Epigenetic and Molecular Alterations in Obesity: Linking CRP and DNA Methylation to Systemic Inflammation
Abstract
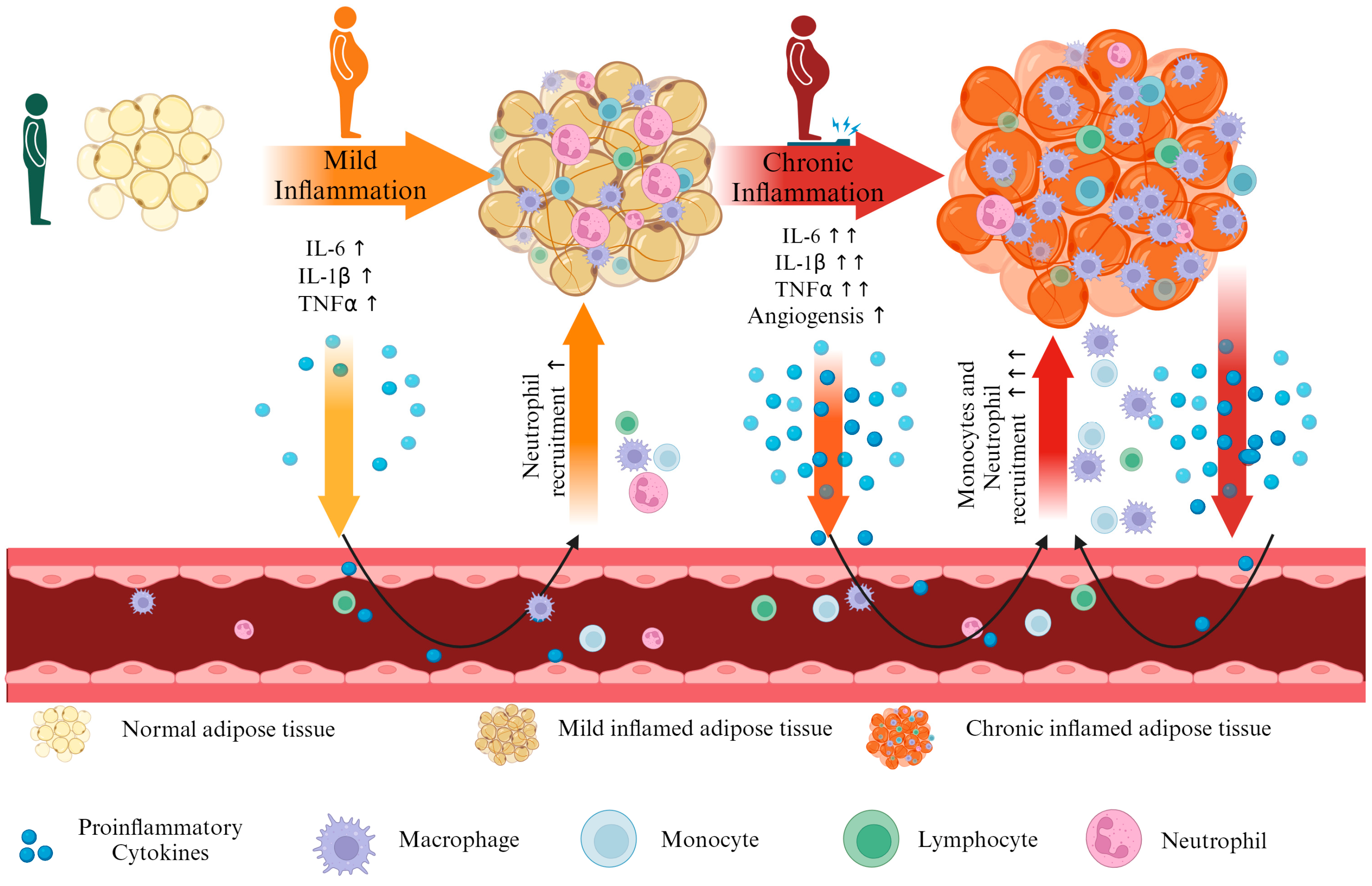
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
2.2. Sample Collection and Nucleic Acid Extraction
2.3. Global DNA Methylation and 5-mC Quantification by ELISA
2.4. Gene Expression Analysis by RT-PCR
2.5. Statistical Analysis
3. Results
3.1. Clinical Parameters
3.2. The DNA Methylation Status
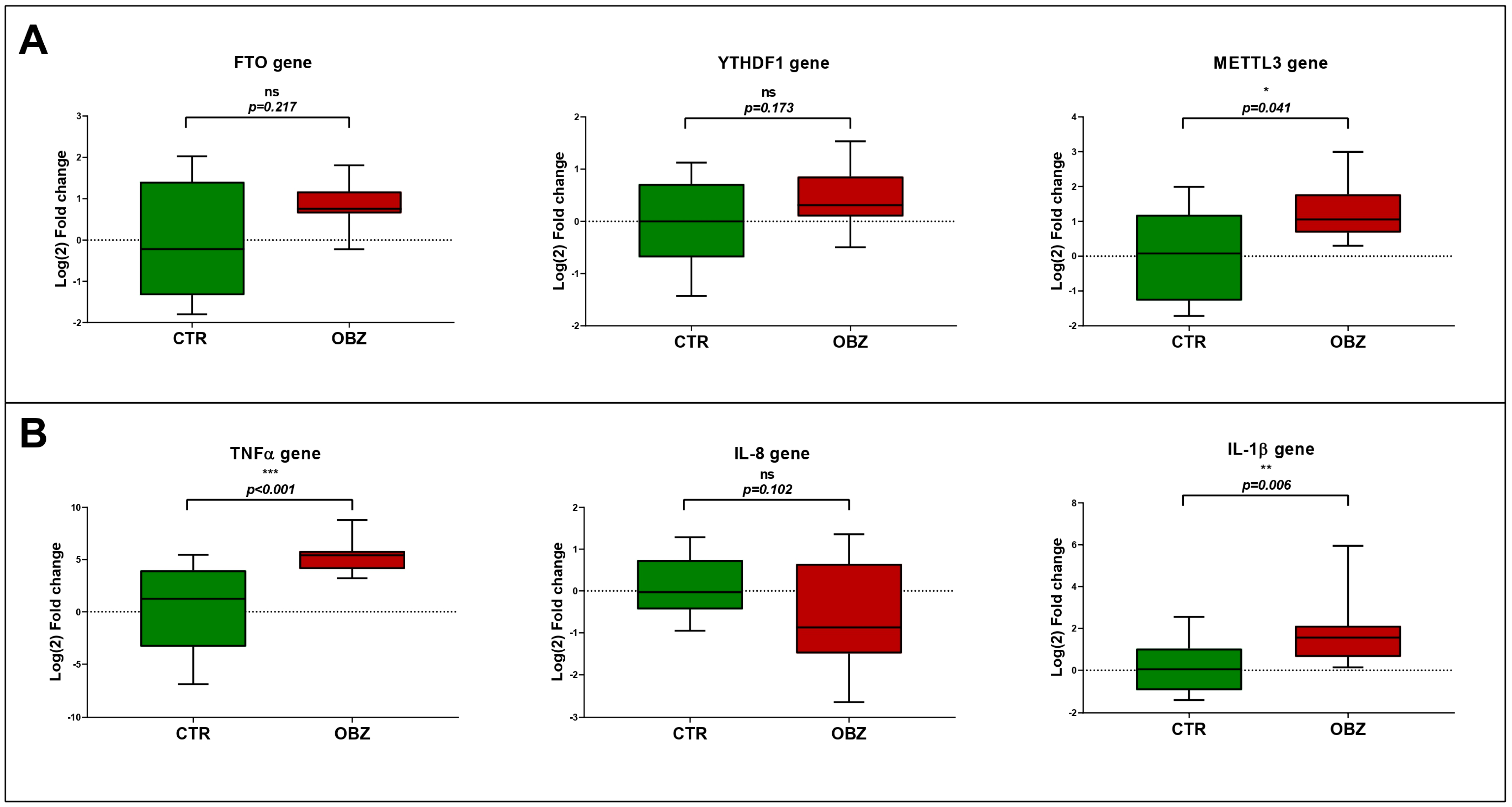
3.3. Gene Expression Analysis
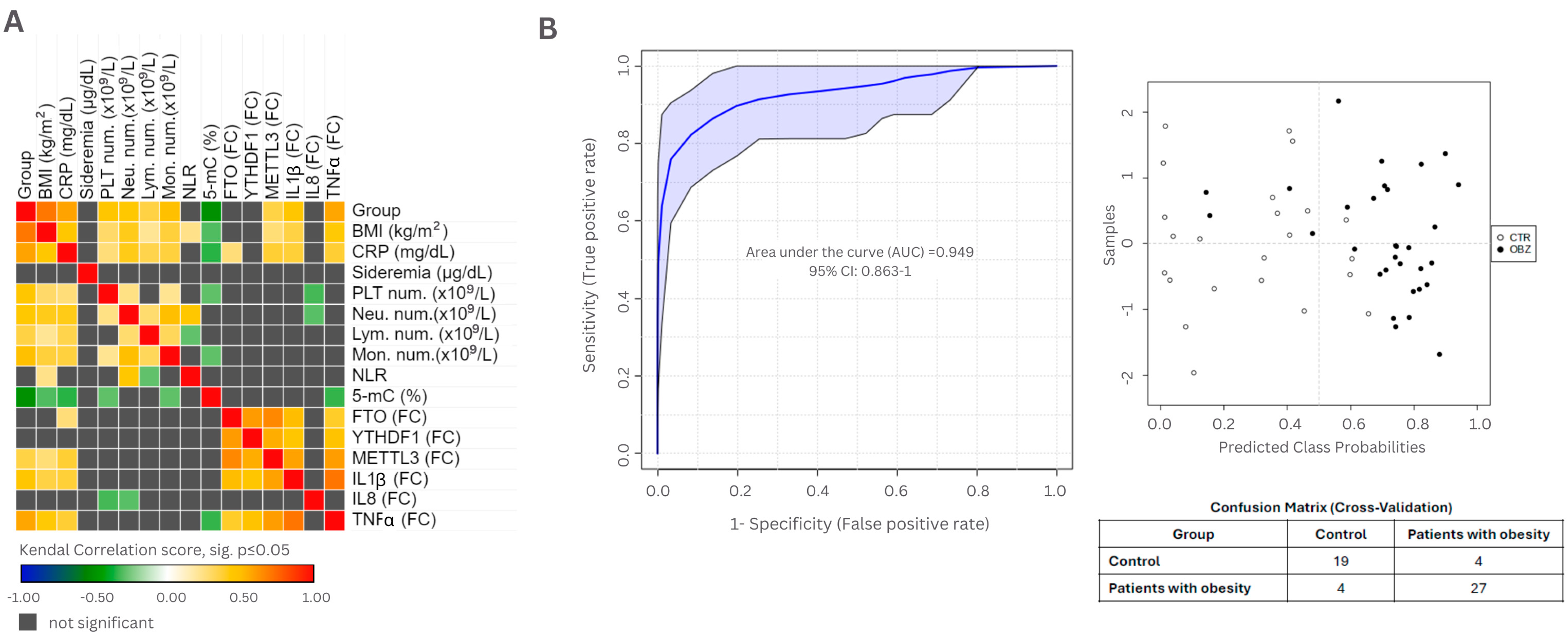
3.4. Evaluation of Clinical Parameters in Epigenetic Context
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| BMI | Body mass index |
| CRP | C-reactive protein |
| DNA | Deoxyribonucleic acid |
| TNF α | Tumor necrosis factor alpha |
| IL-1B | Interleukin-1 beta |
| IL-6 | Interleukin-6 |
| IL-8 | Interleukin-8 |
| NLR | Neutrophil-to-lymphocyte ratio |
| FTO | Fat mass and obesity-associated gene |
| METTL3 | RNA methyltransferase complex methyltransferase-like protein |
| YTHDF1 | YTH domain-containing reader protein 1 |
| YTHDF3 | YTH domain-containing reader protein 3 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| IPOM | Intraperitoneal onlay mesh |
| RNA | Ribonucleic acid |
| 5-mC | 5-methyl-Cytosine |
| cDNA | complementary DNA |
| UDGs | Uracil–DNA glycosylases |
| ROC | Receiver operating characteristic curve |
| CTR | Control group |
| OBZ | Obesity group |
| AUC | Aria under the curve |
| PLT | Platelet |
| Neu num | Neutrophil number |
| Lym num | Lymphocyte number |
| Mon num | Monocyte number |
| FC | Fold change |
| SD | Standard deviation |
| GLP-1 | Glucagon-like peptide-1 |
| GLP-1R | Glucagon-like peptide-1 receptor |
| MAPKs | Mitogen-activated protein kinase |
| PI3K/AKT | Phosphoinositide 3-kinase/AKT |
| JAK/STAT | Janus kinases/signal transducer and activator of transcription protein |
| TGF-β | transforming growth factor beta |
| AMPK | AMP-activated protein kinase |
References
- Voruganti, V.S. Precision Nutrition: Recent Advances in Obesity. Physiology 2023, 38, 42–50. [Google Scholar] [CrossRef]
- Sánchez-Carracedo, D. Obesity Stigma and Its Impact on Health: A Narrative Review. Endocrinol. Diabetes Y Nutr. (Engl. Ed.) 2022, 69, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Rohm, T.V.; Meier, D.T.; Olefsky, J.M.; Donath, M.Y. Inflammation in Obesity, Diabetes, and Related Disorders. Immunity 2022, 55, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Autieri, M.V.; Scalia, R. Adipose Tissue Inflammation and Metabolic Dysfunction in Obesity. Am. J. Physiol. Cell Physiol. 2021, 320, C375–C391. [Google Scholar] [CrossRef]
- Li, H.; Meng, Y.; He, S.; Tan, X.; Zhang, Y.; Zhang, X.; Wang, L.; Zheng, W. Macrophages, Chronic Inflammation, and Insulin Resistance. Cells 2022, 11, 3001. [Google Scholar] [CrossRef]
- Xu, L.; Yan, X.; Zhao, Y.; Wang, J.; Liu, B.; Yu, S.; Fu, J.; Liu, Y.; Su, J. Macrophage Polarization Mediated by Mitochondrial Dysfunction Induces Adipose Tissue Inflammation in Obesity. Int. J. Mol. Sci. 2022, 23, 9252. [Google Scholar] [CrossRef]
- Uribe-Querol, E.; Rosales, C. Neutrophils Actively Contribute to Obesity-Associated Inflammation and Pathological Complications. Cells 2022, 11, 1883. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.; Behl, T.; Kaur, I.; Singh, S.; Sharma, N.; Aleya, L. Targeting NLRP3 Inflammasome as a Chief Instigator of Obesity, Contributing to Local Adipose Tissue Inflammation and Insulin Resistance. Environ. Sci. Pollut. Res. 2021, 28, 43102–43113. [Google Scholar] [CrossRef]
- Qiu, F.; Wu, L.; Yang, G.; Zhang, C.; Liu, X.; Sun, X.; Chen, X.; Wang, N. The Role of Iron Metabolism in Chronic Diseases Related to Obesity. Mol. Med. 2022, 28, 130. [Google Scholar] [CrossRef]
- Zhang, Z.; Funcke, J.B.; Zi, Z.; Zhao, S.; Straub, L.G.; Zhu, Y.; Zhu, Q.; Crewe, C.; An, Y.A.; Chen, S.; et al. Adipocyte Iron Levels Impinge on a Fat-Gut Crosstalk to Regulate Intestinal Lipid Absorption and Mediate Protection from Obesity. Cell Metab. 2021, 33, 1624–1639.e9. [Google Scholar] [CrossRef]
- Ding, X.; Bian, N.; Wang, J.; Chang, X.; An, Y.; Wang, G.; Liu, J. Serum Ferritin Levels Are Associated with Adipose Tissue Dysfunction-Related Indices in Obese Adults. Biol. Trace Elem. Res. 2023, 201, 636–643. [Google Scholar] [CrossRef]
- Szklarz, M.; Gontarz-Nowak, K.; Matuszewski, W.; Bandurska-Stankiewicz, E. “Ferrocrinology”—Iron Is an Important Factor Involved in Gluco- and Lipocrinology. Nutrients 2022, 14, 4693. [Google Scholar] [CrossRef] [PubMed]
- Bjørklund, G.; Peana, M.; Pivina, L.; Dosa, A.; Aaseth, J.; Semenova, Y.; Chirumbolo, S.; Medici, S.; Dadar, M.; Costea, D.O. Iron Deficiency in Obesity and after Bariatric Surgery. Biomolecules 2021, 11, 613. [Google Scholar] [CrossRef] [PubMed]
- Ameka, M.K.; Hasty, A.H. Fat and Iron Don’t Mix. Immunometabolism 2020, 2, e200034. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, N.U.; El-Mallah, C.; Herter-Aeberli, I.; Bissani, N.; Wehbe, N.; Obeid, O.; Zimmermann, M.B. The Effect of Central Obesity on Inflammation, Hepcidin, and Iron Metabolism in Young Women. Int. J. Obes. 2020, 44, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Bondesan, A.; Caroli, D.; Grugni, G.; Sartorio, A. The Neutrophil to Lymphocyte Ratio (NLR) Positively Correlates with the Presence and Severity of Metabolic Syndrome in Obese Adults, but Not in Obese Children/Adolescents. BMC Endocr. Disord. 2023, 23, 121. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Zheng, Y.; Jia, H.; Zheng, X.; Gao, R.; Li, W. Neutrophil-Lymphocyte Ratio as a Predictive Marker for Postoperative Infectious Complications: A Systematic Review and Meta-Analysis. Heliyon 2023, 9, e15586. [Google Scholar] [CrossRef]
- Da Silva, M.; Cleghorn, M.C.; Elnahas, A.; Jackson, T.D.; Okrainec, A.; Quereshy, F.A. Postoperative Day One Neutrophil-to-Lymphocyte Ratio as a Predictor of 30-Day Outcomes in Bariatric Surgery Patients. Surg. Endosc. 2017, 31, 2645–2650. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Pan, Y.; Ning, Y. Predictive Value of IL-6 Combined with NLR in Inflammation and Cancer. Cancer Investig. 2021, 39, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhang, B.; Wu, B.; Xiao, H.; Li, Z.; Li, R.; Xu, X.; Li, T. Correction To: Signaling pathways in obesity: Mechanisms and therapeutic interventions. Sig Transduct. Target. Ther. 2022, 7, 369. [Google Scholar] [CrossRef]
- Jin, X.; Qiu, T.; Li, L.; Yu, R.; Chen, X.; Li, C.; Proud, C.G.; Jiang, T. Pathophysiology of Obesity and Its Associated Diseases. Acta Pharm. Sin. B 2023, 13, 2403–2424. [Google Scholar] [CrossRef] [PubMed]
- Aronica, L.; Levine, A.J.; Brennan, K.; Mi, J.; Gardner, C.; Haile, R.W.; Hitchins, M.P. A Systematic Review of Studies of DNA Methylation in the Context of a Weight Loss Intervention. Epigenomics 2017, 9, 769–787. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, C.F.; Nonino, C.B.; de Oliveira, B.A.P.; Pinhel, M.A.d.S.; Mansego, M.L.; Milagro, F.I.; Zulet, M.A.; Martinez, J.A. DNA Methylation and Hydroxymethylation Levels in Relation to Two Weight Loss Strategies: Energy-Restricted Diet or Bariatric Surgery. Obes. Surg. 2016, 26, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Martin Carli, J.F.; LeDuc, C.A.; Zhang, Y.; Stratigopoulos, G.; Leibel, R.L. FTO Mediates Cell-Autonomous Effects on Adipogenesis and Adipocyte Lipid Content by Regulating Gene Expression via 6mA DNA Modifications 1. J. Lipid Res. 2018, 59, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, W.; Wang, X. Studies on the Fat Mass and Obesity-Associated (FTO) Gene and Its Impact on Obesity-Associated Diseases. Genes. Dis. 2023, 10, 2351–2365. [Google Scholar] [CrossRef] [PubMed]
- Pipek, L.Z.; Moraes, W.A.F.; Nobetani, R.M.; Cortez, V.S.; Condi, A.S.; Taba, J.V.; Nascimento, R.F.V.; Suzuki, M.O.; do Nascimento, F.S.; de Mattos, V.C.; et al. Surgery Is Associated with Better Long-Term Outcomes than Pharmacological Treatment for Obesity: A Systematic Review and Meta-Analysis. Sci. Rep. 2024, 14, 9521. [Google Scholar] [CrossRef]
- Jiang, L.; Qin, J.; Dai, Y.; Zhao, S.; Zhan, Q.; Cui, P.; Ren, L.; Wang, X.; Zhang, R.; Gao, C.; et al. Prospective Observational Study on Biomarkers of Response in Pancreatic Ductal Adenocarcinoma. Nat. Med. 2024, 30, 749–761. [Google Scholar] [CrossRef]
- Ntentakis, D.P.; Correa, V.S.M.C.; Ntentaki, A.M.; Delavogia, E.; Narimatsu, T.; Efstathiou, N.E.; Vavvas, D.G. Effects of Newer-Generation Anti-Diabetics on Diabetic Retinopathy: A Critical Review. Graefe’s Arch. Clin. Exp. Ophthalmol. 2024, 262, 717–752. [Google Scholar] [CrossRef]
- Sodhi, M.; Rezaeianzadeh, R.; Kezouh, A.; Etminan, M. Risk of Gastrointestinal Adverse Events Associated With Glucagon-Like Peptide-1 Receptor Agonists for Weight Loss. JAMA 2023, 330, 1795–1797. [Google Scholar] [CrossRef]
- Hon, G.C.; Hawkins, R.D.; Caballero, O.L.; Lo, C.; Lister, R.; Pelizzola, M.; Valsesia, A.; Ye, Z.; Kuan, S.; Edsall, L.E.; et al. Global DNA Hypomethylation Coupled to Repressive Chromatin Domain Formation and Gene Silencing in Breast Cancer. Genome Res. 2012, 22, 246–258. [Google Scholar] [CrossRef]
- Hansen, K.D.; Timp, W.; Bravo, H.C.; Sabunciyan, S.; Langmead, B.; McDonald, O.G.; Wen, B.; Wu, H.; Liu, Y.; Diep, D.; et al. Increased Methylation Variation in Epigenetic Domains across Cancer Types. Nat. Genet. 2011, 43, 768–775. [Google Scholar] [CrossRef]
- David, L.; Onaciu, A.; Toma, V.; Borșa, R.M.; Moldovan, C.; Țigu, A.B.; Cenariu, D.; Șimon, I.; Știufiuc, G.F.; Carasevici, E.; et al. Understanding DNA Epigenetics by Means of Raman/SERS Analysis for Cancer Detection. Biosensors 2024, 14, 41. [Google Scholar] [CrossRef]
- Chai, V.; Vassilakos, A.; Lee, Y.; Wright, J.A.; Young, A.H. Optimization of the PAXgeneTM Blood RNA Extraction System for Gene Expression Analysis of Clinical Samples. J. Clin. Lab. Anal. 2005, 19, 182–188. [Google Scholar] [CrossRef]
- Hirukawa, A.; Singh, S.; Wang, J.; Rennhack, J.P.; Swiatnicki, M.; Sanguin-Gendreau, V.; Zuo, D.; Daldoul, K.; Lavoie, C.; Park, M.; et al. Reduction of Global H3K27me3 Enhances HER2/ErbB2 Targeted Therapy. Cell Rep. 2019, 29, 249–257.e8. [Google Scholar] [CrossRef] [PubMed]
- Iluta, S.; Pasca, S.; Gafencu, G.; Jurj, A.; Terec, A.; Teodorescu, P.; Selicean, C.; Jitaru, C.; Preda, A.; Cenariu, D.; et al. Azacytidine plus Olaparib for Relapsed Acute Myeloid Leukaemia, Ineligible for Intensive Chemotherapy, Diagnosed with a Synchronous Malignancy. J. Cell Mol. Med. 2021, 25, 6094–6102. [Google Scholar] [CrossRef]
- Blüher, M. Obesity: Global Epidemiology and Pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Cerhan, J.R.; Moore, S.C.; Jacobs, E.J.; Kitahara, C.M.; Rosenberg, P.S.; Adami, H.O.; Ebbert, J.O.; English, D.R.; Gapstur, S.M.; Giles, G.G.; et al. A Pooled Analysis of Waist Circumference and Mortality in 650,000 Adults. Mayo Clin. Proc. 2014, 89, 335–345. [Google Scholar] [CrossRef]
- Alberti, K.G.M.M.; Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z.; Cleeman, J.I.; Donato, K.A.; Fruchart, J.C.; James, W.P.T.; Loria, C.M.; Smith, S.C. Harmonizing the Metabolic Syndrome: A Joint Interim Statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; And International Association for the Study of Obesity. Circulation 2009, 120, 1640–1645. [Google Scholar]
- Caballero, B. Humans against Obesity: Who Will Win? Proc. Adv. Nutr. 2019, 10, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Wahl, S.; Drong, A.; Lehne, B.; Loh, M.; Scott, W.R.; Kunze, S.; Tsai, P.C.; Ried, J.S.; Zhang, W.; Yang, Y.; et al. Epigenome-Wide Association Study of Body Mass Index, and the Adverse Outcomes of Adiposity. Nature 2017, 541, 81–86. [Google Scholar] [CrossRef]
- Shoelson, S.E.; Herrero, L.; Naaz, A. Obesity, Inflammation, and Insulin Resistance. Gastroenterology 2007, 132, 2169–2180. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and Metabolic Health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Illán-Gómez, F.; Gonzálvez-Ortega, M.; Orea-Soler, I.; Alcaraz-Tafalla, M.S.; Aragón-Alonso, A.; Pascual-Díaz, M.; Pérez-Paredes, M.; Lozano-Almela, M.L. Obesity and Inflammation: Change in Adiponectin, C-Reactive Protein, Tumour Necrosis Factor-Alpha and Interleukin-6 after Bariatric Surgery. Obes. Surg. 2012, 22, 950–955. [Google Scholar] [CrossRef] [PubMed]
- Maurizi, G.; Della Guardia, L.; Maurizi, A.; Poloni, A. Adipocytes Properties and Crosstalk with Immune System in Obesity-Related Inflammation. J. Cell Physiol. 2018, 233, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Blaszczak, A.M.; Jalilvand, A.; Hsueh, W.A. Adipocytes, Innate Immunity and Obesity: A Mini-Review. Front. Immunol. 2021, 12, 650768. [Google Scholar] [CrossRef] [PubMed]
- Elgazar-Carmon, V.; Rudich, A.; Hadad, N.; Levy, R. Neutrophils Transiently Infiltrate Intra-Abdominal Fat Early in the Course of High-Fat Feeding. J. Lipid Res. 2008, 49, 1894–1903. [Google Scholar] [CrossRef]
- Xu, X.; Su, S.; Wang, X.; Barnes, V.; De Miguel, C.; Ownby, D.; Pollock, J.; Snieder, H.; Chen, W. Obesity Is Associated with More Activated Neutrophils in African American Male Youth. Int. J. Obes. 2015, 39, 26–32. [Google Scholar] [CrossRef]
- Herrero-Cervera, A.; Soehnlein, O.; Kenne, E. Neutrophils in Chronic Inflammatory Diseases. Cell Mol. Immunol. 2022, 19, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Todosenko, N.; Khaziakhmatova, O.; Malashchenko, V.; Yurova, K.; Bograya, M.; Beletskaya, M.; Vulf, M.; Mikhailova, L.; Minchenko, A.; Soroko, I.; et al. Adipocyte- and Monocyte-Mediated Vicious Circle of Inflammation and Obesity (Review of Cellular and Molecular Mechanisms). Int. J. Mol. Sci. 2023, 24, 12259. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, Y.; Lee, M.N.; Nah, J.; Yun, N.; Wu, D.; Pae, M. Time-Restricted Feeding Reduces Monocyte Production by Controlling Hematopoietic Stem and Progenitor Cells in the Bone Marrow during Obesity. Front. Immunol. 2022, 13, 1054875. [Google Scholar] [CrossRef]
- Breznik, J.A.; Naidoo, A.; Foley, K.P.; Schulz, C.; Lau, T.C.; Loukov, D.; Sloboda, D.M.; Bowdish, D.M.E.; Schertzer, J.D. TNF, but Not Hyperinsulinemia or Hyperglycemia, Is a Key Driver of Obesity-Induced Monocytosis Revealing That Inflammatory Monocytes Correlate with Insulin in Obese Male Mice. Physiol. Rep. 2018, 6, e13937. [Google Scholar] [CrossRef] [PubMed]
- Joffin, N.; Gliniak, C.M.; Funcke, J.B.; Paschoal, V.A.; Crewe, C.; Chen, S.; Gordillo, R.; Kusminski, C.M.; Oh, D.Y.; Geldenhuys, W.J.; et al. Adipose Tissue Macrophages Exert Systemic Metabolic Control by Manipulating Local Iron Concentrations. Nat. Metab. 2022, 4, 1474–1494. [Google Scholar] [CrossRef] [PubMed]
- Samocha-Bonet, D.; Justo, D.; Rogowski, O.; Saar, N.; Abu-Abeid, S.; Shenkerman, G.; Shapira, I.; Berliner, S.; Tomer, A. Platelet Counts and Platelet Activation Markers in Obese Subjects. Mediat. Inflamm. 2008, 2008, 834153. [Google Scholar] [CrossRef] [PubMed]
- Yudkin, J.S.; Kumari, M.; Humphries, S.E.; Mohamed-Ali, V. Inflammation, Obesity, Stress and Coronary Heart Disease: Is Interleukin-6 the Link? Atherosclerosis 2000, 148, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, L.; Henschler, R.; Lecchi, L.; Rebulla, P.; Mertelsmann, R.; Sirchia, G. Interleukin-6 and Interleukin-11 Act Synergistically with Thrombopoietin and Stem Cell Factor to Modulate Ex Vivo Expansion of Human CD41+ and CD61+ Megakaryocytic Cells. Haematologica 2000, 85, 25–30. [Google Scholar] [PubMed]
- Davì, G.; Guagnano, M.T.; Ciabattoni, G.; Basili, S.; Falco, A.; Marinopiccoli, M.; Nutini, M.; Sensi, S.; Patrono, C. Platelet Activation in Obese Women: Role of Inflammation and Oxidant Stress. JAMA 2002, 288, 2008–2014. [Google Scholar] [CrossRef] [PubMed]
- Purdy, J.C.; Shatzel, J.J. The Hematologic Consequences of Obesity. Eur. J. Haematol. 2021, 106, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Sethi, G. Role of Epigenetics in in Flammation-Associated Diseases. Subcell. Biochem. 2013, 61, 1028–1044. [Google Scholar] [CrossRef]
- Ali, M.M.; Phillips, S.A.; Mahmoud, A.M. HIF1α/TET1 Pathway Mediates Hypoxia-Induced Adipocytokine Promoter Hypomethylation in Human Adipocytes. Cells 2020, 9, 134. [Google Scholar] [CrossRef]
- Ali, M.M.; Naquiallah, D.; Qureshi, M.; Mirza, M.I.; Hassan, C.; Masrur, M.; Bianco, F.M.; Frederick, P.; Cristoforo, G.P.; Gangemi, A.; et al. DNA Methylation Profile of Genes Involved in Inflammation and Autoimmunity Correlates with Vascular Function in Morbidly Obese Adults. Epigenetics 2022, 17, 93–109. [Google Scholar] [CrossRef]
- Mirza, I.; Mohamed, A.; Deen, H.; Balaji, S.; Elsabbahi, D.; Munasser, A.; Naquiallah, D.; Abdulbaseer, U.; Hassan, C.; Masrur, M.; et al. Obesity-Associated Vitamin D Deficiency Correlates with Adipose Tissue DNA Hypomethylation, Inflammation, and Vascular Dysfunction. Int. J. Mol. Sci. 2022, 23, 14377. [Google Scholar] [CrossRef] [PubMed]
- Baca, P.; Barajas-Olmos, F.; Mirzaeicheshmeh, E.; Zerrweck, C.; Guilbert, L.; Sánchez, E.C.; Flores-Huacuja, M.; Villafán, R.; Martínez-Hernández, A.; García-Ortiz, H.; et al. DNA Methylation and Gene Expression Analysis in Adipose Tissue to Identify New Loci Associated with T2D Development in Obesity. Nutr. Diabetes 2022, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Aderinto, N.; Olatunji, G.; Kokori, E.; Olaniyi, P.; Isarinade, T.; Yusuf, I.A. Recent Advances in Bariatric Surgery: A Narrative Review of Weight Loss Procedures. Ann. Med. Surg. 2023, 85, 6091–6104. [Google Scholar] [CrossRef] [PubMed]
- Jaacks, L.M.; Vandevijvere, S.; Pan, A.; McGowan, C.J.; Wallace, C.; Imamura, F.; Mozaffarian, D.; Swinburn, B.; Ezzati, M. The Obesity Transition: Stages of the Global Epidemic. Lancet Diabetes Endocrinol. 2019, 7, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Hainer, V.; Toplak, H.; Mitrakou, A. Treatment Modalities of Obesity: What Fits Whom? Diabetes Care 2008, 31 (Suppl. S2), S269–S277. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Brüning, J.C.; Rüther, U. Inactivation of the Fto Gene Protects from Obesity. Nature 2009, 458, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Church, C.; Moir, L.; McMurray, F.; Girard, C.; Banks, G.T.; Teboul, L.; Wells, S.; Brüning, J.C.; Nolan, P.M.; Ashcroft, F.M.; et al. Overexpression of Fto Leads to Increased Food Intake and Results in Obesity. Nat. Genet. 2010, 42, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.L.; Yeo, G.S.H.; O’Rahilly, S.; Coll, A.P. Obesity and FTO: Changing Focus at a Complex Locus. Cell Metab. 2014, 20, 710–718. [Google Scholar] [CrossRef]
- Yan, S.; Zhou, X.; Wu, C.; Gao, Y.; Qian, Y.; Hou, J.; Xie, R.; Han, B.; Chen, Z.; Wei, S.; et al. Adipocyte YTH N(6)-Methyladenosine RNA-Binding Protein 1 Protects against Obesity by Promoting White Adipose Tissue Beiging in Male Mice. Nat. Commun. 2023, 14, 1379. [Google Scholar] [CrossRef]
- Luo, G.; Chen, J.; Ren, Z. Regulation of Methylase METTL3 on Fat Deposition. Diabetes Metab. Syndr. Obes. 2021, 14, 4843–4852. [Google Scholar] [CrossRef]
- Yao, Y.; Bi, Z.; Wu, R.; Zhao, Y.; Liu, Y.; Liu, Q.; Wang, Y.; Wang, X. METTL3 Inhibits BMSC Adipogenic Differentiation by Targeting the JAK1/STAT5/C/EBPβ Pathway via an M6A-YTHDF2–Dependent Manner. FASEB J. 2019, 33, 7529–7544. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Walkey, C.J.; Puigserver, P.; Spiegelman, B.M. Transcriptional Regulation of Adipogenesis. Genes. Dev. 2000, 14, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Petrie, J.R.; Guzik, T.J.; Touyz, R.M. Diabetes, Hypertension, and Cardiovascular Disease: Clinical Insights and Vascular Mechanisms. Can. J. Cardiol. 2018, 34, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Bodhini, D.; Mohan, V. Mediators of Insulin Resistance & Cardiometabolic Risk: Newer Insights. Indian J. Med. Res. 2018, 148, 127–129. [Google Scholar]

| Primer | Sequence |
|---|---|
| FTO Forward | ACT TGG CTC CCT TAT CTG ACC |
| FTO Reverse | TGT GCA GTG TGA GAA AGG CTT |
| METTL3 Forward | CAA GCT GCA CTT CAG ACG AA |
| METTL3 Reverse | GCT TGG CGT GTG GTC TTT |
| YTHDF1 Forward | GCA CAC AAC CTC CAT CTT CG |
| YTHDF1 Reverse | AAC TGG TTC GCC CTC ATT GT |
| TNFα Forward | ATG GGC TAC AGG CTT GTC ACT C |
| TNFα Reverse | CTC TTC TGC CTG CTG CAC TTT G |
| IL-1β Forward | GTG CAG TTC AGT GAT CGT ACA GG |
| IL-1β Reverse | CCA CAG ACC TTC CAG GAG AAT G |
| IL-8 Forward | GAG AGT GAT TGA GAG TGG ACC AC |
| IL-8 Reverse | CAC AAC CCT CTG CAC CCA ATT T |
| GAPDH Forward | AGC CAC ATC GCT CAG ACA C |
| GAPDH Reverse | GCC CAA TAC GAC CAA ATC C |
| Variables | Controls | Patients with Obesity |
|---|---|---|
| Age (years; ±SD) | 46.23 ± 15.21 | 42.41 ± 13.07 |
| BMI (kg/m2) | 23.18 ± 1.82 | 44.12 ± 5.95 *** |
| Median (p25–p75) | 23.49 (22.06–24.65) | 43.00 (39.20–47.43) |
| CRP (mg/dL) | 0.28 ± 0.26 | 0.90 ± 0.68 *** |
| Median (p25–p75) | 0.21 (0.10–0.39) | 0.73 (0.45–1.24) |
| Sideremia (µg/dL) | 86.96 ± 39.56 | 73.39 ± 27.47 * |
| Leucocytes (109/L) | 6.18 ± 1.28 | 8.91 ± 2.24 *** |
| Platelets (109/L) | 239.27 ± 61.86 | 284.49 ± 58.08 *** |
| Neutrophile count (109/L) | 4.16 ± 1.18 | 5.80 ± 1.79 *** |
| Lymphocyte count (109/L) | 1.86 ± 0.52 | 2.44 ± 0.73 *** |
| Median (p25–p75) | 1.79 (1.46–2.21) | 2.36 (1.83–2.71) |
| Monocyte count (109/L) | 0.38 ± 0.14 | 0.52 ± 0.17 *** |
| Median (p25–p75) | 0.38 (0.30–0.45) | 0.48 (0.40–0.61) |
| NLR | 2.36 ± 0.82 | 2.55 ± 1.07 |
| Median (p25–p75) | 2.22 (1.68–3.03) | 2.42 (1.76–3.06) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cucoreanu, C.; Tigu, A.-B.; Nistor, M.; Moldovan, R.-C.; Pralea, I.-E.; Iacobescu, M.; Iuga, C.-A.; Szabo, R.; Dindelegan, G.-C.; Ciuce, C. Epigenetic and Molecular Alterations in Obesity: Linking CRP and DNA Methylation to Systemic Inflammation. Curr. Issues Mol. Biol. 2024, 46, 7430-7446. https://doi.org/10.3390/cimb46070441
Cucoreanu C, Tigu A-B, Nistor M, Moldovan R-C, Pralea I-E, Iacobescu M, Iuga C-A, Szabo R, Dindelegan G-C, Ciuce C. Epigenetic and Molecular Alterations in Obesity: Linking CRP and DNA Methylation to Systemic Inflammation. Current Issues in Molecular Biology. 2024; 46(7):7430-7446. https://doi.org/10.3390/cimb46070441
Chicago/Turabian StyleCucoreanu, Ciprian, Adrian-Bogdan Tigu, Madalina Nistor, Radu-Cristian Moldovan, Ioana-Ecaterina Pralea, Maria Iacobescu, Cristina-Adela Iuga, Robert Szabo, George-Calin Dindelegan, and Constatin Ciuce. 2024. "Epigenetic and Molecular Alterations in Obesity: Linking CRP and DNA Methylation to Systemic Inflammation" Current Issues in Molecular Biology 46, no. 7: 7430-7446. https://doi.org/10.3390/cimb46070441
APA StyleCucoreanu, C., Tigu, A.-B., Nistor, M., Moldovan, R.-C., Pralea, I.-E., Iacobescu, M., Iuga, C.-A., Szabo, R., Dindelegan, G.-C., & Ciuce, C. (2024). Epigenetic and Molecular Alterations in Obesity: Linking CRP and DNA Methylation to Systemic Inflammation. Current Issues in Molecular Biology, 46(7), 7430-7446. https://doi.org/10.3390/cimb46070441









