Serine Metabolism Regulates the Replicative Senescence of Human Dental Pulp Cells through Histone Methylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. Western Blot
2.3. Antibody
2.4. RNA Extraction and Quantitative Reverse Transcription PCR (RT-qPCR)
2.5. S-Adenosylmethionine (SAM) Assay
2.6. Cell Counting Kit-8 (CCK-8) Assay
2.7. 5-Ethynyl-2′-Deoxyuridine (EdU) Incorporation Assay
2.8. Colony Formation Assay
2.9. Osteogenic Induction
2.10. Senescence-Associated β-Galactosidase (SA-β-Gal) Staining
2.11. Immunofluorescence
2.12. Chromatin Immunoprecipitation (ChIP)
2.13. Statistical Analysis
3. Results
3.1. Human Dental Pulp Cells Undergo Senescence during Passage
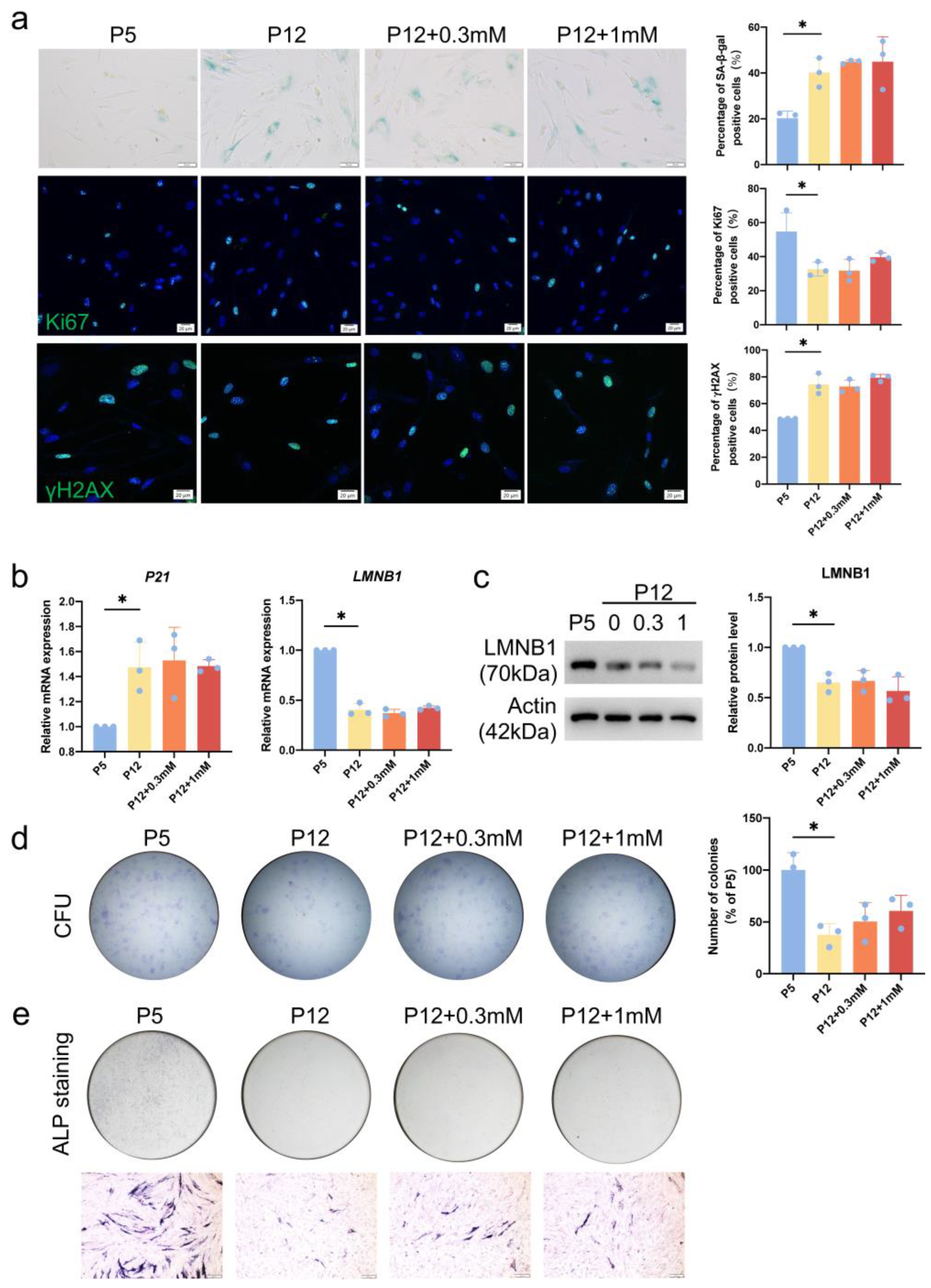
3.2. Serine Metabolism Decreases during Replicative Senescence in Human Dental Pulp Cells
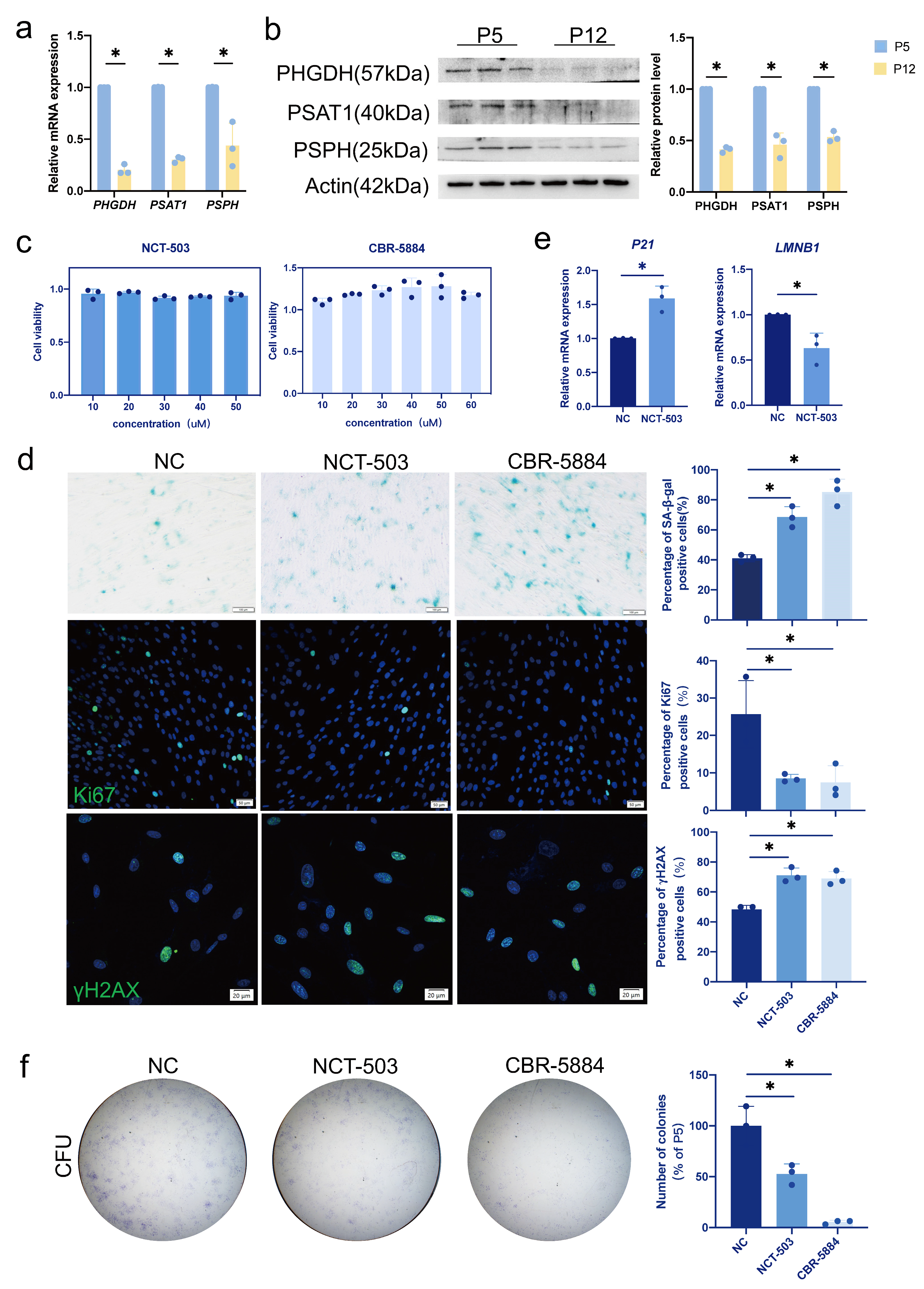
3.3. Inhibition of PHGDH Phenocopies the Replicative Senescence
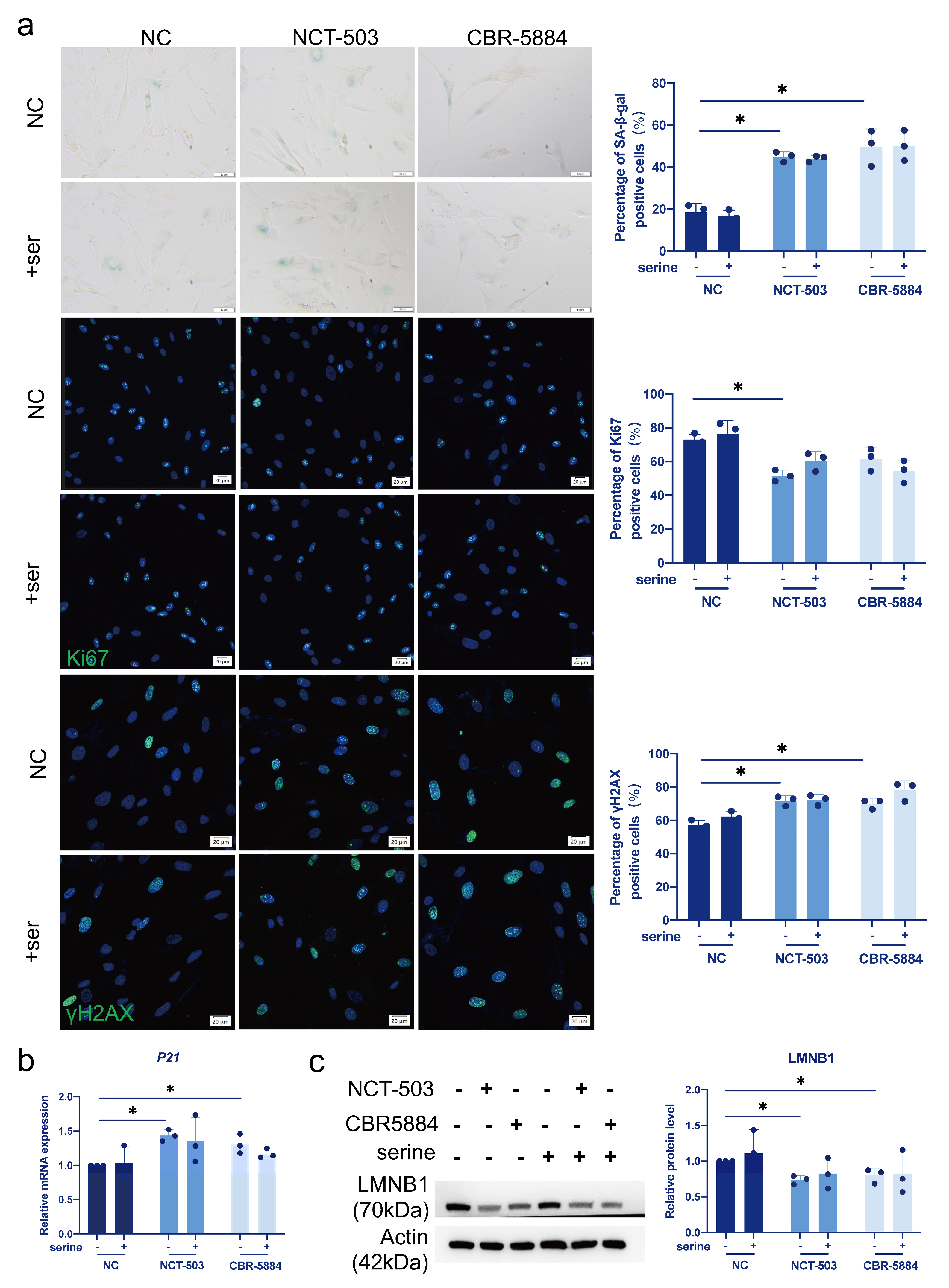
3.4. Serine Supplementation Fails to Rescue Replicative Senescence
3.5. Serine Supplementation Does Not Rescue Cellular Senescence Caused by PHGDH Inhibition
3.6. S-Adenosylmethionine (SAM) Regulates the Recruitment of H3K36me3 in the SIRT1 and RUNX2 Promoter Regions in Senescent Cells
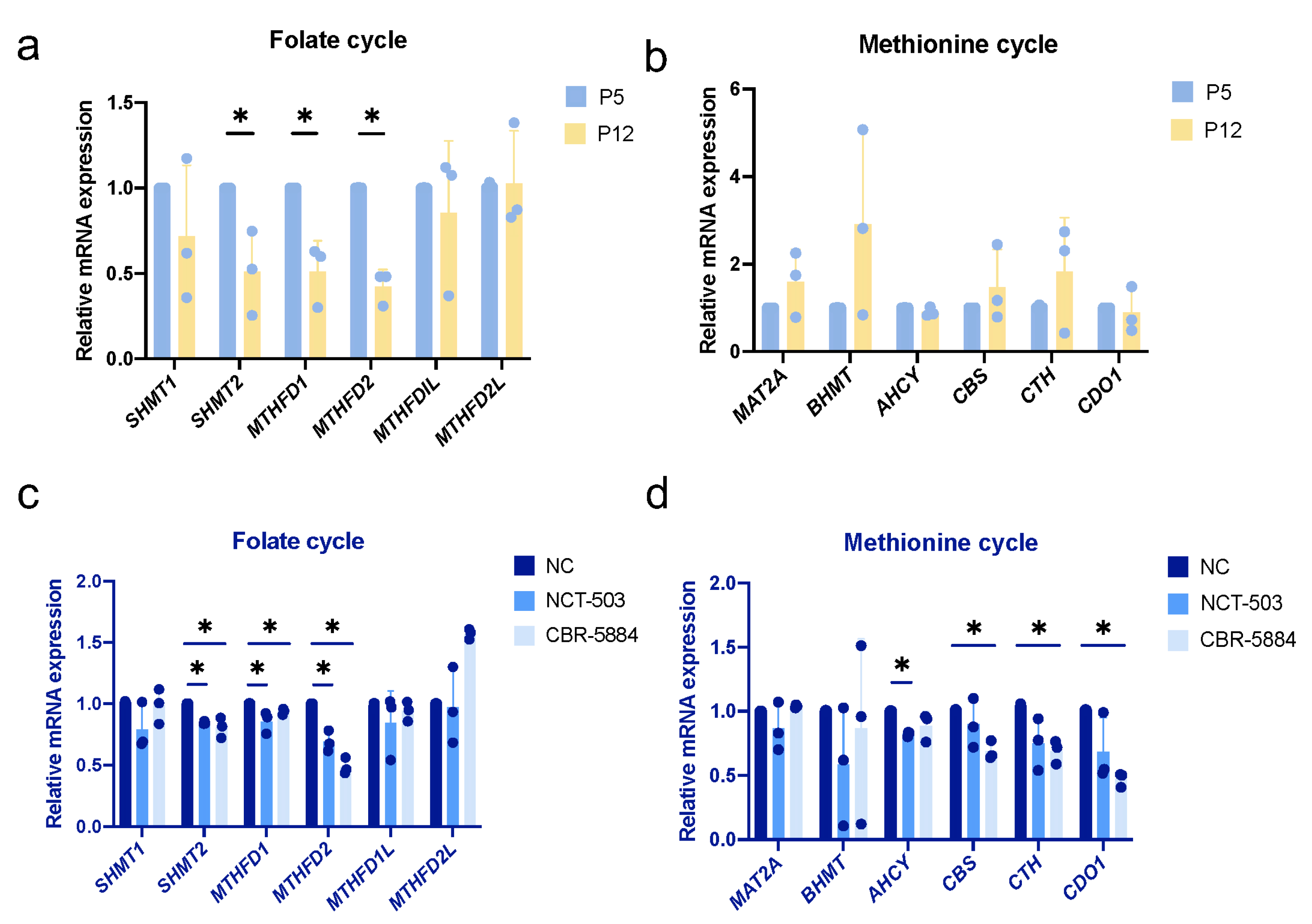
3.7. Folate Metabolism Is Altered during Cellular Senescence
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed]
- Nuti, N.; Corallo, C.; Chan, B.M.; Ferrari, M.; Gerami-Naini, B. Multipotent Differentiation of Human Dental Pulp Stem Cells: A Literature Review. Stem. Cell Rev. Rep. 2016, 12, 511–523. [Google Scholar] [CrossRef]
- Peng, L.; Ye, L.; Zhou, X.D. Mesenchymal stem cells and tooth engineering. Int. J. Oral. Sci. 2009, 1, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef]
- Turinetto, V.; Vitale, E.; Giachino, C. Senescence in Human Mesenchymal Stem Cells: Functional Changes and Implications in Stem Cell-Based Therapy. Int. J. Mol. Sci. 2016, 17, 1164. [Google Scholar] [CrossRef] [PubMed]
- Wiley, C.D.; Campisi, J. The metabolic roots of senescence: Mechanisms and opportunities for intervention. Nat. Metab. 2021, 3, 1290–1301. [Google Scholar] [CrossRef]
- López-Otín, C.; Galluzzi, L.; Freije, J.M.P.; Madeo, F.; Kroemer, G. Metabolic Control of Longevity. Cell 2016, 166, 802–821. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef]
- Kozieł, R.; Ruckenstuhl, C.; Albertini, E.; Neuhaus, M.; Netzberger, C.; Bust, M.; Madeo, F.; Wiesner, R.J.; Jansen-Dürr, P. Methionine restriction slows down senescence in human diploid fibroblasts. Aging Cell 2014, 13, 1038–1048. [Google Scholar] [CrossRef]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef]
- Mentch, S.J.; Mehrmohamadi, M.; Huang, L.; Liu, X.; Gupta, D.; Mattocks, D.; Gómez Padilla, P.; Ables, G.; Bamman, M.M.; Thalacker-Mercer, A.E.; et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015, 22, 861–873. [Google Scholar] [CrossRef]
- Yang, R.L.; Huang, H.M.; Han, C.S.; Cui, S.J.; Zhou, Y.K.; Zhou, Y.H. Serine Metabolism Controls Dental Pulp Stem Cell Aging by Regulating the DNA Methylation of p16. J. Dent. Res. 2021, 100, 90–97. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, L.; Huang, H.; Yu, Q.; Hu, B.; Wang, G.; Ge, F.; Yin, T.; Li, S.; Yu, X. Phosphoglycerate dehydrogenase activates PKM2 to phosphorylate histone H3T11 and attenuate cellular senescence. Nat. Commun. 2023, 14, 1323. [Google Scholar] [CrossRef]
- Shimi, T.; Butin-Israeli, V.; Adam, S.A.; Hamanaka, R.B.; Goldman, A.E.; Lucas, C.A.; Shumaker, D.K.; Kosak, S.T.; Chandel, N.S.; Goldman, R.D. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011, 25, 2579–2593. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Iohara, K.; Imabayashi, K.; Ishizaka, R.; Watanabe, A.; Nabekura, J.; Ito, M.; Matsushita, K.; Nakamura, H.; Nakashima, M. Complete pulp regeneration after pulpectomy by transplantation of CD105+ stem cells with stromal cell-derived factor-1. Tissue Eng. Part A 2011, 17, 1911–1920. [Google Scholar] [CrossRef]
- Ito, K.; Yamada, Y.; Nakamura, S.; Ueda, M. Osteogenic potential of effective bone engineering using dental pulp stem cells, bone marrow stem cells, and periosteal cells for osseointegration of dental implants. Int. J. Oral Maxillofac. Implants 2011, 26, 947–954. [Google Scholar]
- Riccio, M.; Maraldi, T.; Pisciotta, A.; La Sala, G.B.; Ferrari, A.; Bruzzesi, G.; Motta, A.; Migliaresi, C.; De Pol, A. Fibroin scaffold repairs critical-size bone defects in vivo supported by human amniotic fluid and dental pulp stem cells. Tissue Eng. Part A 2012, 18, 1006–1013. [Google Scholar] [CrossRef]
- Aimetti, M.; Ferrarotti, F.; Cricenti, L.; Mariani, G.M.; Romano, F. Autologous dental pulp stem cells in periodontal regeneration: A case report. Int. J. Periodontics Restorative Dent. 2014, 34 (Suppl. S3), s27–s33. [Google Scholar] [CrossRef]
- Li, H.; Fan, X.; Kovi, R.C.; Jo, Y.; Moquin, B.; Konz, R.; Stoicov, C.; Kurt-Jones, E.; Grossman, S.R.; Lyle, S.; et al. Spontaneous expression of embryonic factors and p53 point mutations in aged mesenchymal stem cells: A model of age-related tumorigenesis in mice. Cancer Res. 2007, 67, 10889–10898. [Google Scholar] [CrossRef]
- Ngo, B.; Kim, E.; Osorio-Vasquez, V.; Doll, S.; Bustraan, S.; Liang, R.J.; Luengo, A.; Davidson, S.M.; Ali, A.; Ferraro, G.B.; et al. Limited Environmental Serine and Glycine Confer Brain Metastasis Sensitivity to PHGDH Inhibition. Cancer Discov. 2020, 10, 1352–1373. [Google Scholar] [CrossRef]
- Shan, X.; Hu, P.; Ni, L.; Shen, L.; Zhang, Y.; Ji, Z.; Cui, Y.; Guo, M.; Wang, H.; Ran, L.; et al. Serine metabolism orchestrates macrophage polarization by regulating the IGF1-p38 axis. Cell. Mol. Immunol. 2022, 19, 1263–1278. [Google Scholar] [CrossRef]
- Ye, J.; Mancuso, A.; Tong, X.; Ward, P.S.; Fan, J.; Rabinowitz, J.D.; Thompson, C.B. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl. Acad. Sci. USA 2012, 109, 6904–6909. [Google Scholar] [CrossRef]
- Stegen, S.; Loopmans, S.; Stockmans, I.; Moermans, K.; Carmeliet, P.; Carmeliet, G. De novo serine synthesis regulates chondrocyte proliferation during bone development and repair. Bone Res. 2022, 10, 14. [Google Scholar] [CrossRef]
- Vandekeere, S.; Dubois, C.; Kalucka, J.; Sullivan, M.R.; García-Caballero, M.; Goveia, J.; Chen, R.; Diehl, F.F.; Bar-Lev, L.; Souffreau, J.; et al. Serine Synthesis via PHGDH Is Essential for Heme Production in Endothelial Cells. Cell Metab. 2018, 28, 573–587.e513. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Hu, Q.; Wang, L.; Liu, J.; Zheng, Z.; Zhang, W.; Ren, J.; Zhu, F.; Liu, G.H. Epigenetic regulation of aging: Implications for interventions of aging and diseases. Signal. Transduct. Target. Ther. 2022, 7, 374. [Google Scholar] [CrossRef]
- Sen, P.; Shah, P.P.; Nativio, R.; Berger, S.L. Epigenetic Mechanisms of Longevity and Aging. Cell 2016, 166, 822–839. [Google Scholar] [CrossRef]
- Sen, P.; Dang, W.; Donahue, G.; Dai, J.; Dorsey, J.; Cao, X.; Liu, W.; Cao, K.; Perry, R.; Lee, J.Y.; et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015, 29, 1362–1376. [Google Scholar] [CrossRef]
- Bonkowski, M.S.; Sinclair, D.A. Slowing ageing by design: The rise of NAD (+) and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 2016, 17, 679–690. [Google Scholar] [CrossRef]
- Karsenty, G.; Kronenberg, H.M.; Settembre, C. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 2009, 25, 629–648. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Anderson, O.S.; Sant, K.E.; Dolinoy, D.C. Nutrition and epigenetics: An interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutr. Biochem. 2012, 23, 853–859. [Google Scholar] [CrossRef]
- Li, A.M.; Ye, J. Reprogramming of serine, glycine and one-carbon metabolism in cancer. Biochim. Biophys Acta Mol. Basis. Dis. 2020, 1866, 165841. [Google Scholar] [CrossRef]
- Pacold, M.E.; Brimacombe, K.R.; Chan, S.H.; Rohde, J.M.; Lewis, C.A.; Swier, L.J.; Possemato, R.; Chen, W.W.; Sullivan, L.B.; Fiske, B.P.; et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 2016, 12, 452–458. [Google Scholar] [CrossRef]


| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| qPCR | ||
| P21 | 5′-GATGAGTTGGGAGGAGGCAG-3′ | 5′-CTGAGAGTCTCCAGGTCCAC-3′ |
| LMNB1 | 5′-AAGCATGAAACGCGCTTGG-3′ | 5′-AGTTTGGCATGGTAAGTCTGC-3′ |
| ALP | 5′-GACCTCCTCGGAAGACACTC-3′ | 5′-TGAAGGGCTTCTTGTCTGTG-3′ |
| RUNX2 | 5′-GACTGTGGTTACCGTCATGGC-3′ | 5′-ACTTGGTTTTTCATAACAGCGGA-3′ |
| COL1A1 | 5′-TCTAGACATGTTCAGCTTTGTGGAC-3′ | 5′-TCTGTACGCAGGTGATTGGTG-3′ |
| PHGDH | 5′-GCAAATCTGCGGAAAGTGCT-3′ | 5′-ATAAGGCCTTCACAGTCCTGC-3′ |
| PSAT1 | 5′- AAAAACAATGGAGGTGCCGC-3′ | 5′-GGCTCCACTGGACAAACGTA-3′ |
| PSPH | 5′-GTAGGGCTCTGGATGCTGC-3′ | 5′-TGCAAGTGCTTCTGTAAACTTAAAA-3′ |
| SETD2 | 5′-TGCTTCTAGTCGATTTTTGCCC-3′ | 5′-AGGGTTTGGAGTATCACTTTGC-3′ |
| KDM4A | 5′-CCTCACTGCGCTGTCTGTAT-3′ | 5′-CCAGTCGAAGTGAAGCACAT-3′ |
| SHMT1 | 5′-TACCCGGGCCAGAGATACTA-3′ | 5′-CTGAGTAGGGCTGGACGTTG-3′ |
| SHMT2 | 5′-TTCTCTTTGTTTTGGGCGGC-3′ | 5′-TGTTTGCTTCCCCAGTCTGA-3′ |
| MTHFD1 | 5′-TAGGAACGATGAGCACAATGC-3′ | 5′-AGACACTGGCCAGACTTTCAA-3′ |
| MTHFD2 | 5′-TGGCTGCGACTTCTCTAATG-3′ | 5′-CCTTCCAGAAATGACAACAGC-3′ |
| MTHFD1L | 5′-CCCTTTGGTCGGAACGATGA-3′ | 5′-TGCCGAACACCATACTCCAC-3′ |
| MTHFD2L | 5′-CCAGGAGGTGATGCAACTGT-3′ | 5′-TCCTGTCACTGGATCGTGGA-3′ |
| AHCY | 5′-ATTCCGGTGTATGCCTGGAAG-3′ | 5′-GAGATGCCTCGGATGCCTG-3′ |
| BHMT | 5′-TGGAGAACAGGGGCAACTATG-3′ | 5′-CTGACTCACTCCTCCTGCTAC-3′ |
| MAT2A | 5′-GACATTGGTGCTGGAGACCA-3′ | 5′-ACTCTGATGGGAAGCACAGC-3′ |
| CBS | 5′-GATTATCGAGCCGACATCCG-3′ | 5′-GTCCTCACAATCTCAGCCC-3′ |
| CTH | 5′-TTAGCCTATTGCGTCATTTAAGCA-3′ | 5′-CTGCACCCGGTCATGAGTT-3′ |
| CDO1 | 5′-TCTCTGTTGGGGTGAAGGAC-3′ | 5′-GCCAGGCAAATAATGTCTCC-3′ |
| -actin | 5′-CCTCGCCTTTGCCGATCC-3′ | 5′-CGCGGCGATATCATCATCC-3′ |
| ChIP-qPCR | ||
| SIRT1 | 5′-AGAAACGCTGTGCTCCAGGCAGATG-3′ | 5′-GTAAAACGAGGGGTACCTAGTAGTTC-3′ |
| RUNX2 | 5′-CCCAAGCTCATCTTGTACTCG-3′ | 5′-CCTAGAAGGGGCCTGGAA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, S.; Cui, J.; Shi, Y. Serine Metabolism Regulates the Replicative Senescence of Human Dental Pulp Cells through Histone Methylation. Curr. Issues Mol. Biol. 2024, 46, 2856-2870. https://doi.org/10.3390/cimb46040179
Zhou S, Cui J, Shi Y. Serine Metabolism Regulates the Replicative Senescence of Human Dental Pulp Cells through Histone Methylation. Current Issues in Molecular Biology. 2024; 46(4):2856-2870. https://doi.org/10.3390/cimb46040179
Chicago/Turabian StyleZhou, Shuhan, Jingyao Cui, and Yu Shi. 2024. "Serine Metabolism Regulates the Replicative Senescence of Human Dental Pulp Cells through Histone Methylation" Current Issues in Molecular Biology 46, no. 4: 2856-2870. https://doi.org/10.3390/cimb46040179





