Functional Validation of Different Alternative Splicing Variants of the Chrysanthemum lavandulifolium ClNUM1 Gene in Tobacco
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Vectors and Strains
2.2. Homologous Gene Cloning
2.3. Stress Treatment of Chrysanthemum lavandulifolium
2.4. RNA Extraction and RT-qPCR
2.5. Agrobacterium Transformation Method
2.6. Transgenic Tobacco Resistance Screening and Management and Maintenance Methods
2.7. Abiotic Stress Treatments and Morphometric Measurements
2.7.1. Stress Treatment of Transgenic Tobacco at the Seedling Stage
2.7.2. Stress Treatment of Transgenic Tobacco at the Mature Stage
2.8. Measurement of Physiological Parameters
2.9. Paraffin Sectioning
2.10. Statistical Analysis
3. Results
3.1. Bioinformatics Analysis of the ClNUM1 Splice Variants
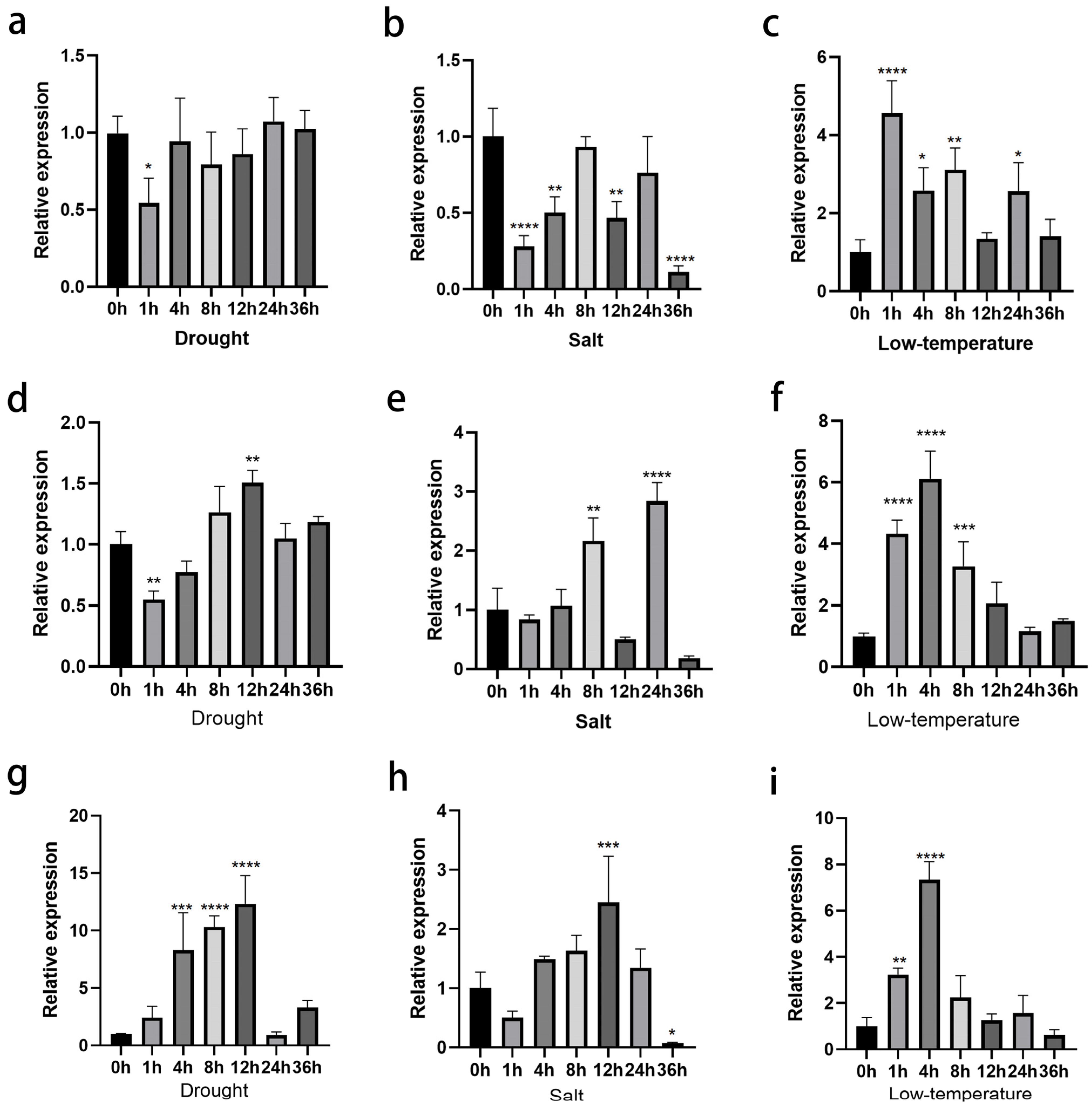
3.2. ClNUM1.1, ClNUM1.2, and ClNUM1.3 in C. lavandulifolium Show Different Responses to Stress Treatments
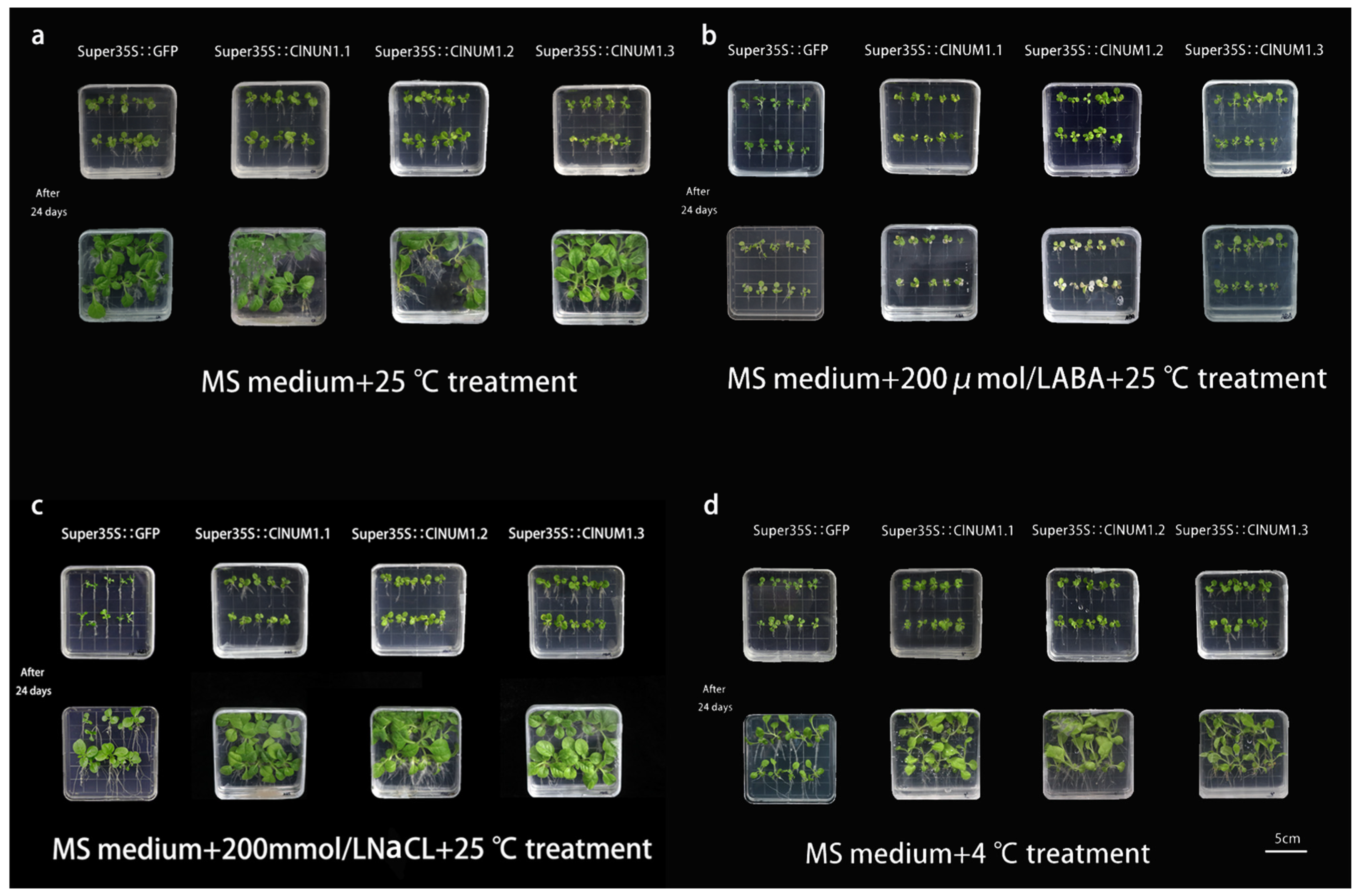
3.3. Phenotypic Analysis of Young ClNUM1 Transgenic Tobacco under Abiotic Stress
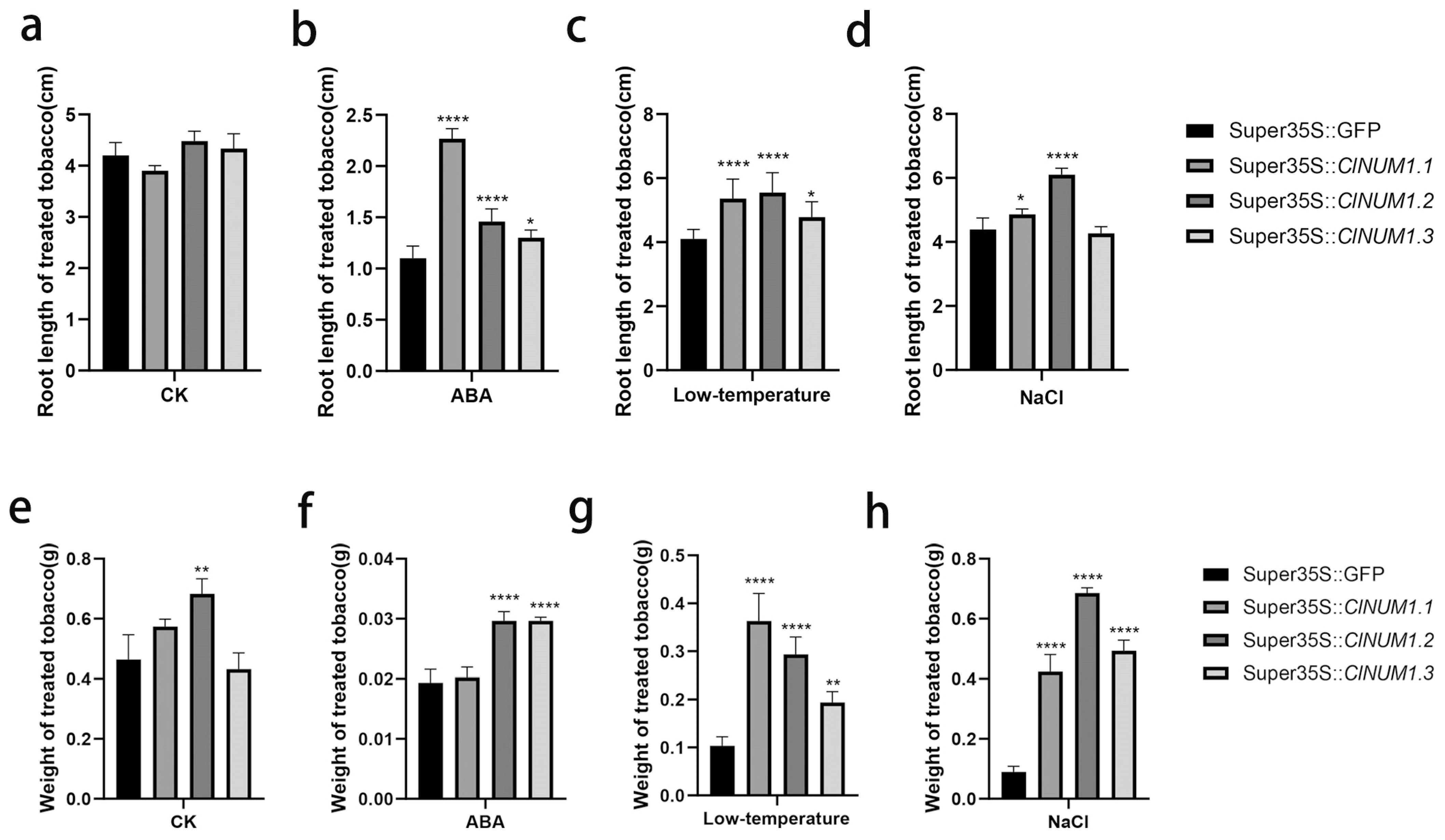
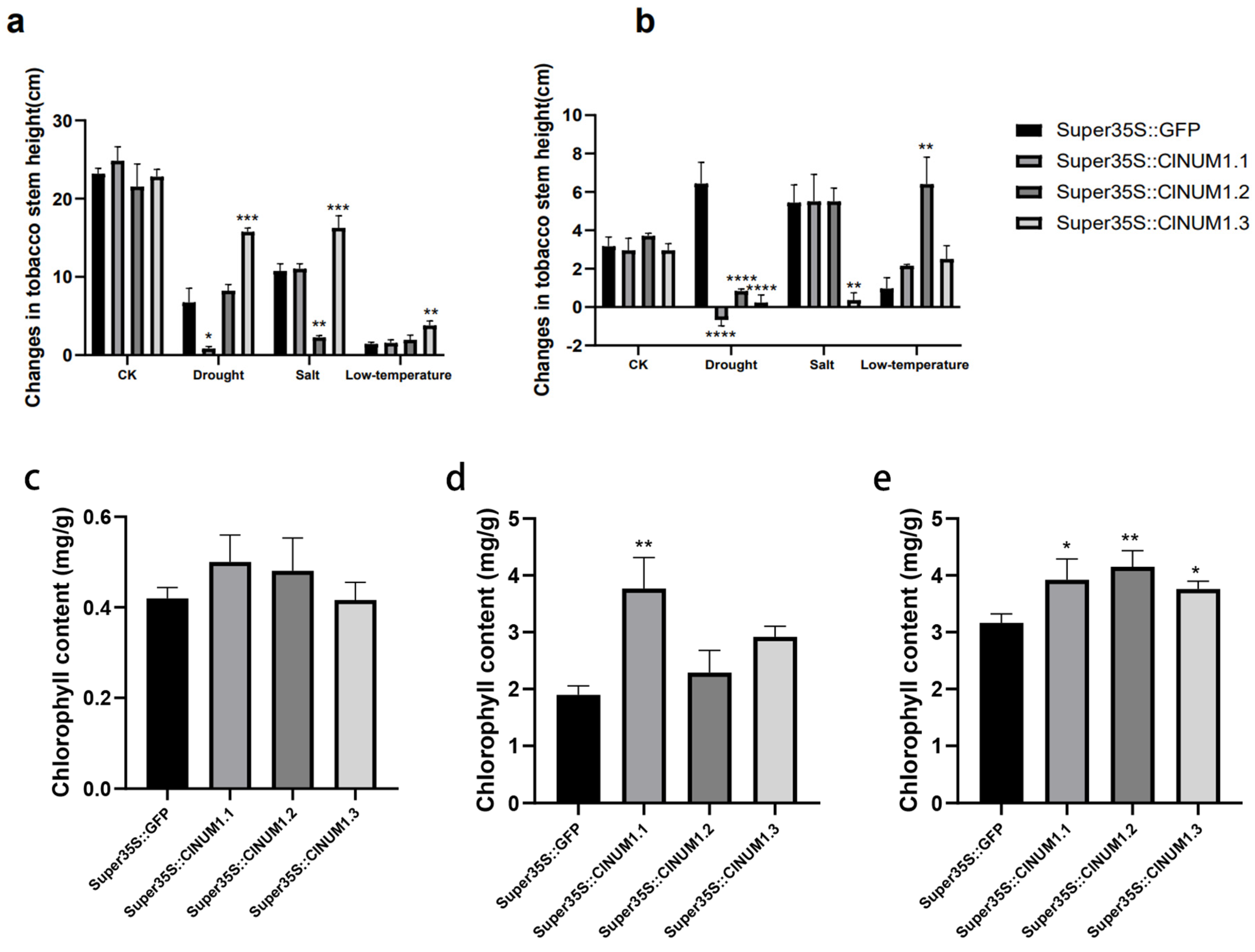
3.4. Phenotypic Study of ClNUM1 Transgenic Tobacco at Maturity under Abiotic Stress and Determination of Physiological Indexes
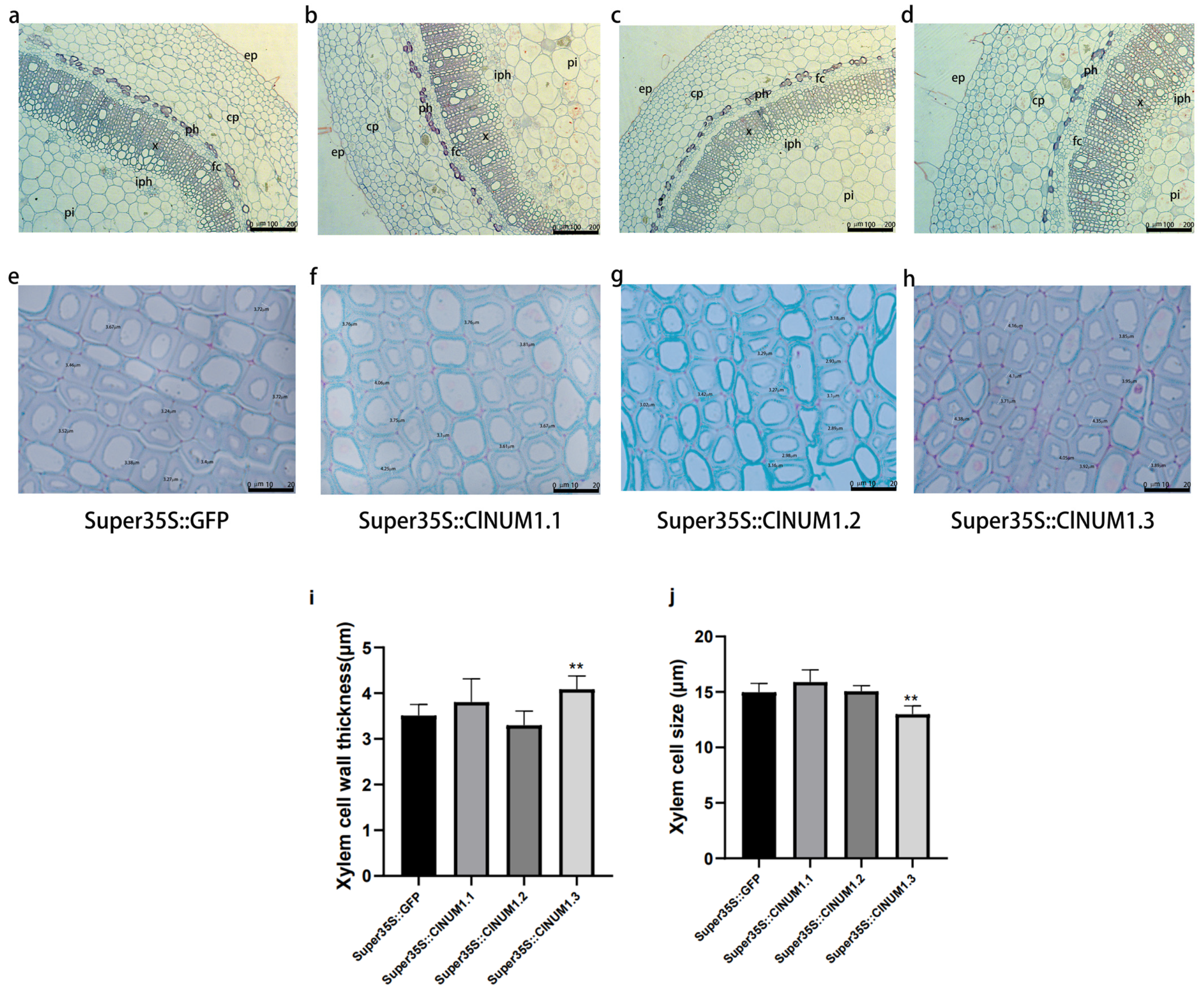
3.5. Anatomical Analysis of Transgenic Tobacco Stalks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, L.; Dang, J.; Liu, X.; Jiang, H. Investigation and Research on the Applied Value of Chrysanthemum. Contemp. Hortic. 2022, 13, 21–23. [Google Scholar] [CrossRef]
- Cheng, X. Interspecific Hybridization between Chrysanthemum Species and Research on Resistant Germplasm Innovation. Master’s Thesis, University of Nanjing Agriculture, Nanjing, China, 2012. [Google Scholar]
- Peng, Y. Salt-Tolerance Analysis of Chrysanthemum lavandulifolium and Transformation of DlBADH Promoters to Chrysanthemum. Master’s Thesis, Beijing Forestry University, Beijing, China, 2011. [Google Scholar]
- An, C.; Sheng, L.P.; Du, X.P.; Wang, Y.J.; Zhang, Y.; Song, A.P.; Jiang, J.F.; Guan, Z.Y.; Fang, W.M.; Chen, F.D.; et al. Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin. Hortic. Res. 2019, 6, 84. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Y.; Wang, S.L.; Wu, X.; Yang, K.; Niu, Y.J.; Dai, S.L. Transcriptome-wide survey and expression analysis of stress-responsive NAC genes in Chrysanthemum lavandulifolium. Plant Sci. 2012, 193, 18–27. [Google Scholar] [CrossRef]
- Wang, K.; Wu, Y.H.; Tian, X.Q.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B. Overexpression of DgWRKY4 Enhances Salt Tolerance in Chrysanthemum Seedlings. Front. Plant Sci. 2017, 8, 1592. [Google Scholar] [CrossRef]
- Jaffar, M.A.; Song, A.P.; Faheem, M.; Chen, S.M.; Jiang, J.F.; Liu, C.; Fan, Q.Q.; Chen, F.D. Involvement of CmWRKY10 in Drought Tolerance of Chrysanthemum through the ABA-Signaling Pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef] [PubMed]
- Li, C.N.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Y.; Tian, C.; Wang, X.H.; Zhou, L.J.; Jiang, J.F.; Wang, L.K.; Chen, F.D.; Chen, S.M. Molecular module of CmMYB15-like-Cm4CL2 regulating lignin biosynthesis of chrysanthemum (Chrysanthemum morifolium) in response to aphid (Macrosiphoniella sanborni) feeding. New Phytol. 2023, 237, 1776–1793. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhong, M.; Wu, Y.H.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B.; Jia, Y.; et al. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Sheng, L.P.; Zhang, H.R.; Du, X.P.; An, C.; Xia, X.L.; Chen, F.D.; Jiang, J.F.; Chen, S.M. CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis. Int. J. Mol. Sci. 2017, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.J.; Geng, Z.Q.; Wang, Y.X.; Wang, Y.G.; Liu, S.H.; Chen, C.W.; Song, A.P.; Jiang, J.F.; Chen, S.M.; Chen, F.D. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic. Res. 2021, 8, 248. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.N.; Hao, X.Y.; Zhang, W.X.; Xu, Y.X.; He, W.T.; Li, Y.X.; Cai, S.Y.; Zhao, X.; Song, X.B. Alternative Splicing for Leucanthemella linearis NST1Contributes to Variable Abiotic Stress Resistance in Transgenic Tobacco. Genes 2023, 14, 1549. [Google Scholar] [CrossRef] [PubMed]
- Gao, G. Chrysanthemum CmWRKY15-1 Enhanced Resistance to Chrysanthemum White Rust by Regulating the Expression of CmNPR1. Master’s Thesis, Shenyang Agricultural University, Shenyang, China, 2022. [Google Scholar]
- Meng, R. Study on the Function of Shennong Chrysanthemum CiMYB32 in Response to Drought Stress. Master’s Thesis, Northeast Forestry University, Harbin, China, 2023. [Google Scholar]
- Sitbon, E.; Pietrokovski, S. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 2003, 28, 473–477. [Google Scholar] [CrossRef]
- Belfort, M.; Roberts, R.J. Homing endonucleases: Keeping the house in order. Nucleic Acids Res. 1997, 25, 3379–3388. [Google Scholar] [CrossRef]
- Lagerback, P.; Carlson, K. Amino Acid Residues in the GIY-YIG Endonuclease II of Phage T4 Affecting Sequence Recognition and Binding as Well as Catalysis. J. Bacteriol. 2008, 190, 5533–5544. [Google Scholar] [CrossRef]
- Andersson, C.E.; Lagerback, P.; Carlson, K. Structure of Bacteriophage T4 Endonuclease II Mutant E118A, a Tetrameric GIY-YIG Enzyme. J. Mol. Biol. 2010, 379, 1003–1016. [Google Scholar] [CrossRef]
- Van Roey, P.; Waddling, C.A.; Fox, K.M.; Belfort, M.; Derbyshire, V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001, 20, 3631–3637. [Google Scholar] [CrossRef]
- Raczynska, K.D.; Simpson, C.G.; Ciesiolka, A.; Szewc, L.; Lewandowska, D.; McNicol, J.; Szweykowska-Kulinska, Z.; Brown, J.W.S.; Jarmolowski, A. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010, 38, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jia, Z.; Pu, Q.; Tian, Y.; Zhu, F.; Liu, Y. ABA Mediates Plant Development and Abiotic Stress via Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 3796. [Google Scholar] [CrossRef]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.S.; Nimmo, H.G. Alternative Splicing Mediates Responses of the Arabidopsis Circadian Clock to Temperature Changes. Plant Cell 2012, 24, 961–981. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Xu, Q.; Wang, X. Regulation of the circadian clock through pre-mRNA splicing in Arabidopsis. J. Exp. Bot. 2014, 65, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.; Riegler, S.; Ayatollahi, Z.; Cavallari, N.; Giono, L.E.; Nimeth, B.A.; Mutanwad, K.V.; Schweighofer, A.; Lucyshyn, D.; Barta, A.; et al. Targeting alternative splicing by RNAi: From the differential impact on splice variants to triggering artificial pre-mRNA splicing. Nucleic Acids Res. 2021, 49, 1133–1151. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Liu, Y.; Kang, D.; Fan, K.; Wang, C.; Wang, G.; Liu, Y. Two alternative splicing variants of maize HKT1;1 confer salt tolerance in transgenic tobacco plants. Plant Cell Tissue Organ Cult. 2015, 123, 569–578. [Google Scholar] [CrossRef]
- Lv, B.; Hu, K.; Tian, T.; Wei, K.; Zhang, F.; Jia, Y.; Tian, H.; Ding, Z. The pre-mRNA splicing factor RDM16 regulates root stem cell maintenance in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 662–675. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Deng, Q.; Wu, M.; Wang, Z.; Wei, D.; Wang, H.; Xiang, H.; Zhang, H.; Tang, Q. Mechanisms of alternative splicing in regulating plant flowering: A review. Chin. J. Biotechnol. 2021, 37, 2991–3004. [Google Scholar] [CrossRef]
- Simpson, C.G.; Fuller, J.; Calixto, C.P.G.; McNicol, J.; Booth, C.; Brown, J.W.S.; Staiger, D. Monitoring Alternative Splicing Changes in Arabidopsis Circadian Clock Genes. Methods Mol. Biol. 2016, 1398, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Jabre, I.; Reddy, A.S.N.; Staiger, D.; Syed, N.H. Perspective on Alternative Splicing and Proteome Complexity in Plants. Trends Plant Sci. 2019, 24, 496–506. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Van Allen, B.; Habben, J.; Li, B.L. Genome-Wide Analysis of Alternative Splicing during Development and Drought Stress in Maize. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Alhabsi, A.; Butt, H.; Kirschner, G.K.; Blilou, I.; Mahfouz, M.M.; Sunkar, R. SCR106 splicing factor modulates abiotic stress responses by maintaining RNA splicing in rice. J. Exp. Bot. 2023, 75, 802–818. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.-X.; Wu, Y.-M.; Zhang, J.-X.; Zhang, J.-X.; Zhou, H.; Zeng, R.-F.; Zheng, W.-X.; Qiu, M.-Q.; Zhou, J.-J.; Xie, Z.-Z.; et al. A bZIP transcription factor CiFD regulates drought- and low-temperature-induced flowering by alternative splicing in citrus. J. Integr. Plant Biol. 2023, 65, 674–691. [Google Scholar] [CrossRef]
- Albaqami, M.; Laluk, K.; Reddy, A.S.N. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 2019, 100, 379–390. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.R.; Tittes, S.; Mendieta, J.P.; Collier-zans, E.; Rowe, H.C.; Rieseberg, L.H.; Kane, N.C. Genetics of alternative splicing evolution during sunflower domestication. Proc. Natl. Acad. Sci. USA 2018, 115, 6768–6773. [Google Scholar] [CrossRef]
- Iñiguez, L.P.; Ramírez, M.; Barbazuk, W.B.; Hernández, G. Identification and analysis of alternative splicing events in Phaseolus vulgaris and Glycine max. BMC Genom. 2017, 18, 650. [Google Scholar] [CrossRef]
- Xu, Y.; Cheng, J.; Hu, H.; Yan, L.; Jia, J.; Wu, B. Genome-Wide Identification of NAC Family Genes in Oat and Functional Characterization of AsNAC109 in Abiotic Stress Tolerance. Plants 2024, 13, 1017. [Google Scholar] [CrossRef]
- Chen, N.; Shao, Q.; Lu, Q.; Li, X.; Gao, Y.; Xiao, Q. Research progress on function of NAC transcription factors in tomato (Solanum lycopersicum L.). Euphytica 2023, 219, 22. [Google Scholar] [CrossRef]
- Takata, N.; Awano, T.; Nakata, M.T.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Luo, F.; Zhong, Y.; He, J.; Li, L. Modulation of NAC transcription factor NST1 activity by XYLEM NAC DOMAIN1 regulates secondary cell wall formation in Arabidopsis. J. Exp. Bot. 2020, 71, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, C.; Wang, Y.; Zhang, Y.; Zhang, Y.; Wang, Y.; Wang, C. Expression profiles of genes regulated by BplMYB46 in Betula platyphylla. J. For. Res. 2019, 30, 2267–2276. [Google Scholar] [CrossRef]
- Malacarne, G.; Lagreze, J.; Martin, B.R.S.; Malnoy, M.; Moretto, M.; Moser, C.; Costa, L.D. Insights into the cell-wall dynamics in grapevine berries during ripening and in response to biotic and abiotic stress. Plant Mol. Biol. 2024, 114, 38. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, W.; Niu, D.; Liu, X. Effects of abiotic stress on chlorophyll metabolism. Plant Sci. 2024, 342, 112030. [Google Scholar] [CrossRef]


| CsNST1 Primer Sequence | |
|---|---|
| NST1-F | 5′-ATGCTGTCTTCTACTTTGTAAGCTC-3′ |
| NST1-R | 5′-TTAATACCCTGTTTCAGAAAATGGATG-3′ |
| Actin7 Primer Sequence | |
|---|---|
| Actin7-F | 5′-TCCGGCTATGTATGTTGCTATTC-3′ |
| Actin7-R | 5′-AATCTTCATCAAGGGATCGGTAAG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Wang, H.; Guo, Y.; Hao, X.; Li, Y.; He, W.; Zhao, X.; Cai, S.; Song, X. Functional Validation of Different Alternative Splicing Variants of the Chrysanthemum lavandulifolium ClNUM1 Gene in Tobacco. Curr. Issues Mol. Biol. 2024, 46, 5242-5256. https://doi.org/10.3390/cimb46060314
Zhang W, Wang H, Guo Y, Hao X, Li Y, He W, Zhao X, Cai S, Song X. Functional Validation of Different Alternative Splicing Variants of the Chrysanthemum lavandulifolium ClNUM1 Gene in Tobacco. Current Issues in Molecular Biology. 2024; 46(6):5242-5256. https://doi.org/10.3390/cimb46060314
Chicago/Turabian StyleZhang, Wenxin, Hai Wang, Yuning Guo, Xueying Hao, Yanxi Li, Wenting He, Xiang Zhao, Shiyi Cai, and Xuebin Song. 2024. "Functional Validation of Different Alternative Splicing Variants of the Chrysanthemum lavandulifolium ClNUM1 Gene in Tobacco" Current Issues in Molecular Biology 46, no. 6: 5242-5256. https://doi.org/10.3390/cimb46060314
APA StyleZhang, W., Wang, H., Guo, Y., Hao, X., Li, Y., He, W., Zhao, X., Cai, S., & Song, X. (2024). Functional Validation of Different Alternative Splicing Variants of the Chrysanthemum lavandulifolium ClNUM1 Gene in Tobacco. Current Issues in Molecular Biology, 46(6), 5242-5256. https://doi.org/10.3390/cimb46060314




