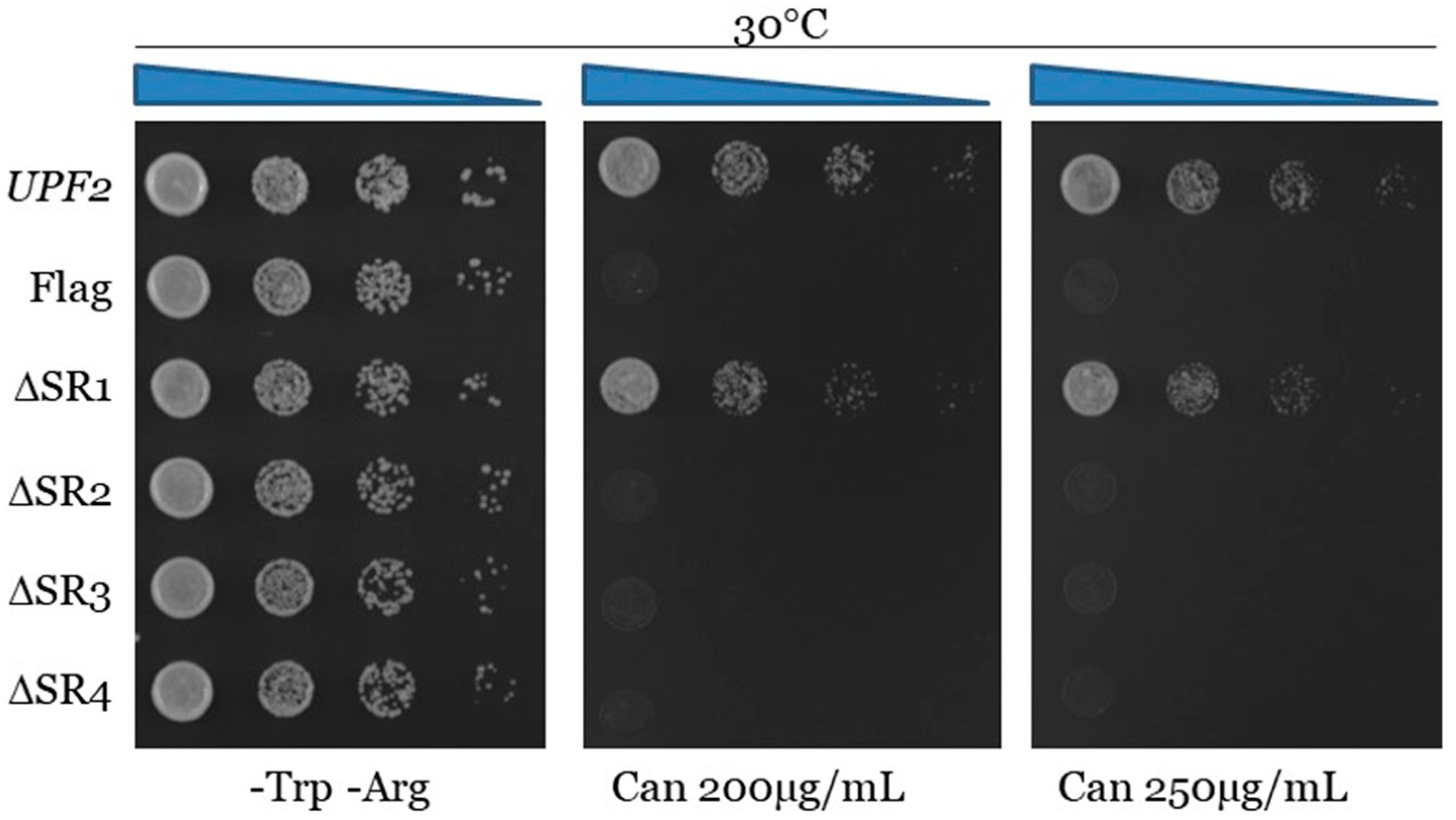
3.2. Upf2p mIF4G Domain Regions Harboring Phosphorylated Residues Are Required for NMD and Translation Termination Accuracy
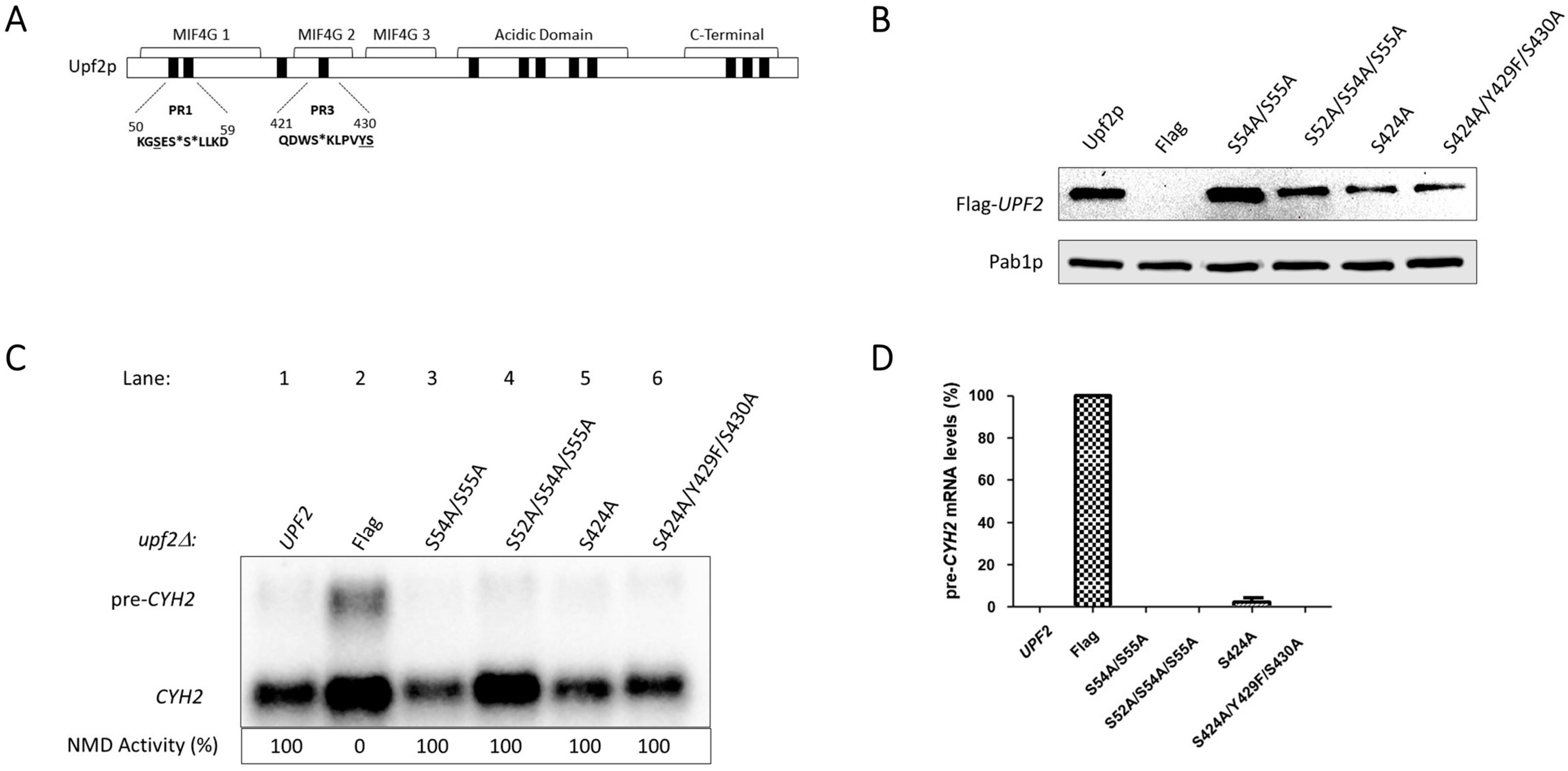
3.3. Upf2p mIF4G-1 S54 and S55, and mIF4G-2 S424, Phosphorylated Residues Are Not Required for NMD and Translation Termination Accuracy
3.4. Unphosphorylated Residues within Upf2p mIF4G-1 and mIF4G-2 Are Essential for NMD and Translation Termination Accuracy
3.5. Upf2p mIF4G-1 Residue D59 Is Required for NMD and Translation Termination Accuracy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gonzalez, C.I.; Wang, W.; Peltz, S.W. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae: A quality control mechanism that degrades transcripts harboring premature termination codons. Cold Spring Harb. Symp. Quant. Biol. 2001, 66, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Amrani, N.; Dong, S.; He, F.; Ganesan, R.; Ghosh, S.; Kervestin, S.; Li, C.; Mangus, D.A.; Spatrick, P.; Jacobson, A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006, 34, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Kashima, I.; Rehwinkel, J.; Sauliere, J.; Wittkopp, N.; Izaurralde, E. mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007, 581, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cajigas, I.J.; Peltz, S.W.; Wilkinson, M.F.; Gonzalez, C.I. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol. Cell Biol. 2006, 26, 3390–3400. [Google Scholar] [CrossRef] [PubMed]
- Kervestin, S.; Jacobson, A. NMD: A multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012, 13, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Schweingruber, C.; Rufener, S.C.; Zund, D.; Yamashita, A.; Muhlemann, O. Nonsense-mediated mRNA decay—Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 2013, 1829, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, L.; Longman, D.; Caceres, J.F. Translation-coupled mRNA quality control mechanisms. EMBO J. 2023, 42, e114378. [Google Scholar] [CrossRef]
- Andjus, S.; Morillon, A.; Wery, M. From Yeast to Mammals, the Nonsense-Mediated mRNA Decay as a Master Regulator of Long Non-Coding RNAs Functional Trajectory. Noncoding RNA 2021, 7, 44. [Google Scholar] [CrossRef]
- Palacios, I.M. Nonsense-mediated mRNA decay: From mechanistic insights to impacts on human health. Brief. Funct. Genom. 2013, 12, 25–36. [Google Scholar] [CrossRef]
- Rebbapragada, I.; Lykke-Andersen, J. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr. Opin. Cell Biol. 2009, 21, 394–402. [Google Scholar] [CrossRef]
- Wittmann, J.; Hol, E.M.; Jack, H.M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell Biol. 2006, 26, 1272–1287. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Zheng, W.; Tang, S.; Liu, X.; Zinkel, R.A.; Tsui, K.W.; Yandell, B.S.; Culbertson, M.R. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006, 2, e203. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.R. Nonsense-mediated RNA decay: A molecular system micromanaging individual gene activities and suppressing genomic noise. BioEssays News Rev. Mol. Cell. Dev. Biol. 2005, 27, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Hug, N.; Longman, D.; Caceres, J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016, 44, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Celik, A.; Baker, R.; He, F.; Jacobson, A. High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA 2017, 23, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Malabat, C.; Feuerbach, F.; Ma, L.; Saveanu, C.; Jacquier, A. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. eLife 2015, 4, e06722. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, J.; Singh, A.; Medina, D.; Chavez, S.; Perez-Ortin, J.E. Enhanced gene regulation by cooperation between mRNA decay and gene transcription. Biochim. Biophys. Acta Gene Regul. Mech. 2023, 1866, 194910. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Shu, M.D.; Steitz, J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef]
- Mendell, J.T.; Medghalchi, S.M.; Lake, R.G.; Noensie, E.N.; Dietz, H.C. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell Biol. 2000, 20, 8944–8957. [Google Scholar] [CrossRef]
- Kadlec, J.; Izaurralde, E.; Cusack, S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 2004, 11, 330–337. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Brown, A.H.; Jacobson, A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell Biol. 1997, 17, 1580–1594. [Google Scholar] [CrossRef] [PubMed]
- Chamieh, H.; Ballut, L.; Bonneau, F.; Le Hir, H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008, 15, 85–93. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Jacobson, A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995, 9, 437–454. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Echevarria, M.J.; Gonzalez, C.I.; Peltz, S.W. Identifying the right stop: Determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998, 17, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.S.; Li, S.; Wilkinson, M.F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996, 15, 5965–5975. [Google Scholar] [CrossRef] [PubMed]
- Kervestin, S.; Li, C.; Buckingham, R.; Jacobson, A. Testing the faux-UTR model for NMD: Analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie 2012, 94, 1560–1571. [Google Scholar] [CrossRef] [PubMed]
- Amrani, N.; Ganesan, R.; Kervestin, S.; Mangus, D.A.; Ghosh, S.; Jacobson, A. A faux 3’-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 2004, 432, 112–118. [Google Scholar] [CrossRef]
- Lejeune, F.; Maquat, L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005, 17, 309–315. [Google Scholar] [CrossRef]
- Gehring, N.H.; Kunz, J.B.; Neu-Yilik, G.; Breit, S.; Viegas, M.H.; Hentze, M.W.; Kulozik, A.E. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 2005, 20, 65–75. [Google Scholar] [CrossRef]
- Hwang, H.J.; Park, Y.; Kim, Y.K. UPF1: From mRNA Surveillance to Protein Quality Control. Biomedicines 2021, 9, 995. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, F. Nonsense-Mediated mRNA Decay, a Finely Regulated Mechanism. Biomedicines 2022, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.A.; Luz, V.C.C.; Targowski, S.; Ramalho, S.S.; Farinha, C.M.; Amaral, M.D. Integrity and Stability of PTC Bearing CFTR mRNA and Relevance to Future Modulator Therapies in Cystic Fibrosis. Genes 2021, 12, 1810. [Google Scholar] [CrossRef] [PubMed]
- Vallverdu-Prats, M.; Brugada, R.; Alcalde, M. Premature Termination Codon in 5’ Region of Desmoplakin and Plakoglobin Genes May Escape Nonsense-Mediated Decay through the Reinitiation of Translation. Int. J. Mol. Sci. 2022, 23, 656. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Jayachandran, U.; Bonneau, F.; Fiorini, F.; Basquin, C.; Domcke, S.; Le Hir, H.; Conti, E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 2011, 41, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Maquat, L.E. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 2019, 25, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Maciej, V.D.; Machado de Amorim, A.; Pak, M.; Jayachandran, U.; Chakrabarti, S. Modulation of RNA-binding properties of the RNA helicase UPF1 by its activator UPF2. RNA 2023, 29, 178–187. [Google Scholar] [CrossRef]
- Clerici, M.; Mourao, A.; Gutsche, I.; Gehring, N.H.; Hentze, M.W.; Kulozik, A.; Kadlec, J.; Sattler, M.; Cusack, S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009, 28, 2293–2306. [Google Scholar] [CrossRef]
- Ponting, C.P. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 2000, 25, 423–426. [Google Scholar] [CrossRef]
- Clerici, M.; Deniaud, A.; Boehm, V.; Gehring, N.H.; Schaffitzel, C.; Cusack, S. Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res. 2014, 42, 2673–2686. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. Eukaryote-specific domains in translation initiation factors: Implications for translation regulation and evolution of the translation system. Genome Res. 2000, 10, 1172–1184. [Google Scholar] [CrossRef] [PubMed]
- Fourati, Z.; Roy, B.; Millan, C.; Coureux, P.D.; Kervestin, S.; van Tilbeurgh, H.; He, F.; Uson, I.; Jacobson, A.; Graille, M. A highly conserved region essential for NMD in the Upf2 N-terminal domain. J. Mol. Biol. 2014, 426, 3689–3702. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wilkinson, M.F. Deletion mutagenesis of large (12-kb) plasmids by a one-step PCR protocol. BioTechniques 2001, 31, 722–724. [Google Scholar] [CrossRef] [PubMed]
- Makarova, O.; Kamberov, E.; Margolis, B. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 2000, 29, 970–972. [Google Scholar] [CrossRef]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]
- Herrick, D.; Parker, R.; Jacobson, A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 1990, 10, 2269–2284. [Google Scholar] [CrossRef]
- Maderazo, A.B.; He, F.; Mangus, D.A.; Jacobson, A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell Biol. 2000, 20, 4591–4603. [Google Scholar] [CrossRef]
- Cui, Y.; Hagan, K.W.; Zhang, S.; Peltz, S.W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995, 9, 423–436. [Google Scholar] [CrossRef]
- Kaufer, N.F.; Fried, H.M.; Schwindinger, W.F.; Jasin, M.; Warner, J.R. Cycloheximide resistance in yeast: The gene and its protein. Nucleic Acids Res. 1983, 11, 3123–3135. [Google Scholar] [CrossRef]
- Ahmad, M.; Bussey, H. Yeast arginine permease: Nucleotide sequence of the CAN1 gene. Curr. Genet. 1986, 10, 587–592. [Google Scholar] [CrossRef]
- He, F.; Peltz, S.W.; Donahue, J.L.; Rosbash, M.; Jacobson, A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. USA 1993, 90, 7034–7038. [Google Scholar] [CrossRef] [PubMed]
- Lasalde, C.; Rivera, A.V.; Leon, A.J.; Gonzalez-Feliciano, J.A.; Estrella, L.A.; Rodriguez-Cruz, E.N.; Correa, M.E.; Cajigas, I.J.; Bracho, D.P.; Vega, I.E.; et al. Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1. Nucleic Acids Res. 2014, 42, 1916–1929. [Google Scholar] [CrossRef] [PubMed]
- Estrella, L.A.; Wilkinson, M.F.; Gonzalez, C.I. The shuttling protein Npl3 promotes translation termination accuracy in Saccharomyces cerevisiae. J. Mol. Biol. 2009, 394, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Ono, B.I.; Ishino, Y.; Shinoda, S. Nonsense mutations in the can1 locus of Saccharomyces cerevisiae. J. Bacteriol. 1983, 154, 1476–1479. [Google Scholar] [CrossRef] [PubMed]
- Serin, G.; Gersappe, A.; Black, J.D.; Aronoff, R.; Maquat, L.E. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell Biol. 2001, 21, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.I.; Ruiz-Echevarria, M.J.; Vasudevan, S.; Henry, M.F.; Peltz, S.W. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 2000, 5, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Henry, M.; Borland, C.Z.; Bossie, M.; Silver, P.A. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics 1996, 142, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef]
- Lopez-Perrote, A.; Castano, R.; Melero, R.; Zamarro, T.; Kurosawa, H.; Ohnishi, T.; Uchiyama, A.; Aoyagi, K.; Buchwald, G.; Kataoka, N.; et al. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res. 2016, 44, 1909–1923. [Google Scholar] [CrossRef]
- Bufton, J.C.; Powers, K.T.; Szeto, J.A.; Toelzer, C.; Berger, I.; Schaffitzel, C. Structures of nonsense-mediated mRNA decay factors UPF3B and UPF3A in complex with UPF2 reveal molecular basis for competitive binding and for neurodevelopmental disorder-causing mutation. Nucleic Acids Res. 2022, 50, 5934–5947. [Google Scholar] [CrossRef]
- Wang, W.; Czaplinski, K.; Rao, Y.; Peltz, S.W. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001, 20, 880–890. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colón, E.M.; Haddock, L.A., III; Lasalde, C.; Lin, Q.; Ramírez-Lugo, J.S.; González, C.I. Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p. Curr. Issues Mol. Biol. 2024, 46, 244-261. https://doi.org/10.3390/cimb46010017
Colón EM, Haddock LA III, Lasalde C, Lin Q, Ramírez-Lugo JS, González CI. Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p. Current Issues in Molecular Biology. 2024; 46(1):244-261. https://doi.org/10.3390/cimb46010017
Chicago/Turabian StyleColón, Edgardo M., Luis A. Haddock, III, Clarivel Lasalde, Qishan Lin, Juan S. Ramírez-Lugo, and Carlos I. González. 2024. "Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p" Current Issues in Molecular Biology 46, no. 1: 244-261. https://doi.org/10.3390/cimb46010017
APA StyleColón, E. M., Haddock, L. A., III, Lasalde, C., Lin, Q., Ramírez-Lugo, J. S., & González, C. I. (2024). Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p. Current Issues in Molecular Biology, 46(1), 244-261. https://doi.org/10.3390/cimb46010017





