Transcriptome and Metabolome Analyses Provide Insight into the Glucose-Induced Adipogenesis in Porcine Adipocytes
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Porcine Preadipocyte Isolation and Cell Culture
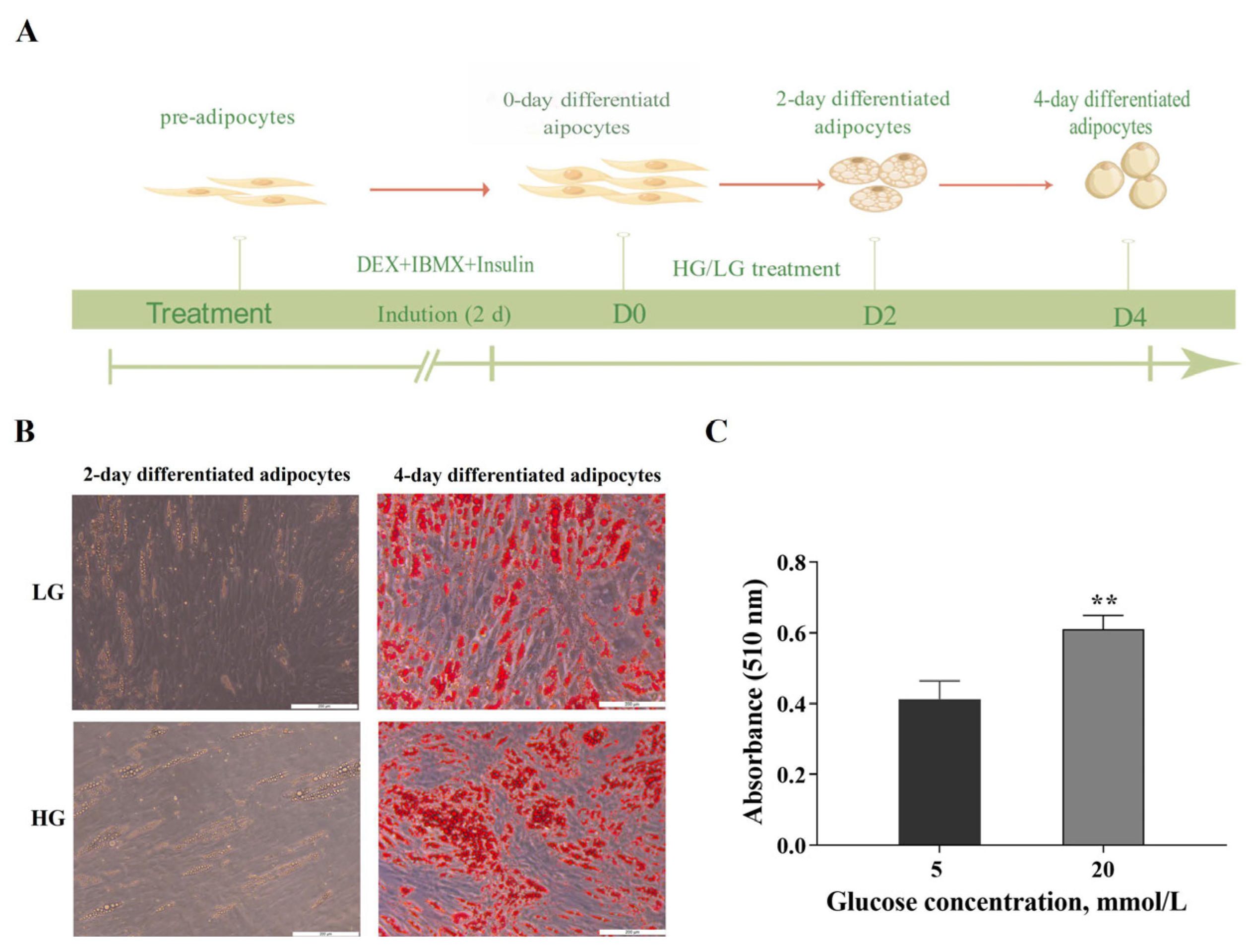
2.3. Oil Red O Staining and Quantification of Lipid Content
2.4. RNA Sequencing and Transcriptome Data Analysis
2.5. Construction of Protein–Protein Interactions (PPI) in Network and Module Analysis
2.6. Metabolite Extraction and Metabolome Analysis by Liquid Chromatography–Mass Spectroscopy (LC–MS)
2.7. Combined Metabolomics and RNA Sequencing Analysis
2.8. Statistical Analysis
3. Results
3.1. HG Promoted Adipogenesis in Porcine Adipocytes
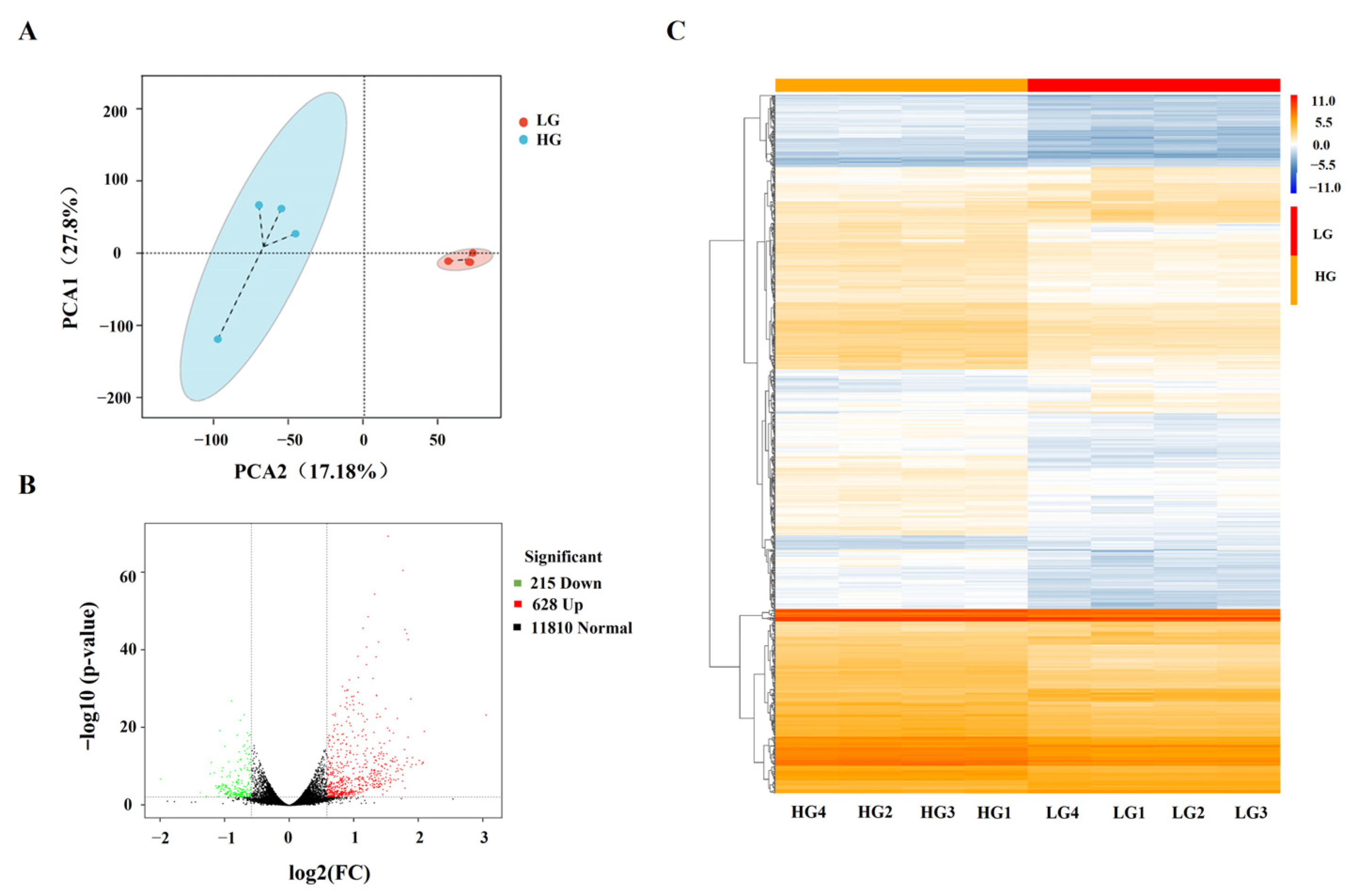
3.2. Identification of DEGs in Porcine Adipocytes
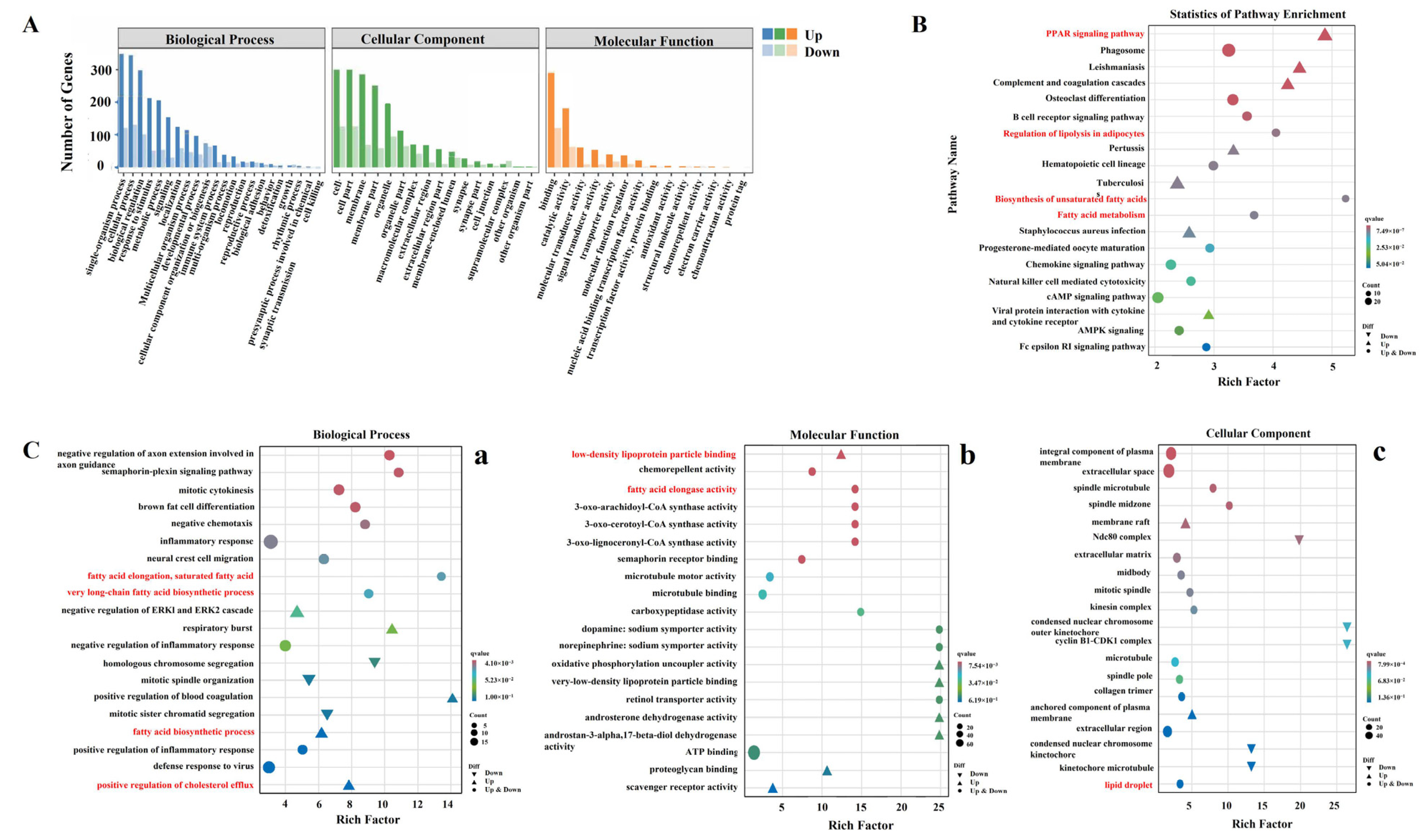
3.3. Enrichment and Functional Annotation of DEGs
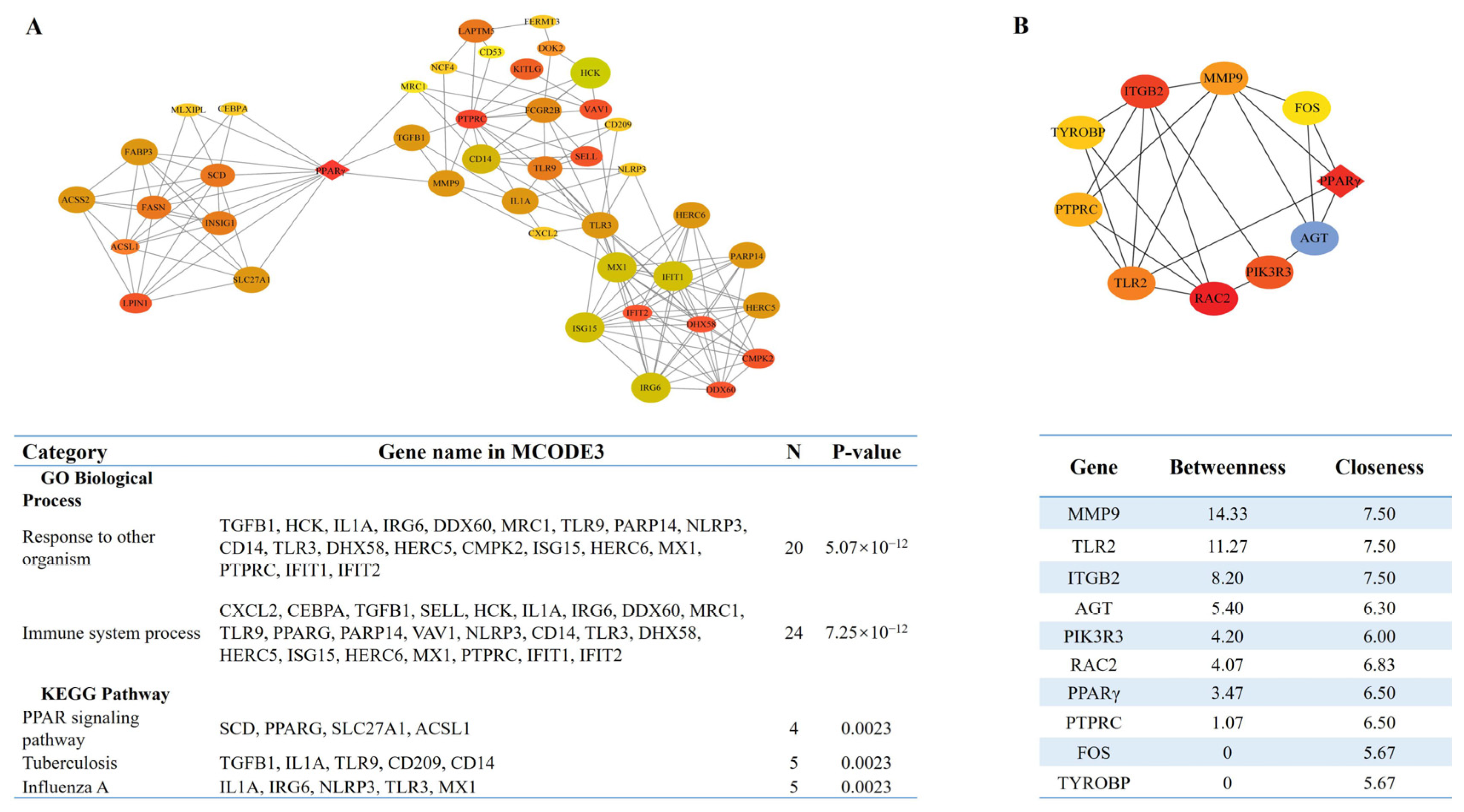
3.4. Identification of the Key Pathways and Hub Genes in the PPI Network
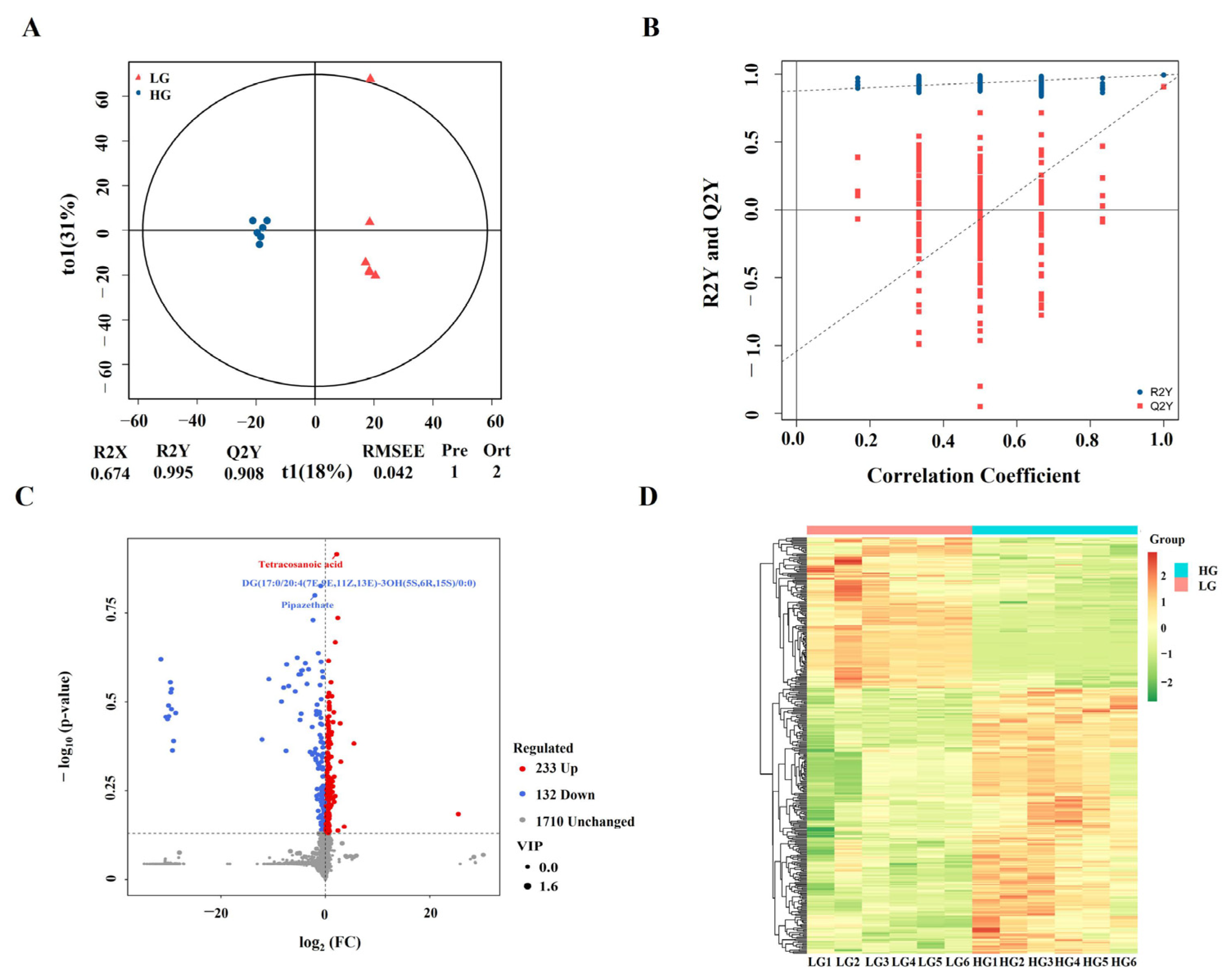
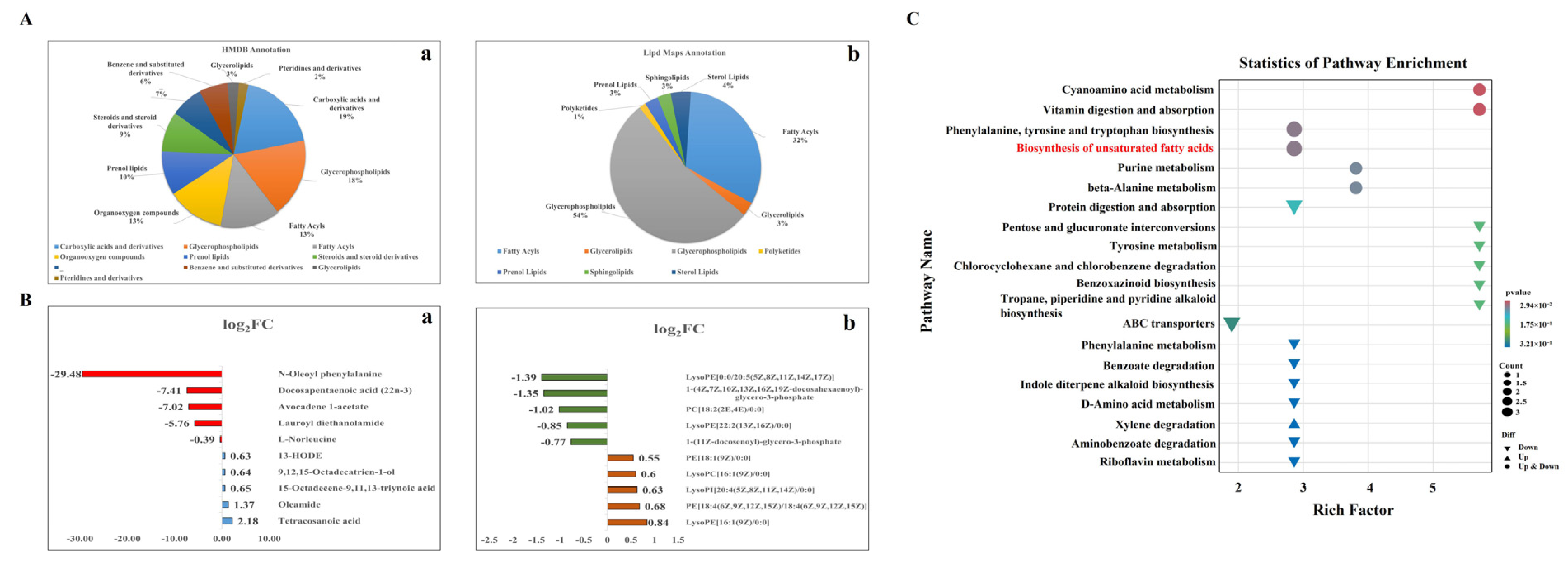
3.5. Identification of Differentially Accumulated Metabolites
3.6. Pathway Enrichment Analysis of DAMs
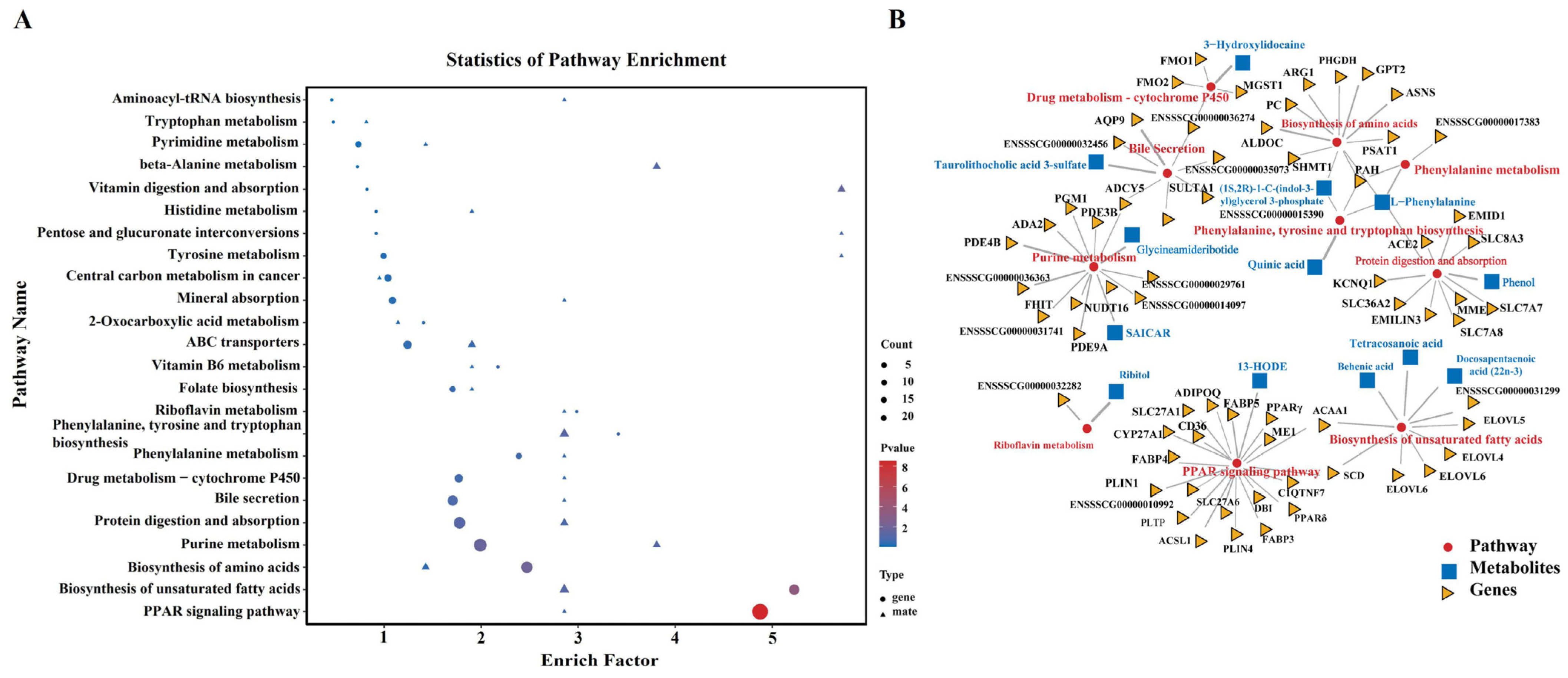
3.7. Integrated Analyses of Transcriptomics and Metabolomics Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Dunshea, F.R.; D’Souza, D.N.; Pethick, D.W.; Harper, G.S.; Warner, R.D. Effects of dietary factors and other metabolic modifiers on quality and nutritional value of meat. Meat Sci. 2005, 71, 8–38. [Google Scholar] [CrossRef]
- Poklukar, K.; Čandek-Potokar, M.; Batorek Lukač, N.; Tomažin, U.; Škrlep, M. Lipid Deposition and Metabolism in Local and Modern Pig Breeds: A Review. Animals 2020, 10, 424. [Google Scholar] [CrossRef]
- Morigny, P.; Boucher, J.; Arner, P.; Langin, D. Lipid and glucose metabolism in white adipocytes: Pathways, dysfunction and therapeutics. Nat. Rev. Endocrinol. 2021, 17, 276–295. [Google Scholar] [CrossRef]
- Ambele, M.A.; Dhanraj, P.; Giles, R.; Pepper, M.S. Adipogenesis: A Complex Interplay of Multiple Molecular Determinants and Pathways. Int. J. Mol. Sci. 2020, 21, 4283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Lu, J.; Chen, Y.; Dai, H.; Zhaxi, Y.; Zhao, Y.; Qiao, Z.; Feng, R.; Wang, Y.; Ma, Z.; et al. Glucose and Insulin Stimulate Lipogenesis in Porcine Adipocytes: Dissimilar and Identical Regulation Pathway for Key Transcription Factors. Mol. Cells 2016, 39, 797–806. [Google Scholar]
- Krishna, M.S.; Revathy, V.M.; Jaleel, A. Adipocytes utilize sucrose as an energy source-Effect of different carbohydrates on adipocyte differentiation. J. Cell. Physiol. 2020, 235, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Pan, S.; Guo, Y.; Xu, P.; Zhai, Y. PPARs as Nuclear Receptors for Nutrient and Energy Metabolism. Molecules 2019, 24, 2545. [Google Scholar] [CrossRef] [PubMed]
- Aprile, M.; Cataldi, S.; Ambrosio, M.R.; D’Esposito, V.; Lim, K.; Dietrich, A.; Blüher, M.; Savage, D.B.; Formisano, P.; Ciccodicola, A.; et al. PPARγΔ5, a Naturally Occurring Dominant-Negative Splice Isoform, Impairs PPARγ Function and Adipocyte Differentiation. Cell Rep. 2018, 25, 1577–1592.e6. [Google Scholar] [CrossRef] [PubMed]
- Umeno, A.; Sakashita, M.; Sugino, S.; Murotomi, K.; Okuzawa, T.; Morita, N.; Tomii, K.; Tsuchiya, Y.; Yamasaki, K.; Horie, M.; et al. Comprehensive analysis of PPARγ agonist activities of stereo-, regio-, and enantio-isomers of hydroxyoctadecadienoic acids. Biosci. Rep. 2020, 40, BSR20193767. [Google Scholar] [CrossRef] [PubMed]
- Havula, E.; Hietakangas, V. Sugar sensing by ChREBP/Mondo-Mlx-new insight into downstream regulatory networks and integration of nutrient-derived signals. Curr. Opin. Cell Biol. 2018, 51, 89–96. [Google Scholar] [CrossRef]
- Vijayakumar, A.; Aryal, P.; Wen, J.; Syed, I.; Vazirani, R.P.; Moraes-Vieira, P.M.; Camporez, J.P.; Gallop, M.R.; Perry, R.J.; Peroni, O.D.; et al. Absence of Carbohydrate Response Element Binding Protein in Adipocytes Causes Systemic Insulin Resistance and Impairs Glucose Transport. Cell Rep. 2017, 21, 1021–1035. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, J.; Chen, Y.; Guo, P.; Qiao, Z.; Feng, R.; Chen, S.; Bai, J.; Huo, S.; Ma, Z.; et al. ChREBP and LXRα mediate synergistically lipogenesis induced by glucose in porcine adipocytes. Gene 2015, 565, 30–38. [Google Scholar] [CrossRef]
- Witte, N.; Muenzner, M.; Rietscher, J.; Knauer, M.; Heidenreich, S.; Nuotio-Antar, A.M.; Graef, F.A.; Fedders, R.; Tolkachov, A.; Goehring, I.; et al. The Glucose Sensor ChREBP Links De Novo Lipogenesis to PPARγ Activity and Adipocyte Differentiation. Endocrinology 2015, 156, 4008–4019. [Google Scholar] [CrossRef] [PubMed]
- Gerlini, R.; Berti, L.; Darr, J.; Lassi, M.; Brandmaier, S.; Fritsche, L.; Scheid, F.; Böhm, A.; Königsrainer, A.; Grallert, K.; et al. Glucose tolerance and insulin sensitivity define adipocyte transcriptional programs in human obesity. Mol. Metab. 2018, 18, 42–50. [Google Scholar] [CrossRef]
- Peshdary, V.; Gagnon, A.; Sorisky, A. Effect of High Glucose Concentration on Human Preadipocytes and Their Response to Macrophage-Conditioned Medium. Can. J. Diabetes. 2016, 40, 411–418. [Google Scholar] [CrossRef]
- Lamichane, S.; Dahal Lamichane, B.; Kwon, S.M. Pivotal Roles of Peroxisome Proliferator-Activated Receptors (PPARs) and Their Signal Cascade for Cellular and Whole-Body Energy Homeostasis. Int. J. Mol. Sci. 2018, 19, 949. [Google Scholar] [CrossRef] [PubMed]
- Herold, J.; Kalucka, J. Angiogenesis in Adipose Tissue: The Interplay Between Adipose and Endothelial Cells. Front. Physiol. 2020, 11, 624903. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, S.; Tan, Z.; Wang, Y.; Zhang, F.; Yang, T.; Liu, Y.; Ao, H.; Xing, K.; Wang, C. Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation. Genes 2019, 10, 552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, B.; Shang, P.; Fu, Y.; Nie, R.; Chamba, Y.; Zhang, H. Comparative Transcriptomic Profiles of Differentiated Adipocytes Provide Insights into Adipogenesis Mechanisms of Subcutaneous and Intramuscular Fat Tissues in Pigs. Cells 2022, 11, 499. [Google Scholar] [CrossRef]
- Boone, S.; Mook-Kanamori, D.; Rosendaal, F.; den Heijer, M.; Lamb, H.; de Roos, A.; le Cessie, S.; Willems van Dijk, K.; de Mutsert, R. Metabolomics: A search for biomarkers of visceral fat and liver fat content. Metabolomics 2019, 15, 139. [Google Scholar] [CrossRef]
- Miehle, F.; Möller, G.; Cecil, A.; Lintelmann, J.; Wabitsch, M.; Tokarz, J.; Adamski, J.; Haid, M. Lipidomic Phenotyping Reveals Extensive Lipid Remodeling during Adipogenesis in Human Adipocytes. Metabolites 2020, 10, 217. [Google Scholar] [CrossRef]
- Liaw, L.; Prudovsky, I.; Koza, R.A.; Anunciado-Koza, R.V.; Siviski, M.E.; Lindner, V.; Friesel, R.E.; Rosen, C.J.; Baker, P.R.; Simons, B.; et al. Lipid Profiling of In Vitro Cell Models of Adipogenic Differentiation: Relationships With Mouse Adipose Tissues. J. Cell. Biochem. 2016, 117, 2182–2193. [Google Scholar] [CrossRef]
- Qiao, Q.; Bouwman, F.G.; Renes, J.; Mariman, E.C.M. An in vitro model for hypertrophic adipocytes: Time-dependent adipocyte proteome and secretome changes under high glucose and high insulin conditions. J. Cell. Mol. Med. 2020, 24, 8662–8673. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic. Acids. Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Yue, J.; Dong, Y.; Liu, S.; Jia, Y.; Li, C.; Wang, Z.; Gong, S. Integrated Proteomic and Metabolomic Analyses Provide Insights Into Acquisition of Embryogenic Ability in Agapanthus praecox. Front. Plant Sci. 2022, 13, 858065. [Google Scholar] [CrossRef]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef]
- Bojic, L.A.; Telford, D.E.; Fullerton, M.D.; Ford, R.J.; Sutherland, B.G.; Edwards, J.Y.; Sawyez, C.G.; Gros, R.; Kemp, B.E.; Steinberg, G.R.; et al. PPARδ activation attenuates hepatic steatosis in Ldlr-/- mice by enhanced fat oxidation, reduced lipogenesis, and improved insulin sensitivity. J. Lipid Res. 2014, 55, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Olson, P.; Hevener, A.; Mehl, I.; Chong, L.W.; Olefsky, J.M.; Gonzalez, F.J.; Ham, J.; Kang, H.; Peters, J.M.; et al. PPARdelta regulates glucose metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. USA 2006, 103, 3444–3449. [Google Scholar] [CrossRef]
- Choi, S.K.; Park, S.; Jang, S.; Cho, H.H.; Lee, S.; You, S.; Kim, S.H.; Moon, H.S. Cascade regulation of PPARγ(2) and C/EBPα signaling pathways by celastrol impairs adipocyte differentiation and stimulates lipolysis in 3T3-L1 adipocytes. Metabolism 2016, 65, 646–654. [Google Scholar] [CrossRef]
- Wang, Q.; Tao, C.; Jiang, L.; Shao, M.; Ye, R.; Zhu, Y.; Gordillo, R.; Ali, A.; Lian, Y.; Holland, W.; et al. Distinct regulatory mechanisms governing embryonic versus adult adipocyte maturation. Nat. Cell. Biol. 2015, 17, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Leblanc, S.E.; Wu, Q.; Konda, S.; Salma, N.; Marfella, C.G.; Ohkawa, Y.; Imbalzano, A.N. Chromatin accessibility and transcription factor binding at the PPARγ2 promoter during adipogenesis is protein kinase A-dependent. J. Cell. Physiol. 2011, 226, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Umeno, A.; Shichiri, M.; Ishida, N.; Hashimoto, Y.; Abe, K.; Kataoka, M.; Yoshino, K.; Hagihara, Y.; Aki, N.; Funaki, M.; et al. Singlet oxygen induced products of linoleates, 10- and 12-(Z,E)-hydroxyoctadecadienoic acids (HODE), can be potential biomarkers for early detection of type 2 diabetes. PLoS ONE 2013, 8, e63542. [Google Scholar] [CrossRef]
- Murotomi, K.; Umeno, A.; Yasunaga, M.; Shichiri, M.; Ishida, N.; Abe, H.; Yoshida, Y.; Nakajima, Y. Switching from singlet-oxygen-mediated oxidation to free-radical-mediated oxidation in the pathogenesis of type 2 diabetes in model mouse. Free Radic. Res. 2015, 49, 133–138. [Google Scholar] [CrossRef]
- Teng, L.; Chen, Y.; Cao, Y.; Wang, W.; Xu, Y.; Wang, Y.; Lv, J.; Li, C.; Su, Y. Overexpression of ATP citrate lyase in renal cell carcinoma tissues and its effect on the human renal carcinoma cells in vitro. Oncol. Lett. 2018, 15, 6967–6974. [Google Scholar] [CrossRef] [PubMed]
- Luong, A.; Hannah, V.C.; Brown, M.S.; Goldstein, J.L. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 2000, 275, 26458–26466. [Google Scholar] [CrossRef]
- Liu, X.; Cooper, D.E.; Cluntun, A.A.; Warmoes, M.O.; Zhao, S.; Reid, M.A.; Liu, J.; Lund, P.J.; Lopes, M.; Garcia, B.A.; et al. Acetate Production from Glucose and Coupling to Mitochondrial Metabolism in Mammals. Cell 2018, 175, e502–e513. [Google Scholar] [CrossRef] [PubMed]
- Sivanand, S.; Viney, I.; Wellen, K.E. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends. Biochem. Sci. 2018, 43, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Lin, S.H.; Ren, F.; Li, J.T.; Chen, J.; Yao, C.; Yang, H.; Jiang, S.; Yan, G.; Wang, D.; et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun. 2016, 7, 11960. [Google Scholar] [CrossRef]
- Zhao, S.; Torres, A.; Henry, R.A.; Trefely, S.; Wallace, M.; Lee, J.V.; Carrer, A.; Sengupta, A.; Campbell, S.L.; Kuo, Y.M.; et al. ATP-Citrate Lyase Controls a Glucose-to-Acetate Metabolic Switch. Cell Rep. 2016, 17, 1037–1052. [Google Scholar] [CrossRef]
- Grzes, M.; Sadkowski, S.; Rzewuska, K.; Szydlowski, M.; Switonski, M. Pig fatness in relation to FASN and INSIG2 genes polymorphism and their transcript level. Mol. Biol. Rep. 2016, 43, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Piazuelo, D.; Criado-Mesas, L.; Revilla, M.; Castelló, A.; Noguera, J.L.; Fernández, A.I.; Ballester, M.; Folch, J.M. Identification of strong candidate genes for backfat and intramuscular fatty acid composition in three crosses based on the Iberian pig. Sci. Rep. 2020, 10, 13962. [Google Scholar] [CrossRef]
- Ros-Freixedes, R.; Gol, S.; Pena, R.N.; Tor, M.; Ibáñez-Escriche, N.; Dekkers, J.C.; Estany, J. Genome-Wide Association Study Singles Out SCD and LEPR as the Two Main Loci Influencing Intramuscular Fat Content and Fatty Acid Composition in Duroc Pigs. PLoS ONE 2016, 11, e0152496. [Google Scholar] [CrossRef]
- Du, J.; Xu, Y.; Zhang, P.; Zhao, X.; Gan, M.; Li, Q.; Ma, J.; Tang, G.; Jiang, Y.; Wang, J.; et al. MicroRNA-125a-5p Affects Adipocytes Proliferation, Differentiation and Fatty Acid Composition of Porcine Intramuscular Fat. Int. J. Mol. Sci. 2018, 19, 501. [Google Scholar] [CrossRef]
- Ribes-Navarro, A.; Navarro, J.C.; Hontoria, F.; Kabeya, N.; Standal, I.B.; Evjemo, J.O.; Monroig, Ó. Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Marine Gammarids: Molecular Cloning and Functional Characterisation of Three Fatty Acyl Elongases. Mar. Drugs 2021, 19, 226. [Google Scholar] [CrossRef]
- Pauter, A.M.; Olsson, P.; Asadi, A.; Herslöf, B.; Csikasz, R.I.; Zadravec, D.; Jacobsson, A. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. J. Lipid Res. 2014, 55, 718–728. [Google Scholar] [CrossRef]
- Pauter, A.M.; Fischer, A.W.; Bengtsson, T.; Asadi, A.; Talamonti, E.; Jacobsson, A. Synergistic Effects of DHA and Sucrose on Body Weight Gain in PUFA-Deficient Elovl2 −/− Mice. Nutrients 2019, 11, 852. [Google Scholar] [CrossRef] [PubMed]
- Morral, N.; Edenberg, H.J.; Witting, S.R.; Altomonte, J.; Chu, T.; Brown, M. Effects of glucose metabolism on the regulation of genes of fatty acid synthesis and triglyceride secretion in the liver. J. Lipid Res. 2007, 48, 1499–1510. [Google Scholar] [CrossRef]
- Gaude, E.; Schmidt, C.; Gammage, P.A.; Dugourd, A.; Blacker, T.; Chew, S.P.; Saez-Rodriguez, J.; O’Neill, J.S.; Szabadkai, G.; Minczuk, M.; et al. NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol. Cell. 2018, 69, 581–593.e587. [Google Scholar] [CrossRef] [PubMed]
- Udhane, S.S.; Legeza, B.; Marti, N.; Hertig, D.; Diserens, G.; Nuoffer, J.M.; Vermathen, P.; Flück, C.E. Combined transcriptome and metabolome analyses of metformin effects reveal novel links between metabolic networks in steroidogenic systems. Sci. Rep. 2017, 7, 8652. [Google Scholar] [CrossRef] [PubMed]
- Juvekar, A.; Hu, H.; Yadegarynia, S.; Lyssiotis, C.A.; Ullas, S.; Lien, E.C.; Bellinger, G.; Son, J.; Hok, R.C.; Seth, P.; et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc. Natl. Acad. Sci. USA 2016, 113, E4338–E4347. [Google Scholar] [CrossRef] [PubMed]
- Chaneton, B.; Hillmann, P.; Zheng, L.; Martin, A.C.L.; Maddocks, O.D.K.; Chokkathukalam, A.; Coyle, J.E.; Jankevics, A.; Holding, F.P.; Vousden, K.H.; et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 2012, 491, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Y.; Zhang, N.; Ding, X.; Zhang, W.; Shen, K.; Huang, L.; Zhou, J.; Cui, S.; Zhu, Z.; et al. Novel impact of the DNMT3A R882H mutation on GSH metabolism in a K562 cell model established by TALENs. Oncotarget 2017, 8, 30395–30409. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, Y.; Liu, D.; Feng, Y.; Yin, J.; Zhou, X. Exogenous and Endogenous Serine Deficiency Exacerbates Hepatic Lipid Accumulation. Oxid. Med. Cell. Longev. 2021, 2021, 4232704. [Google Scholar] [CrossRef]
- Tabatabaie, L.; Klomp, L.W.; Berger, R.; de Koning, T.J. L-serine synthesis in the central nervous system: A review on serine deficiency disorders. Mol. Genet. Metab. 2010, 99, 256–262. [Google Scholar] [CrossRef]
- Gao, X.; Lee, K.; Reid, M.A.; Sanderson, S.M.; Qiu, C.; Li, S.; Liu, J.; Locasale, J.W. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep. 2018, 22, 3507–3520. [Google Scholar] [CrossRef] [PubMed]
- Taha, S.; Volkmer, E.; Haas, E.; Alberton, P.; Straub, T.; David-Rus, D.; Aszodi, A.; Giunta, R.; Saller, M.M. Differences in the Inflammatory Response of White Adipose Tissue and Adipose-Derived Stem Cells. Int. J. Mol. Sci. 2020, 21, 1086. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Sohn, J.H.; Han, S.M.; Park, Y.J.; Huh, J.Y.; Choe, S.S.; Kim, J.B. Adipocytes Are the Control Tower That Manages Adipose Tissue Immunity by Regulating Lipid Metabolism. Front. Immunol. 2020, 11, 598566. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Potter, V.J.; Zidon, T.M.; Padilla, J. Exercise and Estrogen Make Fat Cells “Fit”. Exerc. Sport Sci. Rev. 2015, 43, 172–178. [Google Scholar] [CrossRef]
- Blaszczak, A.M.; Wright, V.P.; Anandani, K.; Liu, J.; Jalilvand, A.; Bergin, S.; Nicoloro, S.M.; Czech, M.P.; Lafuse, W.; Deng, T.; et al. Loss of Antigen Presentation in Adipose Tissue Macrophages or in Adipocytes, but Not Both, Improves Glucose Metabolism. J. Immunol. 2019, 202, 2451–2459. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Zhang, G.; Miao, J.; Wu, D.; Li, X.; Li, J.; Lu, J.; Gun, S. Transcriptome and Metabolome Analyses Provide Insight into the Glucose-Induced Adipogenesis in Porcine Adipocytes. Curr. Issues Mol. Biol. 2024, 46, 2027-2042. https://doi.org/10.3390/cimb46030131
Jiang S, Zhang G, Miao J, Wu D, Li X, Li J, Lu J, Gun S. Transcriptome and Metabolome Analyses Provide Insight into the Glucose-Induced Adipogenesis in Porcine Adipocytes. Current Issues in Molecular Biology. 2024; 46(3):2027-2042. https://doi.org/10.3390/cimb46030131
Chicago/Turabian StyleJiang, Susu, Guohua Zhang, Jian Miao, Dianhu Wu, Ximei Li, Jiawei Li, Jianxiong Lu, and Shuangbao Gun. 2024. "Transcriptome and Metabolome Analyses Provide Insight into the Glucose-Induced Adipogenesis in Porcine Adipocytes" Current Issues in Molecular Biology 46, no. 3: 2027-2042. https://doi.org/10.3390/cimb46030131
APA StyleJiang, S., Zhang, G., Miao, J., Wu, D., Li, X., Li, J., Lu, J., & Gun, S. (2024). Transcriptome and Metabolome Analyses Provide Insight into the Glucose-Induced Adipogenesis in Porcine Adipocytes. Current Issues in Molecular Biology, 46(3), 2027-2042. https://doi.org/10.3390/cimb46030131




