Dysregulation of Transposon Transcription Profiles in Cancer Cells Resembles That of Embryonic Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Bulk RNA-Seq Data Processing
2.2. Single-Cell RNA-Seq Data Processing
3. Results
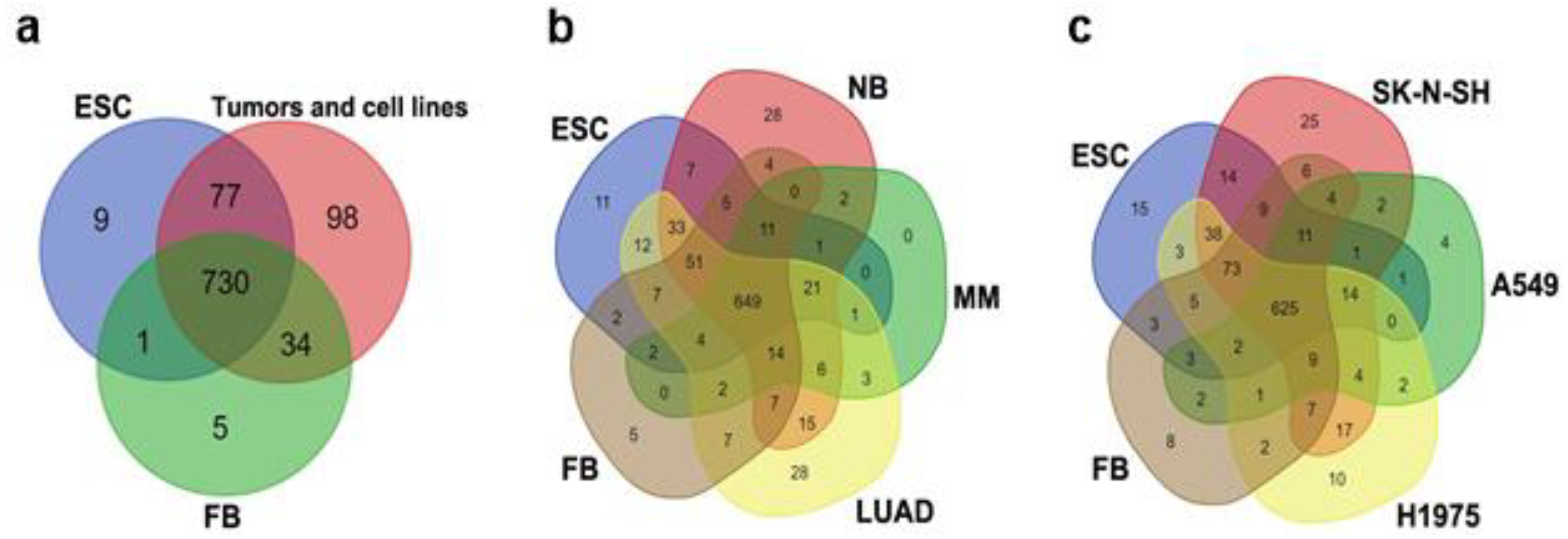
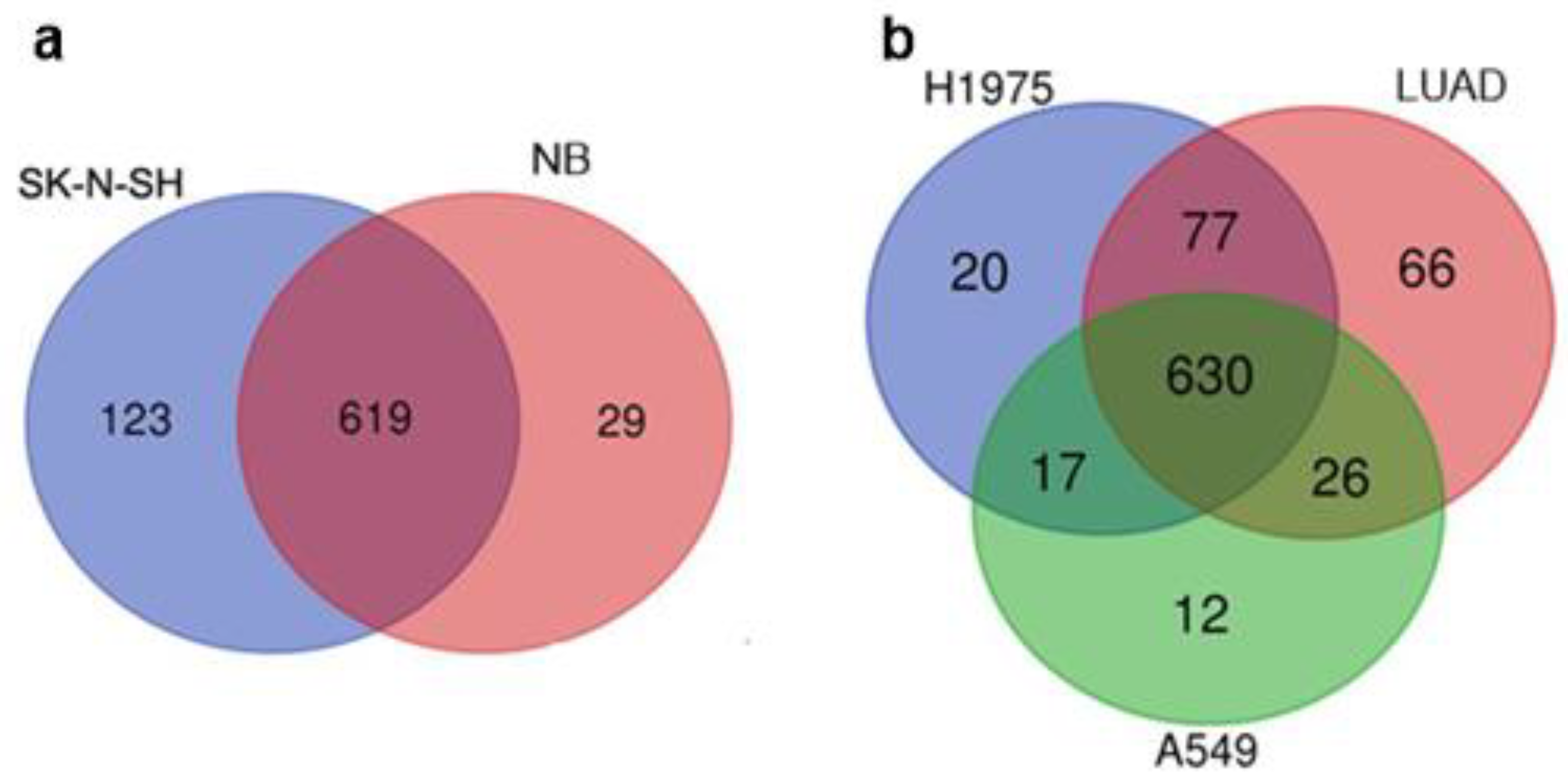
3.1. Comparison of the Datasets of Expressed TEs in ESCs, Tumors, Cancer Cell Lines, and Normal Fibroblasts
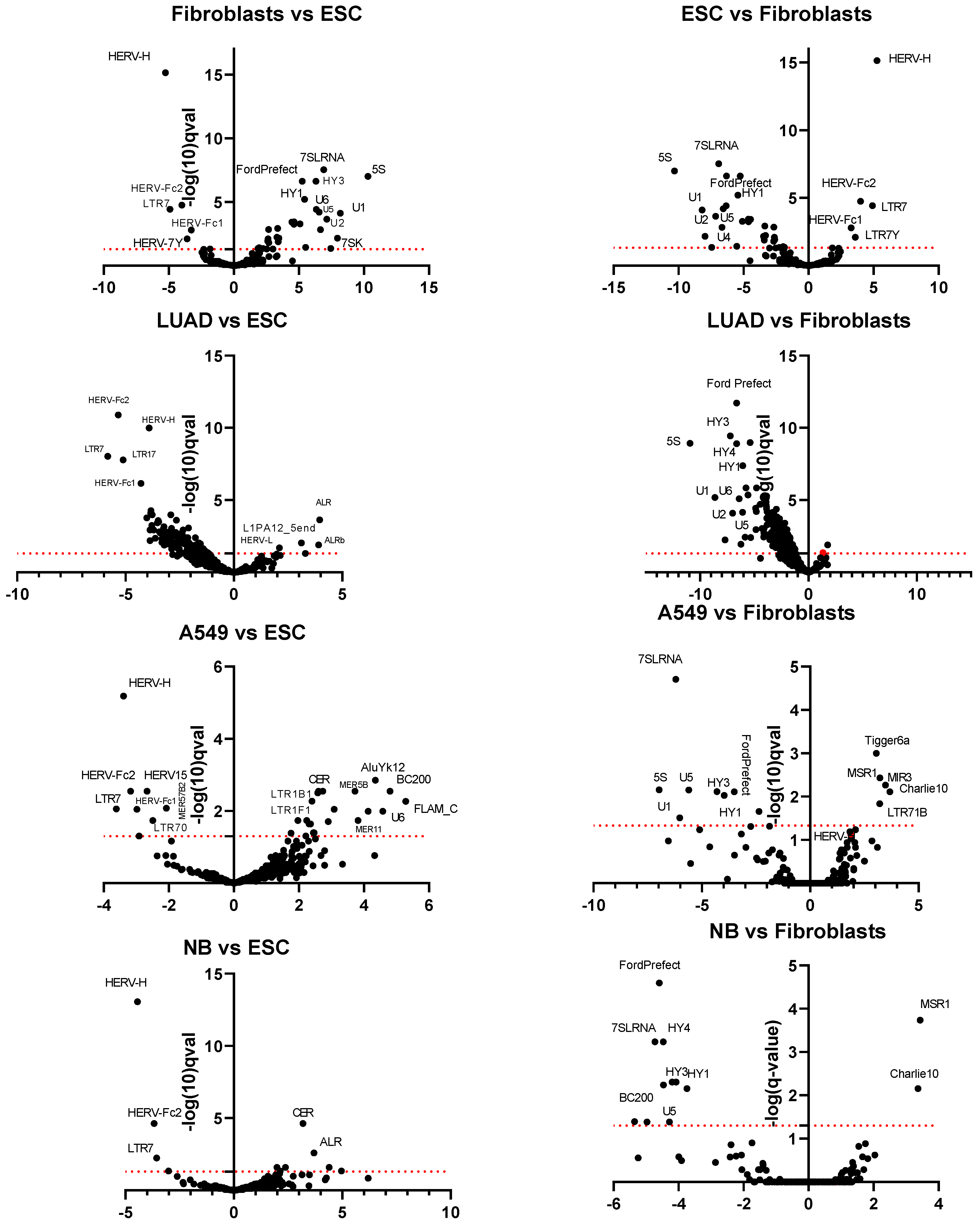
3.2. Differential Expression of TE Transcription in ESC, Normal Fibroblasts, Tumors, and Cell Lines
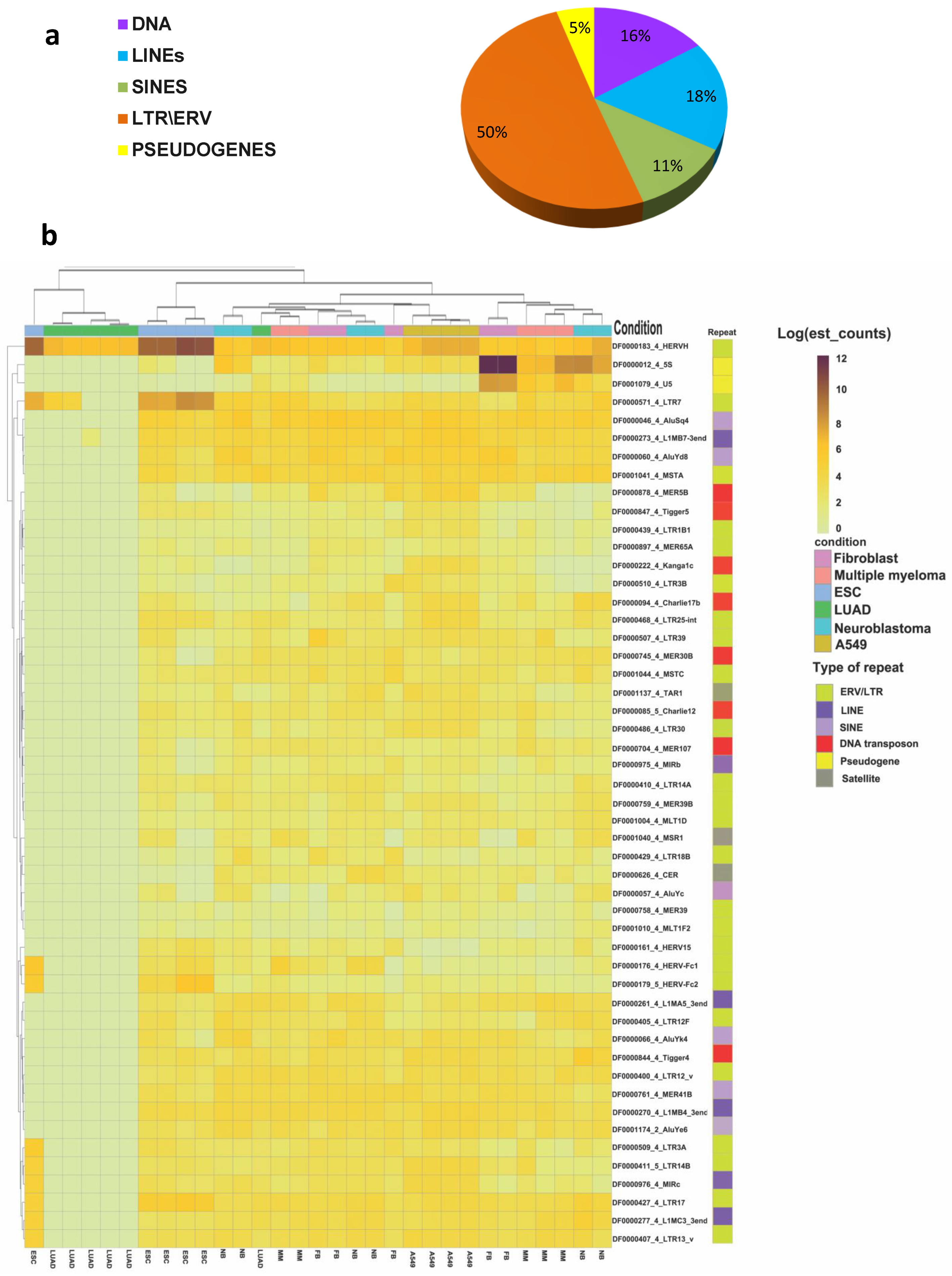
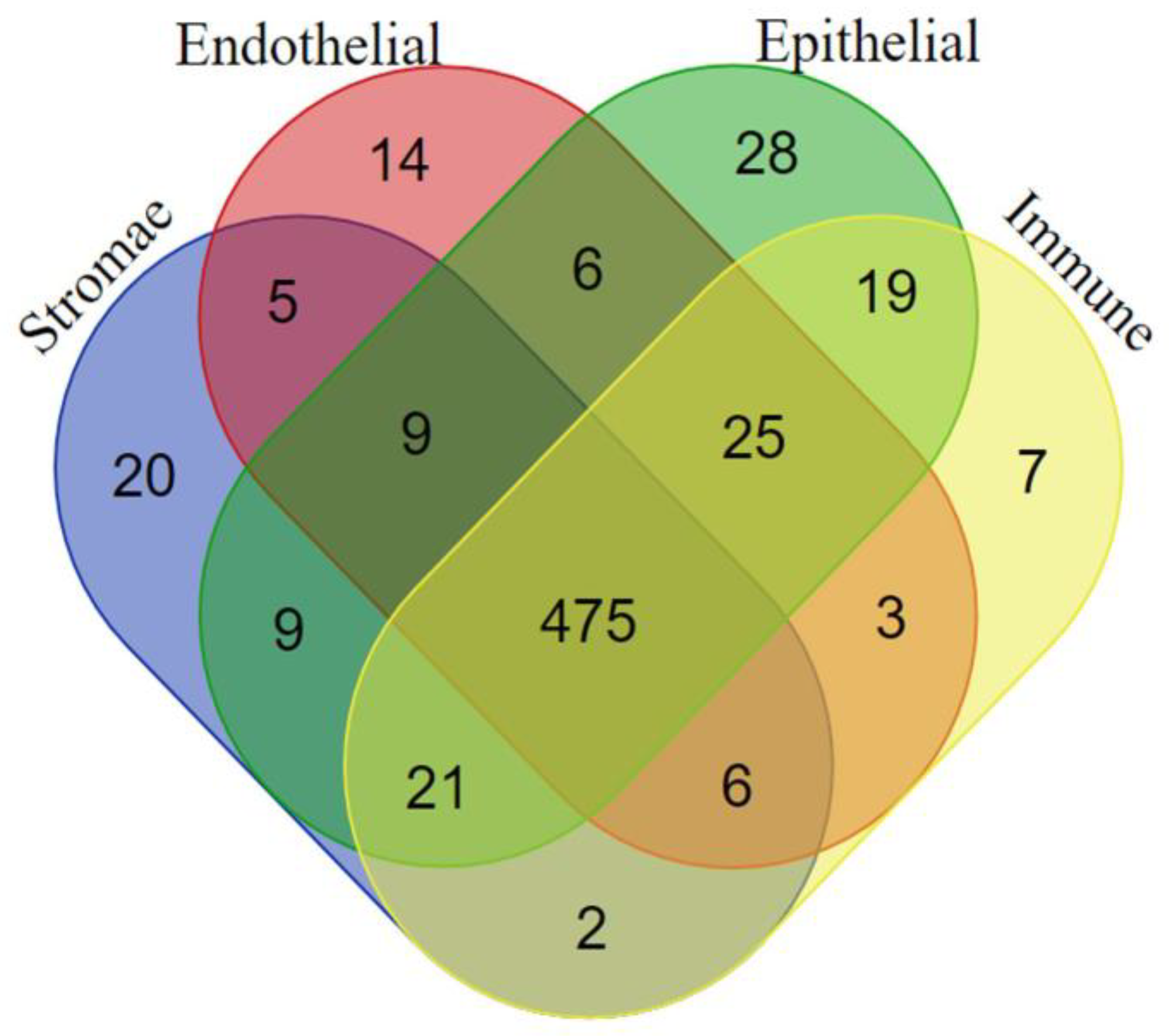
3.3. TE Expression in Cancer Cells and in the TME
4. Discussion
4.1. The Transcription Profile TEs in Tumor Tissue Is Similar to ESCs with the Exception of Four TEs
4.2. The Up-Regulation of Tandem Repeats and Downregulation of Pseudogenes Transcription in Tumors
4.3. The Transcription Profile of TEs in Cancer Cells and the TME
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Glossary
| Autonomous transposon | a transposon that encodes its own enzymes for transposition |
| BLACKJACK, Looper, Zaphod, X and Tigger | ancient DNA transposons. Some X elements are non-autonomous retroelements. |
| DNA transposons | Class II mobile elements that transpose directly from one site to another using a “cut-and-paste” mechanism. DNA transposons have no reverse transcriptase domains and usually have terminal inverted repeats, flanking core-sequence-encoding transposase. |
| Embyonic stem cells (ESCs) | cells of the inner cell mass of the blastocyst, an early stage of the developing embryo that lasts from 4 to 7 days after fertilization. |
| Endogenous retroviruses (ERVSs) | inherited genetic elements derived from exogenous retroviral infections occurring throughout the evolution. |
| Eulor (euteleostomi-conserved low-frequency repeat) | a family of unclassified ancient repeats. |
| EUTREP (eutherian repeat) | a family of ancient repeats that is not attributed to any class of DNA repeats. |
| HERVs—human endogenous retroviruses | a group of viral elements present in the human genome that bear resemblance to contemporary exogenous retroviruses. |
| LINEs, long interspersed nuclear elements | autonomous non-LTR retrotransposons |
| Long non-coding RNA (lncRNA) | transcript that does not encode a protein and is longer than 200 base pairs. lncRNAs are (Pol I)-, Pol II-, and Pol III-transcribed RNAs, as well as RNAs from processed introns |
| LTRs—long terminal repeats | direct simple repeats flanking the core sequence of some types of retrotransposons. |
| MER | medium reiterated frequency repeats—transposons of various families in human genome. |
| MLT | mammalian LTR transposon—a family of ERVs specific to mammals |
| Non-LTR retrotransposons | retrotransposons that lack long terminal repeats. |
| Pseudogene | a DNA segment that has a structural resemblance to a gene; however, it lacks the ability to encode a protein. |
| Retrotransposons | Class I mobile elements that have reverse transcriptase domains. These elements use reverse transcription and replicate themselves in the genome during the transposition process. |
| SINEs, short interspersed nuclear elements | non-autonomous, non-LTR retrotransposons that utilize the enzymatic machinery of LINEs for transposition. |
| The tumor microenvironment (TME) | refers to the dynamic structure surrounding cancer cells and interacting with them. TME includes immune cells, the extracellular matrix, blood vessels, and other cell types, such as fibroblasts. |
| Transcripts per million (TPM) | is a normalization method for RNA-seq. The abbreviation means that “for every 1,000,000 RNA molecules in the RNA-seq sample, TPM value came from this gene/transcript.” |
| Transposable or mobile element (TE) | a sequence of DNA that is capable of changing its location in the genome. |
| Trimming | a process that precedes the assembly of a genome or its analysis. Trimming is a crucial preliminary step in genome assembly or analysis. This process involves the removal of low-quality bases, identified by their high likelihood of incorrect calling, as well as the elimination of adapter sequences |
| UCON (ultraconserved element) | a family of ancient repeats that are not classified yet |
References
- Dandekar, T.; Kunz, M. Genomes: Molecular Maps of Living Organisms. In Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2023; pp. 35–45. ISBN 9783662650356. [Google Scholar]
- Lemerle, E.; Trompouki, E. Transposable Elements in Normal and Malignant Hematopoiesis. DMM Dis. Model. Mech. 2023, 16, dmm050170. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A Unified Classification System for Eukaryotic Transposable Elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Han, K.; Liang, P. Role of Transposable Elements in Gene Regulation in the Human Genome. Life 2021, 11, 118. [Google Scholar] [CrossRef]
- Colonna Romano, N.; Fanti, L. Transposable Elements: Major Players in Shaping Genomic and Evolutionary Patterns. Cells 2022, 11, 1048. [Google Scholar] [CrossRef]
- López-Flores, I.; Garrido-Ramos, M.A. The Repetitive DNA Content of Eukaryotic Genomes. Genome Dyn. 2012, 7, 1–28. [Google Scholar] [CrossRef]
- Britten, R.J. Transposable Element Insertions Have Strongly Affected Human Evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 19945–19948. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.J.; Rosenkrantz, J.L.; Carbone, L.; Chavez, S.L. Endogenous Retroviruses: With Us and against Us. Front. Chem. 2017, 5, 23. [Google Scholar] [CrossRef]
- Solovyeva, A.; Levakin, I.; Zorin, E.; Adonin, L.; Khotimchenko, Y.; Podgornaya, O. Transposons-Based Clonal Diversity in Trematode Involves Parts of Cr1 (Line) in Eu-and Heterochromatin. Genes 2021, 12, 1129. [Google Scholar] [CrossRef]
- Karakülah, G.; Yandim, C. Signature Changes in the Expressions of Protein-Coding Genes, LncRNAs and Repeat Elements in Early and Late Cellular Senescence. Turk. J. Biol. 2020, 44, 356–370. [Google Scholar] [CrossRef]
- Yushkova, E.; Moskalev, A. Transposable Elements and Their Role in Aging. Ageing Res. Rev. 2023, 86, 101881. [Google Scholar] [CrossRef] [PubMed]
- Grundy, E.E.; Diab, N.; Chiappinelli, K.B. Transposable Element Regulation and Expression in Cancer. FEBS J. 2022, 289, 1160–1179. [Google Scholar] [CrossRef] [PubMed]
- Grow, E.J.; Flynn, R.A.; Chavez, S.L.; Bayless, N.L.; Wossidlo, M.; Wesche, D.J.; Martin, L.; Ware, C.B.; Blish, C.A.; Chang, H.Y.; et al. Intrinsic Retroviral Reactivation in Human Preimplantation Embryos and Pluripotent Cells. Nature 2015, 522, 221–246. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.; Xing, X.; Kim, S.; et al. Transposable Elements Drive Widespread Expression of Oncogenes in Human Cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H. Transposable Elements in Cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Köchert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an Endogenous Long Terminal Repeat Activates the CSF1R Proto-Oncogene in Human Lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Babaian, A.; Romanish, M.T.; Gagnier, L.; Kuo, L.Y.; Karimi, M.M.; Steidl, C.; Mager, D.L. Onco-Exaptation of an Endogenous Retroviral LTR Drives IRF5 Expression in Hodgkin Lymphoma. Oncogene 2016, 35, 2542–2546. [Google Scholar] [CrossRef] [PubMed]
- Lynch-Sutherland, C.F.; Chatterjee, A.; Stockwell, P.A.; Eccles, M.R.; Macaulay, E.C. Reawakening the Developmental Origins of Cancer Through Transposable Elements. Front. Oncol. 2020, 10, 468. [Google Scholar] [CrossRef]
- Jansz, N.; Faulkner, G.J. Endogenous Retroviruses in the Origins and Treatment of Cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef]
- Karttunen, K.; Patel, D.; Xia, J.; Fei, L.; Palin, K.; Aaltonen, L.; Sahu, B. Transposable Elements as Tissue-Specific Enhancers in Cancers of Endodermal Lineage. Nat. Commun. 2023, 14, 5313. [Google Scholar] [CrossRef]
- Yavuz, B.G.; Gunaydin, G.; Gedik, M.E.; Kos, K. Cancer Associated Fibroblasts Sculpt Tumour Microenvironment by Recruiting Monocytes and Inducing Immunosuppressive PD-1 + TAMs. Sci. Rep. 2019, 9, 3172. [Google Scholar] [CrossRef]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The Lung Microenvironment: An Important Regulator of Tumour Growth and Metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, W.; Wang, J.; Ao, X.; Xue, J. Non-Coding RNAs in Lung Cancer: Molecular Mechanisms and Clinical Applications. Front. Oncol. 2023, 13, 1256537. [Google Scholar] [CrossRef]
- Enukashvily, N.I.; Ponomartsev, N.V.; Ketkar, A.; Suezov, R.; Chubar, A.V.; Prjibelski, A.D.; Shafranskaya, D.D.; Elmshäuser, S.; Keber, C.U.; Stefanova, V.N.; et al. Pericentromeric Satellite LncRNAs Are Induced in Cancer-Associated Fibroblasts and Regulate Their Functions in Lung Tumorigenesis. Cell Death Dis. 2023, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 May 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential Analysis of RNA-Seq Incorporating Quantification Uncertainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S.; Nittner, D.; Burton, O.; Bassez, A.; Decaluwé, H.; Pircher, A.; Van den Eynde, K.; et al. Phenotype Molding of Stromal Cells in the Lung Tumor Microenvironment. Nat. Med. 2018, 24, 1277–1289. [Google Scholar] [CrossRef]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Sikkema, L.; Ramírez-Suástegui, C.; Strobl, D.C.; Gillett, T.E.; Zappia, L.; Madissoon, E.; Markov, N.S.; Zaragosi, L.-E.; Ji, Y.; Ansari, M.; et al. An Integrated Cell Atlas of the Lung in Health and Disease. Nat. Med. 2023, 29, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K. Human Transposable Elements in Repbase: Genomic Footprints from Fish to Humans. Mob. DNA 2018, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Shvemberger, I.N.; Alexandrova, S.A. PCR-Detected Genome Polymorphism in Malignant Cell Growth. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 2000; pp. 117–159. [Google Scholar]
- Hafez, A.; Mohamed, H. Impacts of Genetic Polymorphisms on Breast Cancer. Int. J. Oncol. Res. 2022, 5, 37. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic Instability—An Evolving Hallmark of Cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.J.; Lu, A.; Moon, C. Role of Genomic Instability in Human Carcinogenesis. Exp. Biol. Med. 2019, 244, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Noberini, R.; Osti, D.; Miccolo, C.; Richichi, C.; Lupia, M.; Corleone, G.; Hong, S.P.; Colombo, P.; Pollo, B.; Fornasari, L.; et al. Extensive and Systematic Rewiring of Histone Post-Translational Modifications in Cancer Model Systems. Nucleic Acids Res. 2018, 46, 3817–3832. [Google Scholar] [CrossRef] [PubMed]
- Nestor, C.E.; Ottaviano, R.; Reinhardt, D.; Cruickshanks, H.A.; Mjoseng, H.K.; McPherson, R.C.; Lentini, A.; Thomson, J.P.; Dunican, D.S.; Pennings, S.; et al. Rapid Reprogramming of Epigenetic and Transcriptional Profiles in Mammalian Culture Systems. Genome Biol. 2015, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Robbez-Masson, L.; Rowe, H.M. Retrotransposons Shape Species-Specific Embryonic Stem Cell Gene Expression. Retrovirology 2015, 12, 45. [Google Scholar] [CrossRef]
- Xiang, X.; Tao, Y.; DiRusso, J.; Hsu, F.M.; Zhang, J.; Xue, Z.; Pontis, J.; Trono, D.; Liu, W.; Clark, A.T. Human Reproduction Is Regulated by Retrotransposons Derived from Ancient Hominidae-Specific Viral Infections. Nat. Commun. 2022, 13, 463. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, T.; Preissl, S.; Amaral, M.L.; Grinstein, J.D.; Farah, E.N.; Destici, E.; Qiu, Y.; Hu, R.; Lee, A.Y.; et al. Transcriptionally Active HERV-H Retrotransposons Demarcate Topologically Associating Domains in Human Pluripotent Stem Cells. Nat. Genet. 2019, 51, 1380–1388. [Google Scholar] [CrossRef]
- Bénit, L.; Calteau, A.; Heidmann, T. Characterization of the Low-Copy HERV-Fc Family: Evidence for Recent Integrations in Primates of Elements with Coding Envelope Genes. Virology 2003, 312, 159–168. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trigiante, G.; Blanes Ruiz, N.; Cerase, A. Emerging Roles of Repetitive and Repeat-Containing RNA in Nuclear and Chromatin Organization and Gene Expression. Front. Cell Dev. Biol. 2021, 9, 735527. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.E.V.; Chivers, M.; Borovska, I.; Monger, S.; Giannoulatou, E.; Kralovicova, J.; Vorechovsky, I. Transposon Clusters as Substrates for Aberrant Splice-Site Activation. RNA Biol. 2021, 18, 354–367. [Google Scholar] [CrossRef]
- Hsieh, F.K.; Ji, F.; Damle, M.; Sadreyev, R.I.; Kingston, R.E. HERVH-Derived LncRNAs Negatively Regulate Chromatin Targeting and Remodeling Mediated by CHD7. Life Sci. Alliance 2022, 5, e202101127. [Google Scholar] [CrossRef]
- Zheng, J.; Wei, Y.; Han, G.Z. The Diversity and Evolution of Retroviruses: Perspectives from Viral “Fossils”. Virol. Sin. 2022, 37, 11–18. [Google Scholar] [CrossRef]
- Fischer, S.; Echeverría, N.; Moratorio, G.; Landoni, A.I.; Dighiero, G.; Cristina, J.; Oppezzo, P.; Moreno, P. Human Endogenous Retrovirus Np9 Gene Is over Expressed in Chronic Lymphocytic Leukemia Patients. Leuk. Res. Rep. 2014, 3, 70–72. [Google Scholar] [CrossRef]
- Saini, S.K.; Ørskov, A.D.; Bjerregaard, A.M.; Unnikrishnan, A.; Holmberg-Thydén, S.; Borch, A.; Jensen, K.V.; Anande, G.; Bentzen, A.K.; Marquard, A.M.; et al. Human Endogenous Retroviruses Form a Reservoir of T Cell Targets in Hematological Cancers. Nat. Commun. 2020, 11, 5660. [Google Scholar] [CrossRef] [PubMed]
- Yandlm, C.; Karakülah, G. Expression Dynamics of Repetitive DNA in Early Human Embryonic Development. BMC Genomics 2019, 20, 439. [Google Scholar] [CrossRef]
- Hackett, J.A.; Kobayashi, T.; Dietmann, S.; Surani, M.A. Activation of Lineage Regulators and Transposable Elements across a Pluripotent Spectrum. Stem Cell Rep. 2017, 8, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Babarinde, I.A.; Zhou, X.; Hutchins, A.P. Transposable Elements in Pluripotent Stem Cells and Human Disease. Front. Genet. 2022, 13, 902541. [Google Scholar] [CrossRef]
- Podgornaya, O.I.; Ostromyshenskii, D.I.; Enukashvily, N.I. Who Needs This Junk, or Genomic Dark Matter. Biochemistry 2018, 83, 450–466. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Sachs, F.; Ramsay, L.A.; Jacques, P.É.; Göke, J.; Bourque, G.; Ng, H.H. The Retrovirus HERVH Is a Long Noncoding RNA Required for Human Embryonic Stem Cell Identity. Nat. Struct. Mol. Biol. 2014, 21, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Chung, N.; Jonaid, G.M.; Quinton, S.; Ross, A.; Sexton, C.E.; Alberto, A.; Clymer, C.; Churchill, D.; Navarro Leija, O.; Han, M.V. Transcriptome Analyses of Tumor-Adjacent Somatic Tissues Reveal Genes Co-Expressed with Transposable Elements. Mob. DNA 2019, 10, 39. [Google Scholar] [CrossRef]
- Ergün, S.; Buschmann, C.; Heukeshoven, J.; Dammann, K.; Schnieders, F.; Lauke, H.; Chalajour, F.; Kilic, N.; Strätling, W.H.; Schumann, G.G. Cell Type-Specific Expression of LINE-1 Open Reading Frames 1 and 2 in Fetal and Adult Human Tissues. J. Biol. Chem. 2004, 279, 27753–27763. [Google Scholar] [CrossRef] [PubMed]
- Santoni, F.A.; Guerra, J.; Luban, J. HERV-H RNA Is Abundant in Human Embryonic Stem Cells and a Precise Marker for Pluripotency. Retrovirology 2012, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Sakashita, A.; Kitano, T.; Ishizu, H.; Guo, Y.; Masuda, H.; Ariura, M.; Murano, K.; Siomi, H. Transcription of MERVL Retrotransposons Is Required for Preimplantation Embryo Development. Nat. Genet. 2023, 55, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Soltanian, S.; Sheikhbahaei, M. Effect of Menadione and Combination of Gemcitabine and Cisplatin on Cancer Stem Cells in Human Non-Small Cell Lung Cancer (Nsclc) Cell Line A549. Iran. J. Pharm. Res. 2021, 20, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Karakülah, G.; Yandim, C. Identification of Differentially Expressed Genomic Repeats in Primary Hepatocellular Carcinoma and Their Potential Links to Biological Processes and Survival. Turk. J. Biol. 2021, 45, 599–612. [Google Scholar] [CrossRef]
- Sin, H.S.; Huh, J.W.; Kim, D.S.; Kim, T.H.; Ha, H.S.; Kim, W.Y.; Park, H.K.; Kim, C.M.; Kim, H.S. Endogenous Retrovirus-Related Sequences Provide an Alternative Transcript of MCJ Genes in Human Tissues and Cancer Cells. Genes Genet. Syst. 2006, 81, 333–339. [Google Scholar] [CrossRef]
- Stricker, E.; Peckham-Gregory, E.C.; Scheurer, M.E. HERVs and Cancer—A Comprehensive Review of the Relationship of Human Endogenous Retroviruses and Human Cancers. Biomedicines 2023, 11, 936. [Google Scholar] [CrossRef]
- Yamada, Y.; Haga, H.; Yamada, Y. Concise Review: Dedifferentiation Meets Cancer Development: Proof of Concept for Epigenetic Cancer. Stem Cells Transl. Med. 2014, 3, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Han, Z.; Zhu, Y.; Chen, J.; Li, W. The Role and Specific Mechanism of OCT4 in Cancer Stem Cells: A Review. Int. J. Stem Cells 2020, 13, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Mullins, C.S.; Linnebacher, M. Endogenous Retrovirus Sequences as a Novel Class of Tumor-Specific Antigens: An Example of HERV-H Env Encoding Strong CTL Epitopes. Cancer Immunol. Immunother. 2012, 61, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.; Wieland, L.; Krüger, A.; Volkmer, I.; Cynis, H.; Emmer, A.; Staege, M.S. Identification of Differentially Expressed Human Endogenous Retrovirus Families in Human Leukemia and Lymphoma Cell Lines and Stem Cells. Front. Oncol. 2021, 11, 637981. [Google Scholar] [CrossRef] [PubMed]
- Carter, T.A.; Singh, M.; Dumbović, G.; Chobirko, J.D.; Rinn, J.L.; Feschotte, C. Mosaic Cis-Regulatory Evolution Drives Transcriptional Partitioning of HERVH Endogenous Retrovirus in the Human Embryo. eLife 2022, 11, e76257. [Google Scholar] [CrossRef] [PubMed]
- Römer, C.; Singh, M.; Hurst, L.D.; Izsvák, Z. How to Tame an Endogenous Retrovirus: HERVH and the Evolution of Human Pluripotency. Curr. Opin. Virol. 2017, 25, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zheng, Y.; Liu, H.; Zhu, X.; Gu, Y.; Lan, Y.; Tan, J.; Xu, H.; Guo, R. A Non-Invasive Monitoring of USPIO Labeled Silk Fibroin/Hydroxyapatite Scaffold Loaded DPSCs for Dental Pulp Regeneration. Mater. Sci. Eng. C 2019, 103, 109736. [Google Scholar] [CrossRef] [PubMed]
- Ohnuki, M.; Tanabe, K.; Sutou, K.; Teramoto, I.; Sawamura, Y.; Narita, M.; Nakamura, M.; Tokunaga, Y.; Nakamura, M.; Watanabe, A.; et al. Dynamic Regulation of Human Endogenous Retroviruses Mediates Factor-Induced Reprogramming and Differentiation Potential. Proc. Natl. Acad. Sci. USA 2014, 111, 12426–12431. [Google Scholar] [CrossRef]
- Yi, J.M.; Kim, H.M.; Kim, H.S. Human Endogenous Retrovirus HERV-H Family in Human Tissues and Cancer Cells: Expression, Identification, and Phylogeny. Cancer Lett. 2006, 231, 228–239. [Google Scholar] [CrossRef]
- Ito, J.; Kimura, I.; Soper, A.; Coudray, A.; Koyanagi, Y.; Nakaoka, H.; Inoue, I.; Turelli, P.; Trono, D.; Sato, K. Endogenous Retroviruses Drive KRAB Zinc-Finger Protein Family Expression for Tumor Suppression. Sci. Adv. 2020, 6, eabc3020. [Google Scholar] [CrossRef]
- Xie, W.; Schultz, M.D.; Lister, R.; Hou, Z.; Rajagopal, N.; Ray, P.; Whitaker, J.W.; Tian, S.; Hawkins, R.D.; Leung, D.; et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell 2013, 153, 1134–1148. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.M.; Kim, H.S. Expression Analysis of Endogenous Retroviral Elements Belonging to the HERV-F Family from Human Tissues and Cancer Cells. Cancer Lett. 2004, 211, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Laska, M.J.; Brudek, T.; Nissen, K.K.; Christensen, T.; Møller-Larsen, A.; Petersen, T.; Nexø, B.A. Expression of HERV-Fc1, a Human Endogenous Retrovirus, Is Increased in Patients with Active Multiple Sclerosis. J. Virol. 2012, 86, 3713–3722. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Walichiewicz, J.; Milosavljevic, A. Prototypic Sequences for Human Repetitive DNA. J. Mol. Evol. 1992, 35, 286–291. [Google Scholar] [CrossRef]
- Rose, A.M.; Krishan, A.; Chakarova, C.F.; Moya, L.; Chambers, S.K.; Hollands, M.; Illingworth, J.C.; Williams, S.M.G.; McCabe, H.E.; Shah, A.Z.; et al. MSR1 Repeats Modulate Gene Expression and Affect Risk of Breast and Prostate Cancer. Ann. Oncol. 2018, 29, 1292–1303. [Google Scholar] [CrossRef]
- Onishi-Seebacher, M.; Erikson, G.; Sawitzki, Z.; Ryan, D.; Greve, G.; Lübbert, M.; Jenuwein, T. Repeat to Gene Expression Ratios in Leukemic Blast Cells Can Stratify Risk Prediction in Acute Myeloid Leukemia. BMC Med. Genom. 2021, 14, 166. [Google Scholar] [CrossRef] [PubMed]
- Grapotte, M.; Saraswat, M.; Bessière, C.; Menichelli, C.; Ramilowski, J.A.; Severin, J.; Hayashizaki, Y.; Itoh, M.; Tagami, M.; Murata, M.; et al. Discovery of Widespread Transcription Initiation at Microsatellites Predictable by Sequence-Based Deep Neural Network. Nat. Commun. 2021, 12, 3297. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Suh, A.; Feschotte, C. The Hidden Elasticity of Avian and Mammalian Genomes. bioRxiv 2016, 081307. [Google Scholar] [CrossRef]
- Perreault, J.; Noël, J.F.; Brière, F.; Cousineau, B.; Lucier, J.F.; Perreault, J.P.; Boire, G. Retropseudogenes Derived from the Human Ro/SS-A Autoantigen-Associated HY RNAs. Nucleic Acids Res. 2005, 33, 2032–2041. [Google Scholar] [CrossRef]
- Yang, D.; Liu, J.; Qian, H.; Zhuang, Q. Cancer-Associated Fibroblasts: From Basic Science to Anticancer Therapy. Exp. Mol. Med. 2023, 55, 1322–1332. [Google Scholar] [CrossRef]
- De Veirman, K.; Rao, L.; De Bruyne, E.; Menu, E.; Van Valckenborgh, E.; Van Riet, I.; Frassanito, M.A.; Di Marzo, L.; Vacca, A.; Vanderkerken, K. Cancer Associated Fibroblasts and Tumor Growth: Focus on Multiple Myeloma. Cancers 2014, 6, 1363–1381. [Google Scholar] [CrossRef] [PubMed]
- Brichkina, A.; Polo, P.; Sharma, S.D.; Visestamkul, N.; Lauth, M. A Quick Guide to CAF Subtypes in Pancreatic Cancer. Cancers 2023, 15, 2614. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.-J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef]
- Evdokimova, V.; Ruzanov, P.; Heisler, L.E.; Mcpherson, J.D.; Orlic-milacic, M.; Specht, K.; Steiger, K.; Delattre, O.; Burdach, S.; Stein, L.D.; et al. Exosomes Transmit Retroelement RNAs to Drive Inflammation and Immunosuppression in Ewing Sarcoma. bioRxiv 2019, 806851. [Google Scholar] [CrossRef]
- Evdokimova, V.; Gassmann, H.; Radvanyi, L.; Burdach, S.E.G. Current State of Immunotherapy and Mechanisms of Immune Evasion in Ewing Sarcoma and Osteosarcoma. Cancers 2023, 15, 272. [Google Scholar] [CrossRef]
- Cañadas, I.; Thummalapalli, R.; Kim, J.W.; Kitajima, S.; Jenkins, R.W.; Christensen, C.L.; Campisi, M.; Kuang, Y.; Zhang, Y.; Gjini, E.; et al. Tumor Innate Immunity Primed by Specific Interferon-Stimulated Endogenous Retroviruses. Nat. Med. 2018, 24, 1143–1150. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, W.; Qin, C.; Zhang, L.; Deng, J.; Liu, S.; Qin, Z. Responsiveness of Stromal Fibroblasts to IFN-γ Blocks Tumor Growth via Angiostasis. J. Immunol. 2009, 183, 6413–6421. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Li, X.; Mao, Y.; Wei, C.; Huang, Z.; Li, G.; Yin, J.; Liang, X.; Liu, Z. Interferon-Dependent SLC14A1+ Cancer-Associated Fibroblasts Promote Cancer Stemness via WNT5A in Bladder Cancer. Cancer Cell 2022, 40, 1550–1565.e7. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, L.; Li, Y.; Deng, P.; Pan, P.; Hu, C.; Yang, H. Pirfenidone Promotes the Levels of Exosomal MiR-200 to down-Regulate ZEB1 and Represses the Epithelial-Mesenchymal Transition of Non-Small Cell Lung Cancer Cells. Hum. Cell 2022, 35, 1813–1823. [Google Scholar] [CrossRef]
- Zhu, X.; Fang, H.; Gladysz, K.; Barbour, J.A.; Wong, J.W.H. Overexpression of Transposable Elements Is Associated with Immune Evasion and Poor Outcome in Colorectal Cancer. Eur. J. Cancer 2021, 157, 94–107. [Google Scholar] [CrossRef]

| SRA | Cell Type | Cell Line | Instrument | Selection | Layout | Analysis * |
|---|---|---|---|---|---|---|
| SRR11802228 | Undifferentiated hESCs | WA09 | Illumina HiSeq 4000 | cDNA | SINGLE | Venn/DE |
| SRR11802260 | Undifferentiated hESCs | WA09 | Illumina HiSeq 4000 | cDNA | SINGLE | Venn/DE |
| SRR19763997 | Human embryonic stem cells (hESCs), | H9 | Illumina HiSeq 4000 | cDNA | PAIRED | Venn/DE |
| SRR19763998 | Human embryonic stem cells (hESCs) | H9 | Illumina HiSeq 4000 | cDNA | PAIRED | Venn/DE |
| SRR19763999 | Human embryonic stem cells (hESCs) | H9 | Illumina HiSeq 4000 | cDNA | PAIRED | Venn/DE |
| SRR17011020 | Human neuroblastoma cells | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR17011019 | Human neuroblastoma cells | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR26588672 | Human neuroblastoma cells | SK-N-SH | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR26588673 | Human neuroblastoma cells | SK-N-SH | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR21143000 | Human neuroblastoma cells | - | Illumina HiSeq 2000 | cDNA | PAIRED | Venn/DE |
| SRR21142995 | Human neuroblastoma cells | - | Illumina HiSeq 2000 | cDNA | PAIRED | Venn/DE |
| SRR24600381 | Lung adenocarcinoma | H1975 | Illumina HiSeq 2500 | cDNA | PAIRED | Venn/DE |
| SRR24600382 | Lung adeno-carcinoma | H1975 | Illumina HiSeq 2500 | cDNA | PAIRED | Venn/DE |
| SRR24600383 | Lung adeno-carcinoma | H1975 | Illumina HiSeq 2500 | cDNA | PAIRED | Venn/DE |
| SRR24166172 | Lung adeno-carcinoma | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR24166174 | Lung adeno-carcinoma | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR24166176 | Lung adeno-carcinoma | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR12924510 | Multiple myeloma | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR12924509 | Multiple myeloma | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRR2497378 | Multiple myeloma | - | Illumina HiSeq 2000 | Random | PAIRED | Venn/DE |
| SRR2497389 | Multiple myeloma | - | Illumina HiSeq 2000 | Random | PAIRED | Venn/DE |
| SRR2497395 | Multiple myeloma | - | Illumina HiSeq 2000 | Random | PAIRED | Venn/DE |
| ERR1406030 | Lung adenocarcinoma | A549 | Illumina HiSeq 2500 | cDNA | PAIRED | Venn/DE |
| SRR15410445 | Lung adeno-carcinoma | A549 | Illumina HiSeq 2500 | cDNA | PAIRED | Venn/DE |
| SRR15410446 | Lung adeno-carcinoma | A549 | Illumina HiSeq 2500 | cDNA | PAIRED | Venn/DE |
| SRR15410447 | Lung adeno-carcinoma | A549 | Illumina HiSeq 2500 | cDNA | PAIRED | Venn/DE |
| SRR26713630 | Fibroblast, primary cells from lung, 2D culture | - | Illumina NovaSeq 6000 | cDNA | PAIRED | Venn/DE |
| SRS9826564 | Fibroblast, untreated normal cells | - | NextSeq 500 | cDNA | PAIRED | Venn/DE |
| SRS9826571 | Fibroblast, untreated normal cells | - | NextSeq 500 | cDNA | PAIRED | Venn |
| ERR12530401 | Fibroblast, normal cells | - | Illumina HiSeq 2500 | PCR | PAIRED | DE |
| SRR27235867 | Fibroblast, normal cells from skin | - | NextSeq 550 | Size fractionation | PAIRED | DE |
| SRR27235866 | Fibroblast, normal cells from skin | - | NextSeq 550 | Size fractionation | PAIRED | DE |
| Cell Types | DNA Transposons | Retroelements * | Pseudogenes | Unknown |
|---|---|---|---|---|
| Expressed in ESCs, tumor tissues, and cancer cell lines but not in fibroblasts | Charlie2a, Charlie2b, Tigger17b, Tigger5b, X4bDNA, Merlin1HS, Ricksha, Arthur1B, Arthur2, BLACKJACK, DNA1_Mam, Eulor11, Looper, MER113A, MER121B, MER45R, MER63A, MER97a, | L1M2a_5end, L1M2b_5end, L1M3c_5end, L1M3d_5end, L1M3e_5end, L1M4a1_5end, L1MC4_5end, L1MC5a_3end, L1MD2_5end, L1ME3C_3end, L1MEb_5end, L1MEd_5end, L1MEg_5end, L1MEi_5end, L1P4b_5end, L1P4c_5end, L1P4d_5end, L4B, CR1_Mam, HAL1b, PlatL3, HERV16, HERV30, HERVK11D, HERVL40, HUERS-P2, ERV24_Prim, ERVL47, LTR10B2, LTR21C, LTR26B, LTR26C, LTR2752, LTR27E, LTR37B, LTR38-int, LTR39-int, LTR48B, LTR53B, LTR53-int, MamGypsy2-I, MLT1F-int, MLT1G, MLT1J1, MLT1J2, PABLB-int, LTR37-int, MER34-int, MER70-int, MER83A-int, MER84-int, MER92B, MER92-int, MER66-int, MamRep1151, MER110A, MER67D | SSU-rRNACel | REP522 |
| Expressed in tumor tissues and cancer cell lines | Arthur1A, Charlie11, Charlie16, Charlie17, Charlie7, Charlie7a, EuthAT-2, EutTc1-N2, hAT-N1_Mam, Helitron1Nb_Mam, Kanga1b, Tigger12A, Tigger15a, Tigger16a, Tigger6, MER97b, MER97d, MER81, MER96, MER20B, MER46C, MER58C, MER112, MER106A, MamTip3, MER99, UCON33, UCON9, X1DNA, X11DNA, X26DNA, Zaphod, Zaphod3 | CR1Amni-1, CR1-16AMi, L1M2a1_5end, L1M7_5end, L1M8_5end, L1MCc_5end, L1ME3F_3end, L1ME4c_3end, L1MEa_5end, L1PA17_5end, L2b_3end, L3, L4C, L5 ERV3-16A3I, HERV-Fc1, LTR3, HERVL32, LTR16, LTR16A, LTR16B1, LTR33B, LTR40A1, LTR47B, LTR50, LTR52-int, LTR55, LTR58, LTR68, LTR69, LTR85c, LTR87, LTR91, MER110, MER110-int, MER76-int, MER76, MER89-int, MLT1H2, MLT1H-int, MLT1J-int, MLT1L, PrimAX-int, MER70B, MER70C, MER74C, MER90, MER92A, MER45A, MamRep605b | tRNA-Ala-GCY tRNA-Ala-GCY tRNA-Gly-GGG tRNA-Leu-CTA tRNA-Thr-ACG tRNA-Val-GTA | MER125 UCON20 UCON21 UCON23 UCON28a UCON64 UCON68 |
| Expressed only in ESCs | Tigger13a, Tigger8, Arthur1C, Charlie14a, EutTc1-N1, X6a_DNA | LTR33A, LTR82B, MLT2E, MER110A | ||
| Expressed only in fibroblasts | MER94, MER102a, MER123 | LTR75, LTR16A2 |
| Cell Types | DNA Transposons | Retroelements | Pseudogenes | Unknown |
|---|---|---|---|---|
| LUAD tissue | Tigger9a, Charlie1, Tigger5b, Charlie2b, Charlie7a, Tigger15a Kanga1d UCON21 MER63C MER46C MER119, UCON9 Looper, MER97a, UCON33, UCON23 MER99 MER125 | MER66-int, L1M3e_5end,, LTR16, L1MEa_5end, L1M3b_5end, L1MEg_5end, L1ME3C_3end, LTR16A, L1MEb_5end, L1MC5a_3end, LTR53, MER68B, L1M3de_5end, LTR67B, HERVL32, MER110, LTR87, LTR55, MER76, HERVK11D, MER74B, L1M4a2_5end, L1ME3Cz_3end, MER34,LTR47B3, HERV1_LTRe, L1ME5_3end,, MER131, MER73, LTR16E1, LTR1C3, LTR27D, HERV-Fc1_LTR3, LTR53-int, LTR39-int, LTR58, LTR26C, MER101B, LTR34, MER21-int, MLT1L, LTR40A1 | tRNA-Tyr-TAT | UCON64, UCON28a |
| LUAD cell line H1975 | Charlie18a Merlin1_HS MamTip3 MER45A MER104 | MamRep1151 LTR65 L1M2a1_5end.LTR16B1 LTR68 MER92A L1P4c_5end MER41G HERV1_I L1ME3G_3end, LTR10B2 MER20B LTR69 | ||
| LUAD cell line A549 | MER112 | HERVL74, LTR43-int, MLT1G MLT1J2, AluSg | tRNA-Pro-CCA, tRNA-Pro-CCY, tRNA-Ala-GCG, tRNA-Thr-ACG, tRNA-Ala-GCY_v, | |
| NB tissue | Arthur1A, Tigger7, Charlie7, Arthur1B, MER121B, MER115, Ricksha_0 | L1M3b_5end, L1MDb_5endб MER68B, LTR75_1, MER84-int, MLT1G, HERV15, MLT1E L1MEi_5end, L1P4c_5end, LTR39-int, L4_B_Mam, L1M3d_5end, MER21-int, LTR48B | tRNA-Pro-CCA, tRNA-Ala-GCY_v, tRNA-Leu-CTA, tRNA-Glu-GAG_v, tRNA-Leu-TTG | UCON68 |
| NB cell line SK-N-SH | Charlie18a, Tigger15a, Tigger6b, Charlie4z, Kanga2_a, Kanga1c, hAT-N1_Mam, Tigger2b_Pri, Tigger12c, Charlie5, Tigger9b, MER44D, FordPrefect, MER46C, MER63B, MER96, UCON21, hAT-5_Mam, MER113A, MER81, Eulor11, MER58D, EutTc1-N2, MER63D, DNA1_Mam ORSL, MER47B, MER125, Tigger14a | LTR72,, L1PREC2_5end, MER65B, MER57E1, MER61C, MER57C2,MLT2B1, L1ME3D_3end, LTR38A1,LTR82A,, LTR41, MLT1E1, LTR35B, MER68-int, MER72B, MLT2B5, MLT-int, L1M2c_5end, HERVL74,LTR26B, L1ME3C_3end, MLT1G3,HERV1_LTRb, LTR16A, MLT1E2, LTR31, AmnSINE2, LTR40b, LTR1B0, MLT1E1A, LTR23-int, LTR53B, LTR38, MER67A, L1MEf_5end, LTR2752, MER66C, LTR33B, MER5C1, LTR29, LTR52, MER50C, MER66D, MER76, L1ME4a_3end, MER61B, LTR21C, LTR1C1, LTR18A, L1ME3Cz_3end, LTR24C, MER61D, MER74C, MER73, MER92B, LTR1C3, LTR27D, L1MCb_5end, L1MEc_5end, LTR27C, HERVIP10B3, HERVFH19, MER52-int, L1ME3G_3end, LTR34, LTR10B2, LTR10B, MER88, MER89, MamRep605, LTR1F2, LTR16C, MLT2F, MLT1F, L1P4a_5end, MER67B, MER77B, L2c_3end | tRNA-Pro-CCY, tRNA-Val-GTY, tRNA-Ser-TCG, tRNA-Thr-ACG, tRNA-Leu-TTA, tRNA-Tyr-TAT, tRNA-Gln-CAA, tRNA-Pro-CCG, tRNA-Met_v, tRNA-Phe-TTY, tRNA-Leu-CTYtRNA, -Ala-GCY |
| Cell Type | DNA Transposons | Retroelements | Pseudogenes | Unknown |
|---|---|---|---|---|
| Epithelial | LTR28C, LTR1D1, LTR26B, MER4CL34, HERVL66, LTR57, LTR41C, L1M2a_5end, HERVP71A, MER67A, LTR52, L1PA17_5end, HERV15, LTR21C, L1ME3Cz_3end, HERV-Fc1, MER92-int, LTR22, MER34C_v, HERVIP10B3, MER61F, MER77, L2, HERV3 | REP522 | ||
| Endothe- lial | X11_DNA, Tigger17d, Tigger5b, MER106AMER47B | MER51D MER61C L1M3b_5end L1MEb_5end MER34C LTR16A2, MER83C, MLT1-int | UCON64 | |
| Immune | MER101, LTR72, LTR26E, HERV9, LTR28, LTR47B2, LTR48 | |||
| Stromal | Charlie18a, Ricksha, MER45B, UCON23, MER103C | L1MC5_3end, L1ME3D_3end, LTR38A1, L1ME3, B_3end, MER67D, LTR28B, L1MEg_5end, LTR18C, HERVL32, LTR59, HERV-Fc1-LTR2, L2b_3end, LTR19C, LTR27C, MLT1I |
| Cell Type (Number of TEs) | DNA Transposons | Retroelements | Pseudogenes | Unknown |
|---|---|---|---|---|
| LUAD airway EP(24) | MER66-int, MER65B, LTR26B, MLT1G3, HERVL66, MER34A1, LTR52, LTR45, L1PA17_5end, HAL1M8, HERVK11D, HERV15, LTR21C, L1ME3Cz_3end, HERV4_I, MER73, LTR22, MER83B, MLT1F, LTR1E, HERV3, L2c_3end | REP522 | ||
| LUAD alveolar EP (6) | MER105 | LTR18A, MER92-int, MER88, MER89 | ||
| Blood vessels (13) | Tigger5b, Tigger17d, UCON9 | MER51D, MER61C, L1MEb_5end, LTR31, LTR37B, LTR16A2, MER83C, MLT1-int | UCON64 | |
| Fibroblast lineage (11) | Ricksha, Charlie18a, MER103C, UCON23 | L1ME3D_3end, L1ME3B_3end, L1MEg_5end, HERV-Fc1_LTR2, L2b_3end, LTR19C, MLT1I | ||
| Lymphatic cells (8) | Ricksha_b, MER58D | L1MC5_3end, MLT1H, L1MD2_5end, LTR2752, HAL1, MER74B | ||
| Lymphoid cells (3) | LTR72, MLT1E3, LTR40c | |||
| Mesothelium (5) | Ricksha_a | MER66A, LTR1B0, LTR23-int, MER101-int | ||
| Myeloid (5) | - | LTR47B4, LTR54B, MER54B, MLT1G, MER52A | ||
| Smooth muscle (6) | Zaphod3, MER8 | MLT1E1, L1M3c_5end, MLT1E, LTR39-int |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solovyeva, A.I.; Afanasev, R.V.; Popova, M.A.; Enukashvily, N.I. Dysregulation of Transposon Transcription Profiles in Cancer Cells Resembles That of Embryonic Stem Cells. Curr. Issues Mol. Biol. 2024, 46, 8576-8599. https://doi.org/10.3390/cimb46080505
Solovyeva AI, Afanasev RV, Popova MA, Enukashvily NI. Dysregulation of Transposon Transcription Profiles in Cancer Cells Resembles That of Embryonic Stem Cells. Current Issues in Molecular Biology. 2024; 46(8):8576-8599. https://doi.org/10.3390/cimb46080505
Chicago/Turabian StyleSolovyeva, Anna I., Roman V. Afanasev, Marina A. Popova, and Natella I. Enukashvily. 2024. "Dysregulation of Transposon Transcription Profiles in Cancer Cells Resembles That of Embryonic Stem Cells" Current Issues in Molecular Biology 46, no. 8: 8576-8599. https://doi.org/10.3390/cimb46080505
APA StyleSolovyeva, A. I., Afanasev, R. V., Popova, M. A., & Enukashvily, N. I. (2024). Dysregulation of Transposon Transcription Profiles in Cancer Cells Resembles That of Embryonic Stem Cells. Current Issues in Molecular Biology, 46(8), 8576-8599. https://doi.org/10.3390/cimb46080505







