Comparison of the Proteome of Huh7 Cells Transfected with Hepatitis B Virus Subgenotype A1, with or without G1862T
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs
2.2. Cell Line and Transfections
2.3. Mass Spectrometry Analysis
2.3.1. Cell Lysis and Protein Preparation
2.3.2. Sample Clean-Up and Digestion
2.3.3. LC-MS Data Acquisition
2.3.4. SWATH-MS Data Processing
2.3.5. Mass Spectrometry Biological Data Annotation
2.4. RNA Expression Levels
2.5. Viral Loads
2.6. HBeAg Expression
2.7. Immunofluorescence
2.8. Microscopy and Image Analysis
2.9. Data Statistical Analysis
3. Results
3.1. Comparison of Viral Load and HBeAg Expression
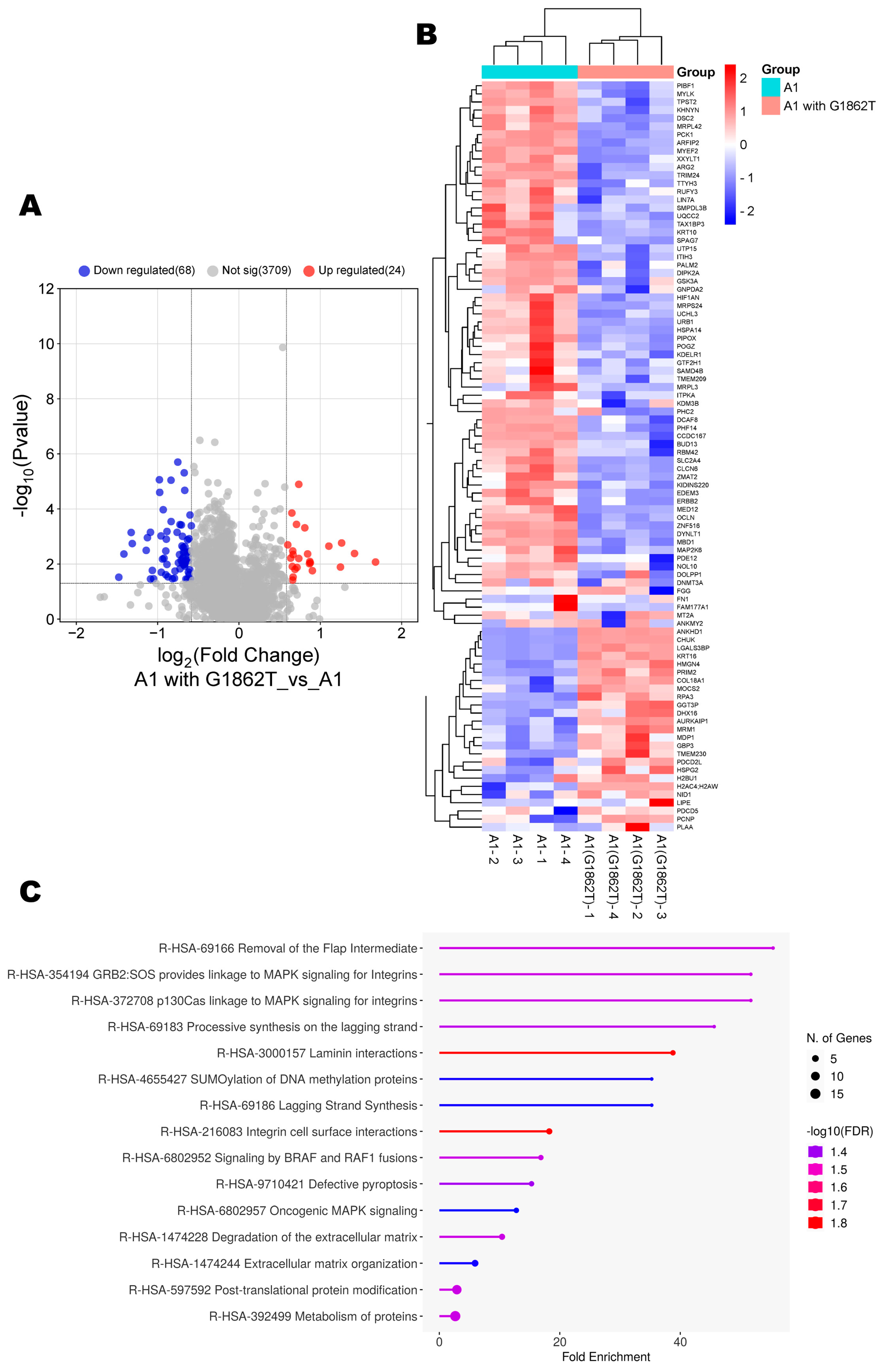
3.2. Comparison of the Proteome of Huh7 Cells Transfected with Wildtype and Mutant Subgenotype A1 Compared to the Vector Control
3.3. Proteomic Analysis of Huh7 Cells Transfected with Wildtype Compared to the Mutant
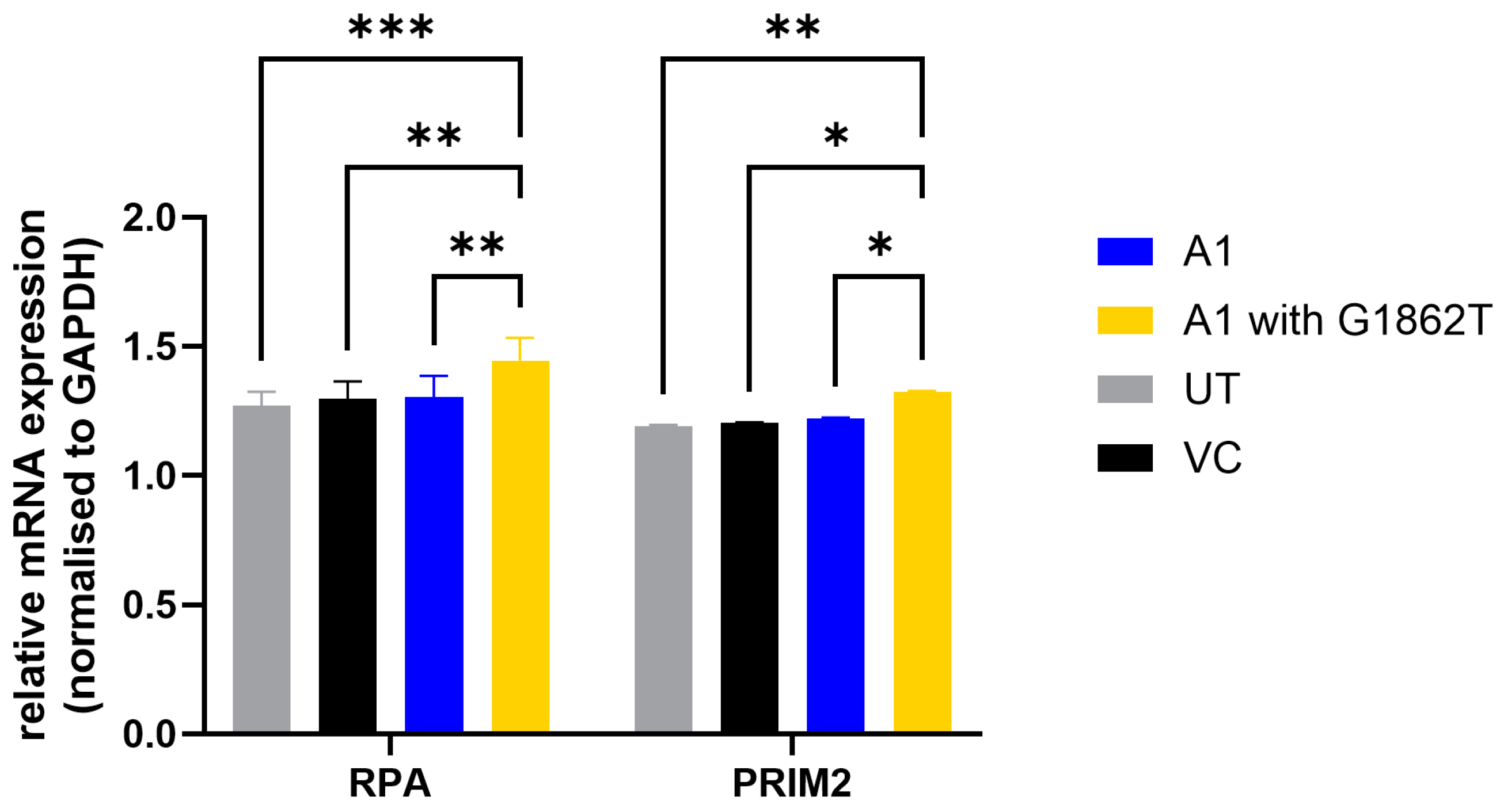
3.4. Increased Transcription of DNA Primase Subunit 2 (PRIM2) and RPA in G1862T-Transfected Cells Compared to WT-Transfected Cells
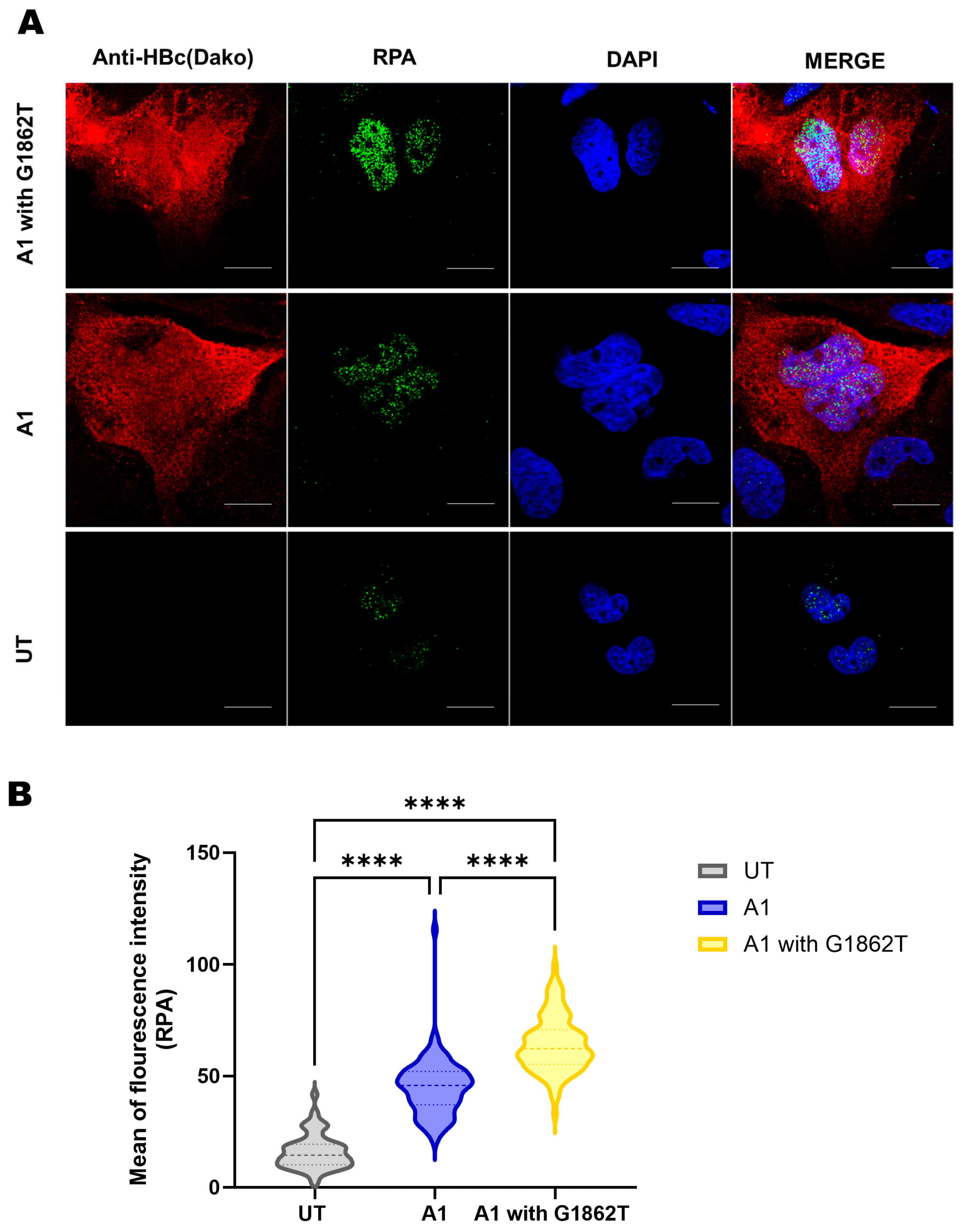
3.5. Immunofluorescence Reveals Increased Expression of RPA in G1862T-Transfected Cells Compared to WT-Transfected Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Hepatitis Report 2024: Action for Access in Low- and Middle-Income Countries; WHO: Geneva, Switzerland, 2024; ISBN 978-92-4-009167-2. [Google Scholar]
- Chauhan, R.; Kazim, S.N.; Bhattacharjee, J.; Sakhuja, P.; Sarin, S.K. Basal core promoter, precore region mutations of HBV and their association with e antigen, genotype, and severity of liver disease in patients with chronic hepatitis B in India. J. Med. Virol. 2006, 78, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Yuen, M.F.; Sablon, E.; Yuan, H.J.; Hui, C.K.; Wong, D.K.H.; Doutreloigne, J.; Wong, B.C.Y.; Chan, A.O.O.; Lai, C.L. Relationship between the development of precore and core promoter mutations and hepatitis B e antigen seroconversion in patients with chronic hepatitis B virus. J. Infect. Dis. 2002, 186, 1335–1338. [Google Scholar] [CrossRef]
- Milich, D.R. Is the function of the HBeAg really unknown? Hum. Vaccines Immunother. 2019, 15, 2187–2191. [Google Scholar] [CrossRef] [PubMed]
- Kamijo, N.; Matsumoto, A.; Umemura, T.; Shibata, S.; Ichikawa, Y.; Kimura, T.; Komatsu, M.; Tanaka, E. Mutations of pre-core and basal core promoter before and after hepatitis B e antigen seroconversion. World J. Gastroenterol. 2015, 21, 541–548. [Google Scholar] [CrossRef]
- Alexopoulou, A.; Karayiannis, P. HBeAg negative variants and their role in the natural history of chronic hepatitis B virus infection. World J. Gastroenterol. 2014, 20, 7644–7652. [Google Scholar] [CrossRef]
- Ghosh, S.; Mondal, R.K.; Banerjee, P.; Nandi, M.; Sarkar, S.; Das, K.; Santra, A.; Banerjee, S.; Chowdhury, A.; Datta, S. Tracking the naturally occurring mutations across the full-length genome of hepatitis B virus of genotype D in different phases of chronic e-antigen-negative infection. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, E412–E418. [Google Scholar] [CrossRef]
- Belyhun, Y.; Liebert, U.G.; Maier, M. Analysis of HBV basal core promoter/precore gene variability in patients with HBV drug resistance and HIV co-infection in Northwest Ethiopia. PLoS ONE 2018, 13, e0191970. [Google Scholar] [CrossRef] [PubMed]
- Mayaphi, S.H.; Martin, D.J.; Mphahlele, M.J.; Blackard, J.T.; Bowyer, S.M.; Nachega, J.B. Variability of the preC/C region of hepatitis B virus genotype A from a South African cohort predominantly infected with HIV. J. Med. Virol. 2013, 85, 1883–1892. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A. Genotypes and Genetic Variability of Hepatitis B Virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef]
- Kew, M.C.; Kramvis, A.; Yu, M.C.; Arakawa, K.; Hodkinson, J. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-saharan Africans. J. Med. Virol. 2005, 75, 513–521. [Google Scholar] [CrossRef]
- Kramvis, A. Molecular characteristics and clinical relevance of African genotypes and subgenotypes of hepatitis B virus. S. Afr. Med. J. 2018, 108, 17–21. [Google Scholar] [PubMed]
- Kramvis, A.; Kew, M.C.; Bukofzer, S. Hepatitis B virus precore mutants in serum and liver of Southern African Blacks with hepatocellular carcinoma. J. Hepatol. 1998, 28, 132–141. [Google Scholar] [CrossRef]
- Bhoola, N.H.; Kramvis, A. Hepatitis B e Antigen Expression by Hepatitis B Virus Subgenotype A1 Relative to Subgenotypes A2 and D3 in Cultured Hepatocellular Carcinoma (Huh7) Cells. Intervirology 2016, 59, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Engelbrecht, J.; Brunak, S.; von Heijne, G.; Pa, M.; Earnest-Silveira, L. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Crowther, C.; Kew, M.; Kramvis, A. A valine to phenylalanine mutation in the precore region of hepatitis B virus causes intracellular retention and impaired secretion of HBe-antigen. Hepatol. Res. Off. J. Jpn. Soc. Hepatol. 2008, 38, 580–592. [Google Scholar] [CrossRef]
- Bhoola, N.H.; Kramvis, A. Expression of wild-type or G1862T mutant HBe antigen of subgenotype A1 of hepatitis B virus and the unfolded protein response in Huh7 cells. J. Gen. Virol. 2017, 98, 1422–1433. [Google Scholar] [CrossRef] [PubMed]
- Deroubaix, A.; Moahla, B.; Penny, C. Monitoring of intracellular localization of Hepatitis B virus P22 protein using Laser Scanning Confocal Microscopy and Airyscan. Microsc. Res. Tech. 2020, 83, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shi, C.; He, M.; Xiong, S.; Xia, X. Endoplasmic reticulum stress: Molecular mechanism and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 352. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhu, H.; Dong, L.; Shi, W.; Chen, R.; Song, Z.; Huang, C.; Li, J.; Dong, X.; Zhou, Y.; et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 2019, 179, 561–577. [Google Scholar] [CrossRef]
- Hannivoort, R.; Hernandez-Gea, V.; Friedman, S. Genomics and proteomics in liver fibrosis and cirrhosis. Fibrogenesis Tissue Repair 2012, 5, 1. [Google Scholar] [CrossRef]
- Wei, D.; Zeng, Y.; Xing, X.; Liu, H.; Lin, M.; Han, X.; Liu, X.; Liu, J. Proteome differences between hepatitis B virus genotype-B-and genotype-C-induced hepatocellular carcinoma revealed by iTRAQ-based quantitative proteomics. J. Proteome Res. 2016, 15, 487–498. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, J.; Lin, C.; Wang, H.; Jiang, Y.; Wang, J.; Yang, P.; He, F. Peroxiredoxin 2: A potential biomarker for early diagnosis of hepatitis B virus related liver fibrosis identified by proteomic analysis of the plasma. BMC Gastroenterol. 2010, 10, 115. [Google Scholar] [CrossRef]
- Bhoola; Reumann, K.; Kew, M.C.; Will, H.; Kramvis, A.; Tabak, F. Construction of replication competent plasmids of hepatitis B virus subgenotypes A1, A2 and D3 with authentic endogenous promoters. J. Virol. Methods 2014, 203, 54–64. [Google Scholar] [CrossRef]
- Olubayo, L.A.I. Construction and Functional Characterization of Genotype E of Hepatitis B Virus Replication Competent Plasmids with Endogenous Promoters. Ph.D. Thesis, University of Witwatersrand Wits WIReDspace, Witwatersrand, South Africa, 2022. [Google Scholar]
- Padarath, K.; Deroubaix, A.; Naicker, P.; Stoychev, S.; Kramvis, A. Comparative Proteomic Analysis of Huh7 Cells Transfected with Sub-Saharan African Hepatitis B Virus (Sub)genotypes Reveals Potential Oncogenic Factors. Viruses 2024, 16, 1052. [Google Scholar] [CrossRef]
- Nweke, E.E.; Naicker, P.; Aron, S.; Stoychev, S.; Devar, J.; Tabb, D.L.; Omoshoro-Jones, J.; Smith, M.; Candy, G. SWATH-MS based proteomic profiling of pancreatic ductal adenocarcinoma tumours reveals the interplay between the extracellular matrix and related intracellular pathways. PLoS ONE 2020, 15, e0240453. [Google Scholar] [CrossRef]
- Viana, R.; Wang, R.; Yu, M.C.; Welschinger, R.; Chen, C.-Y.; Kew, M.C. Hepatitis B viral loads in southern African Blacks with hepatocellular carcinoma. J. Med. Virol. 2009, 81, 1525–1530. [Google Scholar] [CrossRef]
- Duriez, M.; Rossignol, J.M.; Sitterlin, D. The hepatitis B virus precore protein is retrotransported from endoplasmic reticulum (ER) to cytosol through the ER-associated degradation pathway. J. Biol. Chem. 2008, 283, 32352–32360. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Lam, F.C. The DNA damage response—From cell biology to human disease. J. Transl. Genet. Genom. 2022, 6, 204–222. [Google Scholar] [CrossRef]
- Mitra, B.; Wang, J.; Kim, E.S.; Mao, R.; Dong, M.; Liu, Y.; Zhang, J.; Guo, H. Hepatitis B Virus Precore Protein p22 Inhibits Alpha Interferon Signaling by Blocking STAT Nuclear Translocation. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wei, Z.; Cheng, B.; Li, J.; He, Y.; Lan, T.; Kemper, T.; Lin, Y.; Jiang, B.; Jiang, Y.; et al. Endoplasmic reticulum stress promotes HBV production by enhancing use of the autophagosome/multivesicular body axis. Hepatology 2022, 75, 438–454. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, D.T.; Kaufman, R.J. A trip to the ER: Coping with stress. Trends Cell Biol. 2004, 14, 20–28. [Google Scholar] [CrossRef]
- Shore, G.C.; Papa, F.R.; Oakes, S.A. Signaling cell death from the endoplasmic reticulum stress response. Curr. Opin. Cell Biol. 2011, 23, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, R.J. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 2002, 110, 1389–1398. [Google Scholar] [CrossRef]
- Locarnini, S.; Shaw, T.; Dean, J.; Colledge, D.; Thompson, A.; Li, K.; Lemon, S.M.; Lau, G.G.K.; Beard, M.R. Cellular response to conditional expression of the hepatitis B virus precore and core proteins in cultured hepatoma (Huh-7) cells. J. Clin. Virol. 2005, 32, 113–121. [Google Scholar] [CrossRef]
- Jiang, Y.; Han, Q.; Zhao, H.; Zhang, J. The Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.W.; Su, I.J.; Chang, W.T.; Huang, W.; Lei, H.Y. Suppression of p38 mitogen-activated protein kinase inhibits hepatitis B virus replication in human hepatoma cell: The antiviral role of nitric oxide. J. Viral Hepat. 2008, 15, 490–497. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, H.; Kim, S.W.; Lee, N.R.; Yi, C.M.; Heo, J.; Kim, B.J.; Kim, N.J.; Inn, K.S. An Effective Antiviral Approach Targeting Hepatitis B Virus with NJK14047, a Novel and Selective Biphenyl Amide p38 Mitogen-Activated Protein Kinase Inhibitor. Antimicrob. Agents Chemother. 2017, 61, 10–1128. [Google Scholar] [CrossRef]
- Visvanathan, K.; Skinner, N.A.; Thompson, A.J.; Riordan, S.M.; Sozzi, V.; Edwards, R.; Rodgers, S.; Kurtovic, J.; Chang, J.; Lewin, S.; et al. Regulation of Toll-like receptor-2 expression in chronic hepatitis B by the precore protein. Hepatology 2007, 45, 102–110. [Google Scholar] [CrossRef]
- Bolukbas, C.; Bolukbas, F.F.; Horoz, M.; Aslan, M.; Celik, H.; Erel, O. Increased oxidative stress associated with the severity of the liver disease in various forms of hepatitis B virus infection. BMC Infect. Dis. 2005, 5, 95. [Google Scholar] [CrossRef]
- Görlach, A.; Klappa, P.; Kietzmann, D.T. The endoplasmic reticulum: Folding, calcium homeostasis, signaling, and redox control. Antioxid. Redox Signal. 2006, 8, 1391–1418. [Google Scholar] [CrossRef]
- Abdollahi, M.; Ranjbar, A.; Shadnia, S.; Nikfar, S.; Rezaie, A. Pesticides and oxidative stress: A review. Med. Sci. Monit. 2004, 10, 141–147. [Google Scholar]
- Duygu, F.; Karsen, H.; Aksoy, N.; Taskin, A. Relationship of oxidative stress in hepatitis B infection activity with HBV DNA and fibrosis. Ann. Lab. Med. 2012, 32, 113–118. [Google Scholar] [CrossRef]
- Deroubaix, A.; Kramvis, A. In vitro expression of precore proteins of hepatitis B virus subgenotype A1 is affected by HBcAg, and can affect HBsAg secretion. Sci. Rep. 2021, 11, 8167. [Google Scholar] [CrossRef] [PubMed]
- Padarath, K.; Deroubaix, A.; Kramvis, A. The Complex Role of HBeAg and Its Precursors in the Pathway to Hepatocellular Carcinoma. Viruses 2023, 15, 857. [Google Scholar] [CrossRef]
- Huang, R.-X.; Zhou, P.-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Schneider, A.; Smith, R.W.; Kautz, A.R.; Weisshart, K.; Grosse, F.; Nasheuer, H.P. Primase activity of human DNA polymerase alpha-primase. Divalent cations stabilize the enzyme activity of the p48 subunit. J. Biol. Chem. 1998, 273, 21608–21615. [Google Scholar] [CrossRef] [PubMed]
- Weiner, B.E.; Huang, H.; Dattilo, B.M.; Nilges, M.J.; Fanning, E.; Chazin, W.J. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 2007, 282, 33444–33451. [Google Scholar] [CrossRef]
- Baranovskiy, A.G.; Zhang, Y.; Suwa, Y.; Babayeva, N.D.; Gu, J.; Pavlov, Y.I.; Tahirov, T.H. Crystal structure of the human primase. J. Biol. Chem. 2015, 290, 5635–5646. [Google Scholar] [CrossRef]
- Lin, Y.L.; Shivji, M.K.; Chen, C.; Kolodner, R.; Wood, R.D.; Dutta, A. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J. Biol. Chem. 1998, 273, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Algethami, M.; Toss, M.S.; Woodcock, C.L.; Jaipal, C.; Brownlie, J.; Shoqafi, A.; Alblihy, A.; Mesquita, K.A.; Green, A.R.; Mongan, N.P.; et al. Unravelling the clinicopathological and functional significance of replication protein A (RPA) heterotrimeric complex in breast cancers. NPJ Breast Cancer 2023, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Shao, X.; Akter, F.; Zahid, K.R.; Yao, S.; Ma, L.; Xu, G. PRIM2: A Marker of MYC-driven Hyper-proliferation, Disease Progression, Tumor Aggressiveness and Poor Survival in Glioma Patients. Cancer Genom. Proteom. 2024, 21, 186–202. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.C.; Hunter, C.L.; Liu, Y.; Schilling, B.; Rosenberger, G.; Bader, S.L.; Chan, D.W.; Gibson, B.W.; Gingras, A.C.; Held, J.M.; et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, X.; Lyu, L.; Jiang, H.; Zhu, H.J. Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: Focusing on drug-metabolizing enzymes. Drug Metab. Pharmacokinet. 2018, 33, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Wose Kinge, C.N.; Bhoola, N.H.; Kramvis, A. In Vitro Systems for Studying Different Genotypes/Sub-Genotypes of Hepatitis B Virus: Strengths and Limitations. Viruses 2020, 12, 353. [Google Scholar] [CrossRef] [PubMed]
- Lamontagne, J.; Mell, J.C.; Bouchard, M.J. Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression. PLoS Pathog. 2016, 12, e1005438. [Google Scholar] [CrossRef]
- Chang, C.; Jeng, K.-S.; Hu, C.-P.; Lo, S.; Su, T.-S.; Ting, L.; Chou, C.-K.; Han, S.; Pfaff, E.; Salfeld, J. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987, 6, 675–680. [Google Scholar] [CrossRef]


| Gene | Direction | Sequence |
|---|---|---|
| PRIM2 | F | 5′-CTTCAGCCTCTGCTCAATCACC-3′ |
| R | 5′-GTAACTGACGCATGCAAGGTGG-3′ | |
| RPA | F | 5′-GAGCACCTATCAGCAATCCAGG-3′ |
| R | 5′-CCTTCAGGTCTTGGACAAGCCT-3′ | |
| GAPDH | F | 5′-GAAGGTGAAGGTCGGAGT-3′ |
| R | 5′-GAAGATGGTGATGGGATTTC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padarath, K.; Deroubaix, A.; Naicker, P.; Stoychev, S.; Kramvis, A. Comparison of the Proteome of Huh7 Cells Transfected with Hepatitis B Virus Subgenotype A1, with or without G1862T. Curr. Issues Mol. Biol. 2024, 46, 7032-7047. https://doi.org/10.3390/cimb46070419
Padarath K, Deroubaix A, Naicker P, Stoychev S, Kramvis A. Comparison of the Proteome of Huh7 Cells Transfected with Hepatitis B Virus Subgenotype A1, with or without G1862T. Current Issues in Molecular Biology. 2024; 46(7):7032-7047. https://doi.org/10.3390/cimb46070419
Chicago/Turabian StylePadarath, Kiyasha, Aurélie Deroubaix, Previn Naicker, Stoyan Stoychev, and Anna Kramvis. 2024. "Comparison of the Proteome of Huh7 Cells Transfected with Hepatitis B Virus Subgenotype A1, with or without G1862T" Current Issues in Molecular Biology 46, no. 7: 7032-7047. https://doi.org/10.3390/cimb46070419
APA StylePadarath, K., Deroubaix, A., Naicker, P., Stoychev, S., & Kramvis, A. (2024). Comparison of the Proteome of Huh7 Cells Transfected with Hepatitis B Virus Subgenotype A1, with or without G1862T. Current Issues in Molecular Biology, 46(7), 7032-7047. https://doi.org/10.3390/cimb46070419





