The Role of Sirtuin-1 Isoforms in Regulating Mitochondrial Function
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and SIRT1 Plasmid Constructs
2.2. Transfections
2.3. Analysis of Mitochondrial Membrane Potential
2.4. Determination of Mitochondrial Respiration Rate, Glycolytic Rate, and ATP Production Rate
2.5. Assessing the Mitochondrial Electron Transport Chain Activity in Intact Cells Using Oroboros High-Resolution Respirometry
2.6. Quantitative Reverse-Transcriptase PCR
2.7. Immunoprecipitation and Western Blot Analysis
2.8. Statistical Analysis
3. Results
3.1. The SIRT1 Isoforms and Their Impact on Mitochondrial Membrane Potential
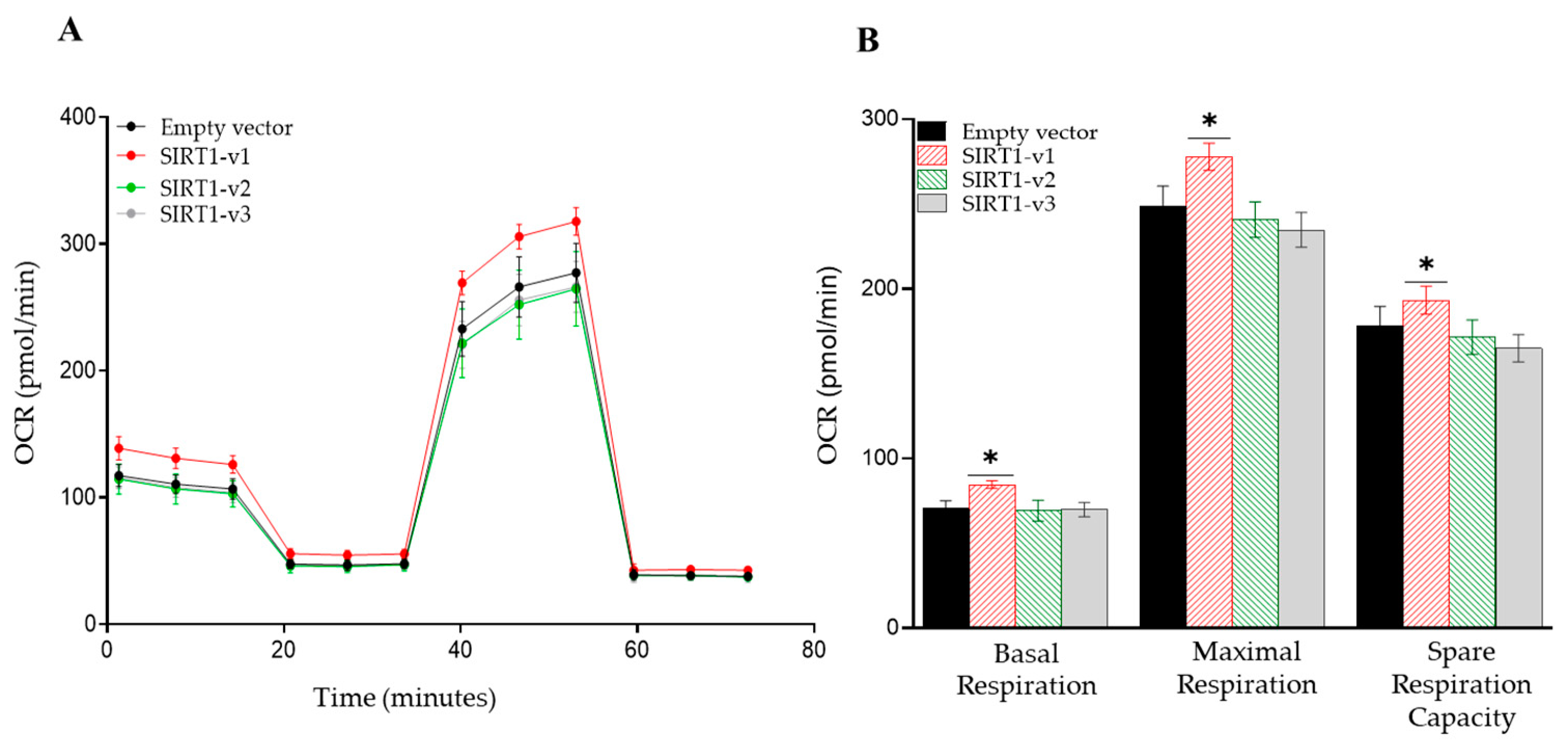
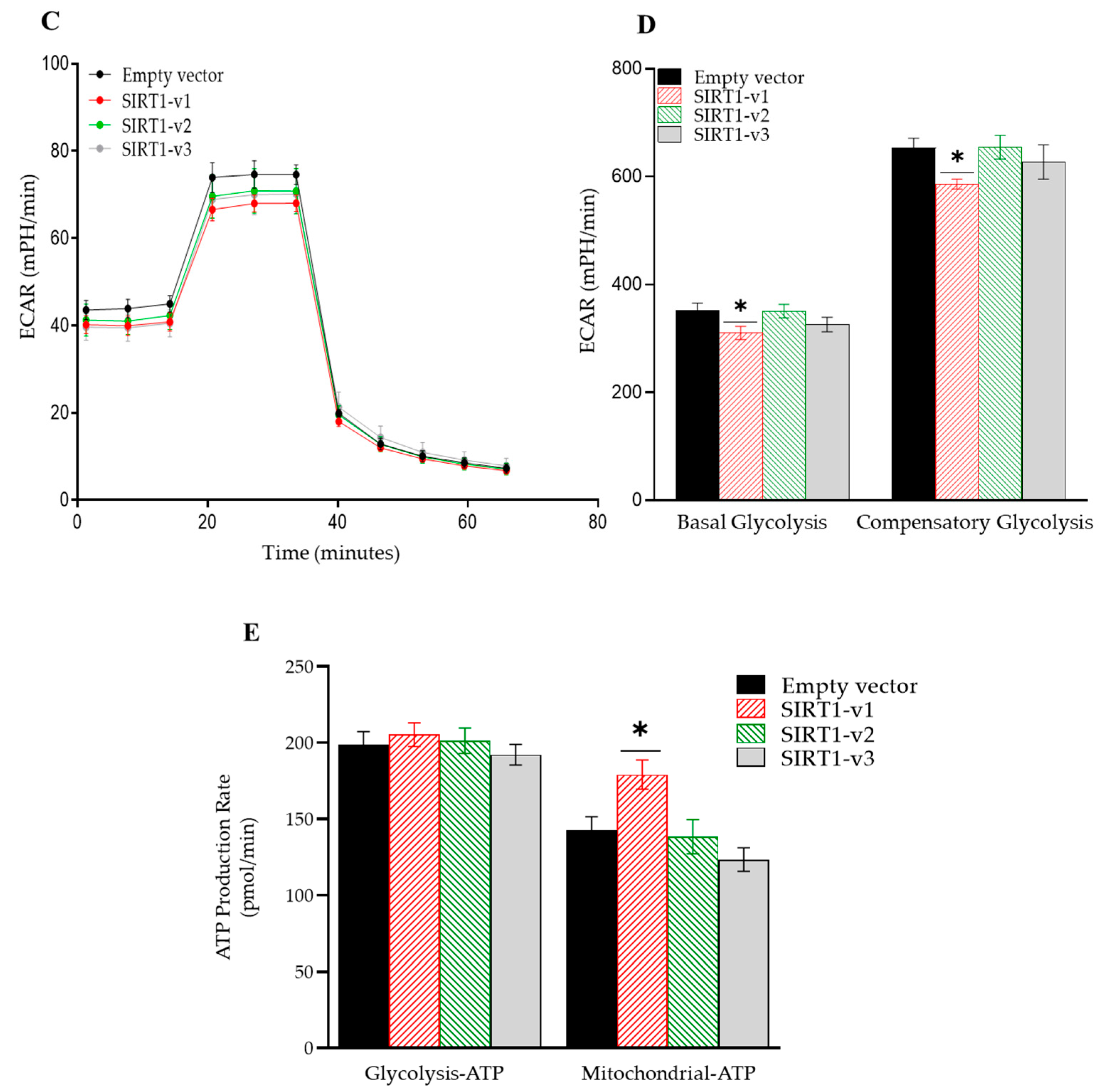
3.2. Loss of N-Terminal Region in SIRT1 Isoforms Affected the Regulation of Cellular Metabolism
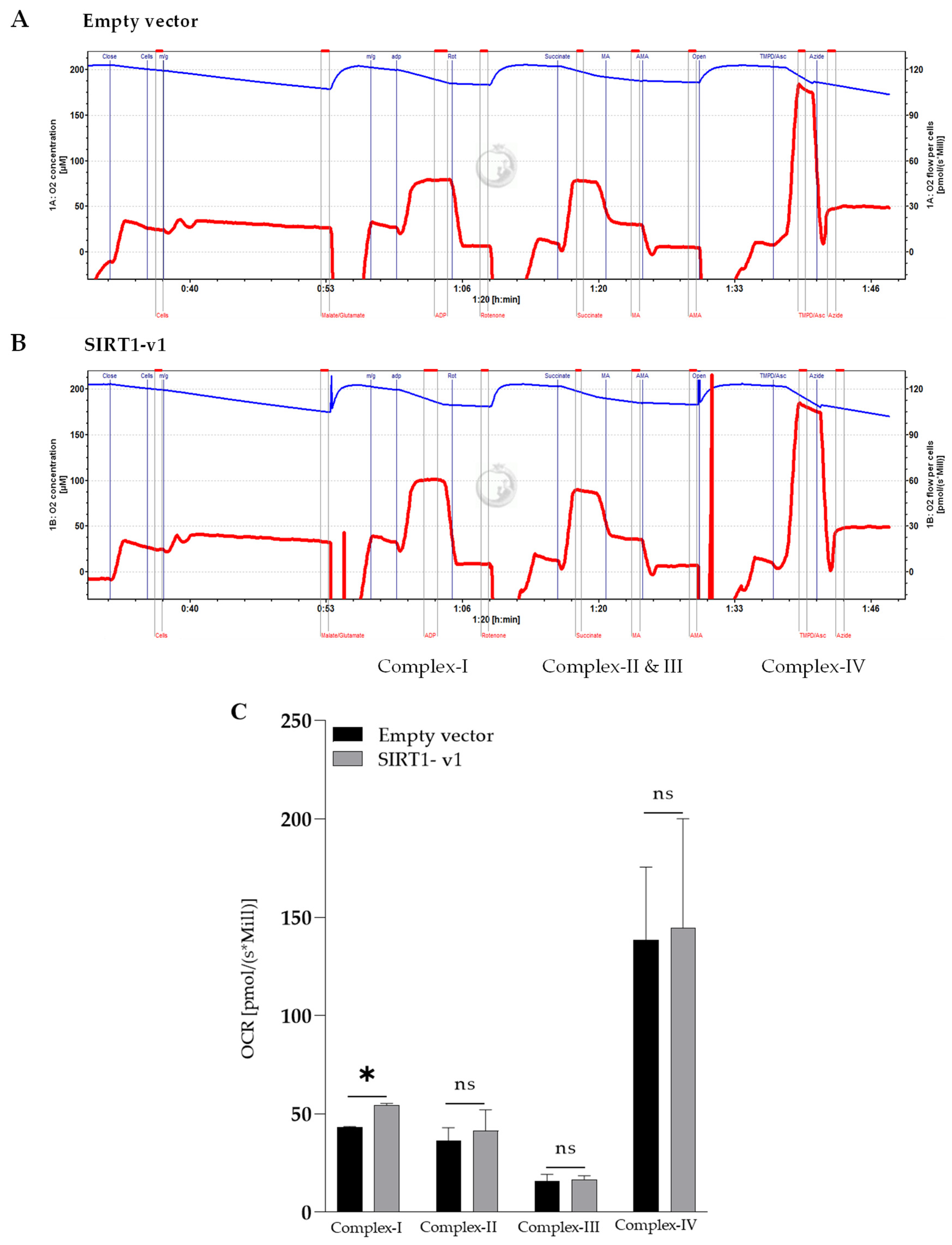
3.3. Loss of N-Terminal Region in SIRT1 Isoforms Affected the Regulation of Mitochondrial Respiratory Complex I
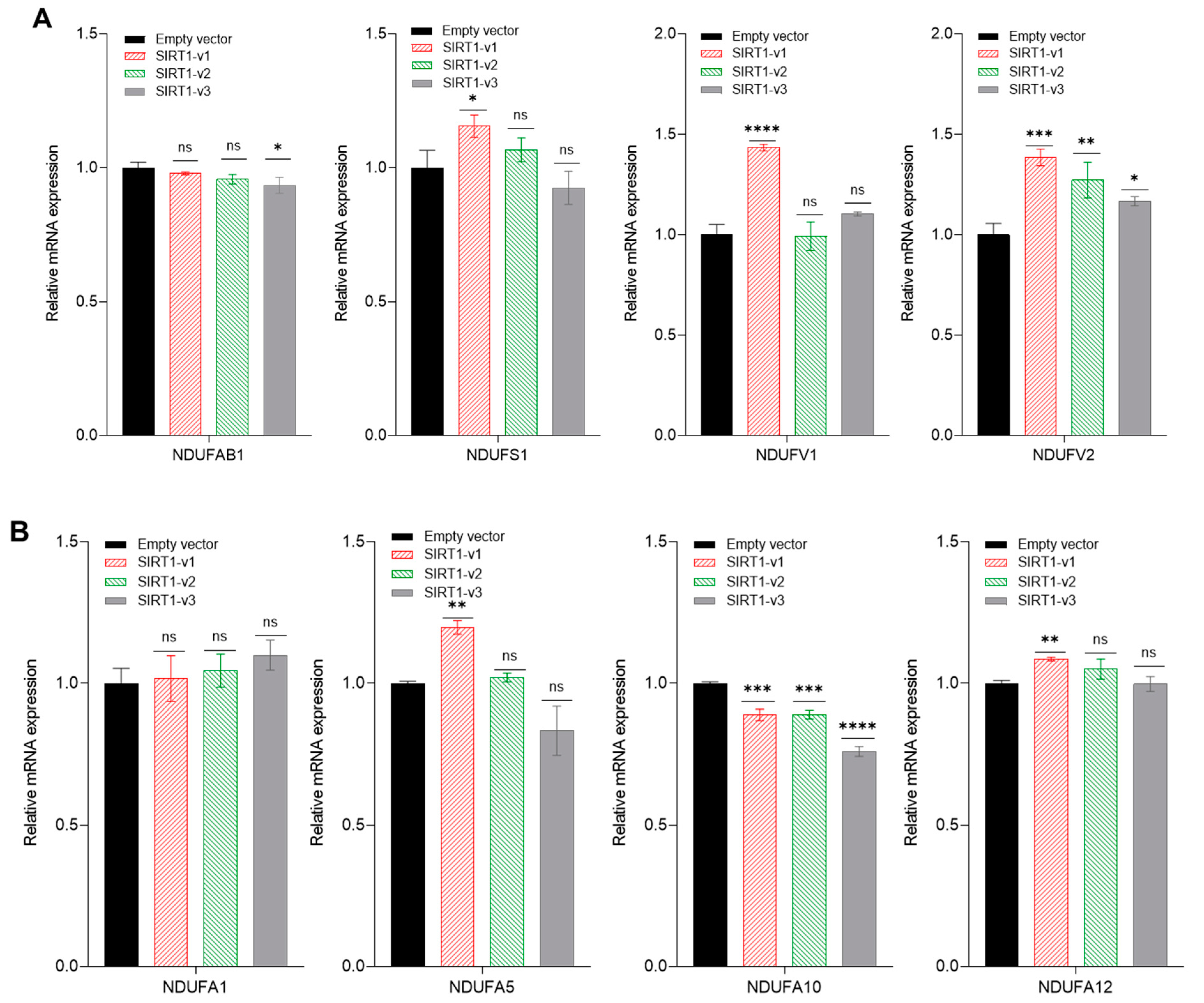
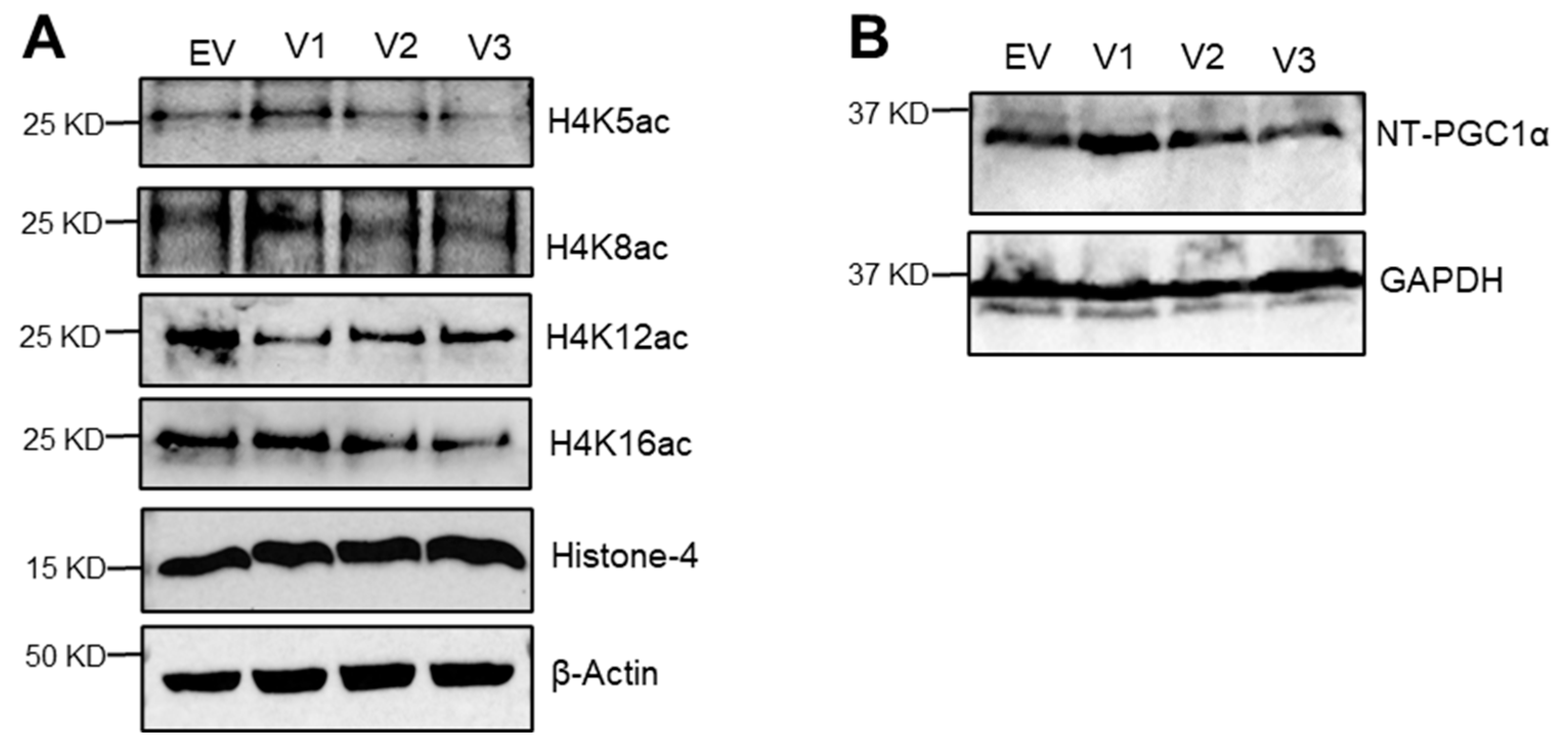
3.4. Loss of N-Terminal Region in SIRT1 Isoforms Affected the Regulation of Mitochondrial Genes and Histone H4 Acetylation
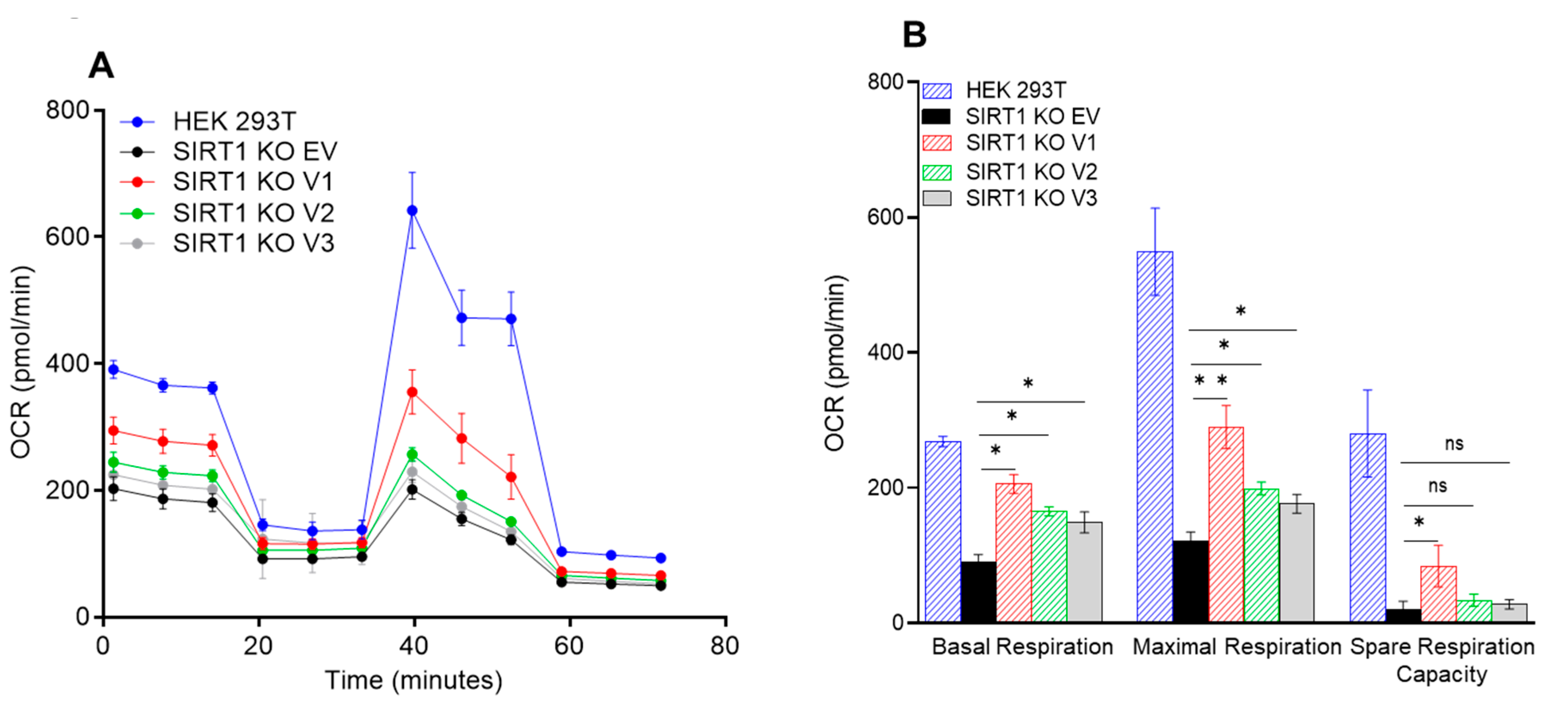
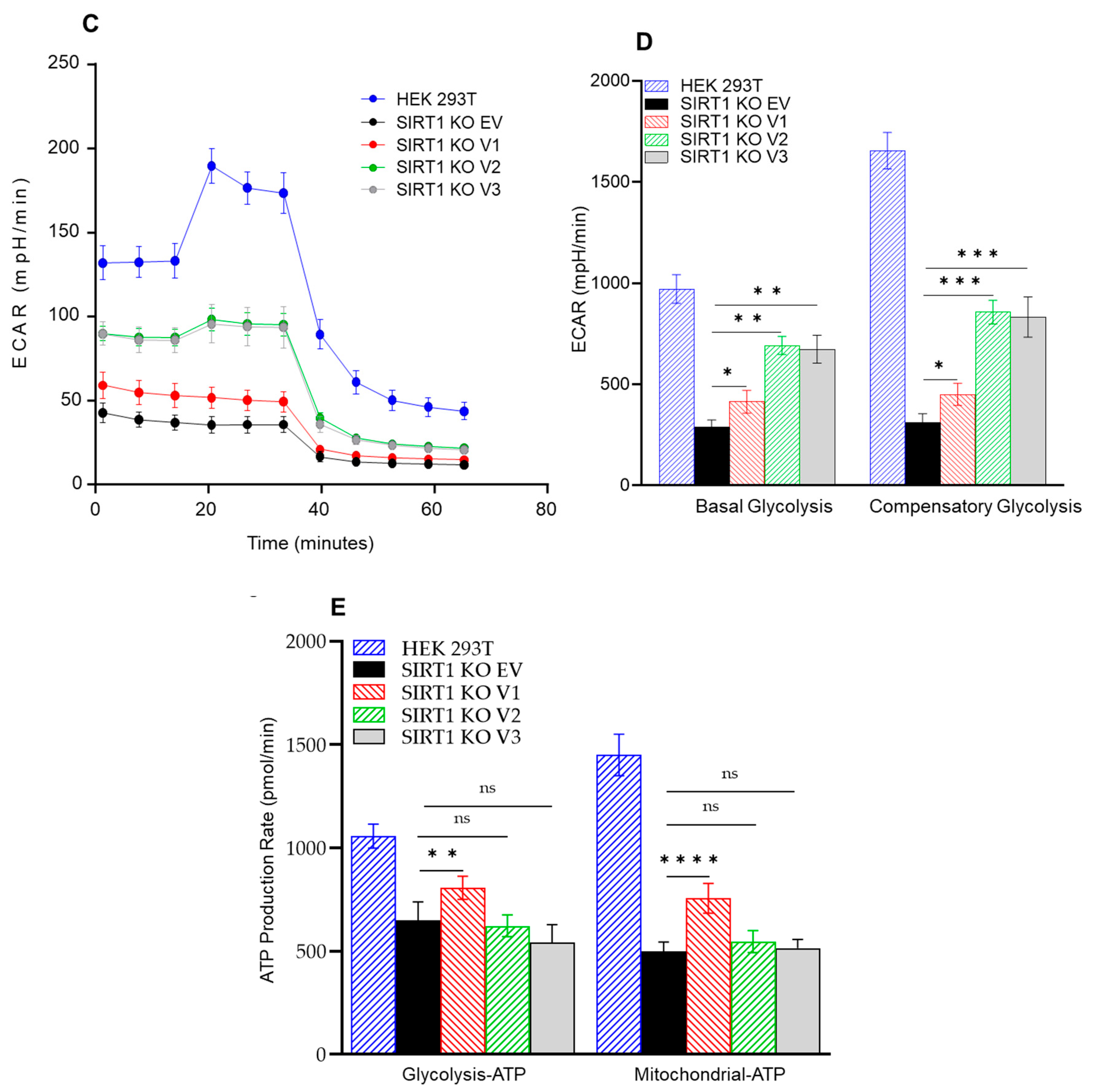
3.5. The Impact of SIRT1 Isoforms on Cellular Respiration in the SIRT1-Knockout Cells
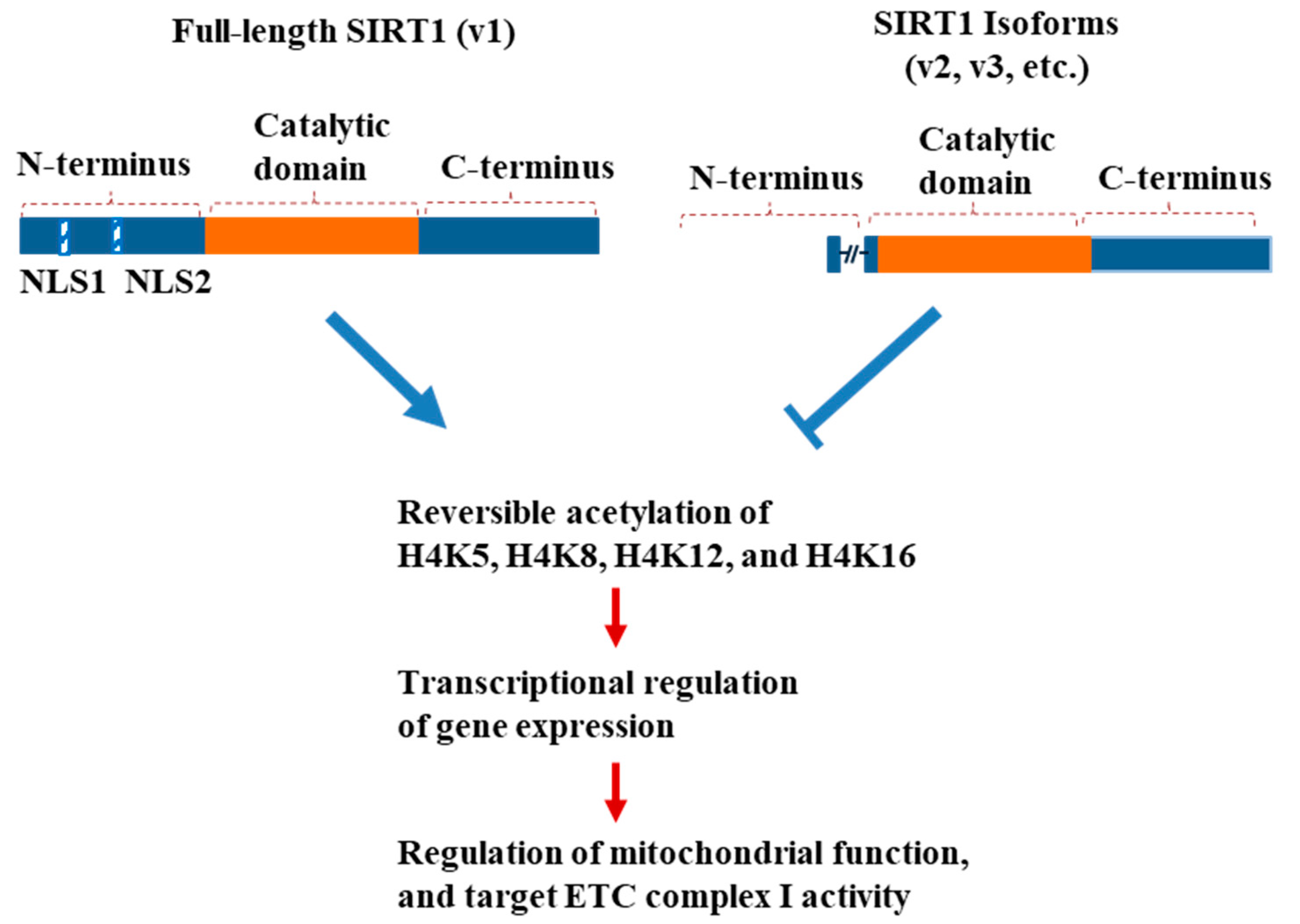
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marasco, L.E.; Kornblihtt, A.R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 2023, 24, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.J.; Smith, C.W.J.; Jiggins, C.D. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. 2022, 23, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Elowitz, M.B. Constitutive splicing and economies of scale in gene expression. Nat. Struct. Mol. Biol. 2019, 26, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Rogalska, M.E.; Vivori, C.; Valcarcel, J. Regulation of pre-mRNA splicing: Roles in physiology and disease, and therapeutic prospects. Nat. Rev. Genet. 2023, 24, 251–269. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015, 84, 165–198. [Google Scholar] [CrossRef]
- Tumasian, R.A., 3rd; Harish, A.; Kundu, G.; Yang, J.H.; Ubaida-Mohien, C.; Gonzalez-Freire, M.; Kaileh, M.; Zukley, L.M.; Chia, C.W.; Lyashkov, A.; et al. Skeletal muscle transcriptome in healthy aging. Nat. Commun. 2021, 12, 2014. [Google Scholar] [CrossRef]
- Debes, C.; Papadakis, A.; Gronke, S.; Karalay, O.; Tain, L.S.; Mizi, A.; Nakamura, S.; Hahn, O.; Weigelt, C.; Josipovic, N.; et al. Ageing-associated changes in transcriptional elongation influence longevity. Nature 2023, 616, 814–821. [Google Scholar] [CrossRef]
- Grootaert, M.O.J.; Bennett, M.R. Sirtuins in atherosclerosis: Guardians of healthspan and therapeutic targets. Nat. Rev. Cardiol. 2022, 19, 668–683. [Google Scholar] [CrossRef]
- Chang, N.; Li, J.; Lin, S.; Zhang, J.; Zeng, W.; Ma, G.; Wang, Y. Emerging roles of SIRT1 activator, SRT2104, in disease treatment. Sci. Rep. 2024, 14, 5521. [Google Scholar] [CrossRef]
- Higgins, C.B.; Mayer, A.L.; Zhang, Y.; Franczyk, M.; Ballentine, S.; Yoshino, J.; DeBosch, B.J. SIRT1 selectively exerts the metabolic protective effects of hepatocyte nicotinamide phosphoribosyltransferase. Nat. Commun. 2022, 13, 1074. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.C.; Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014, 25, 138–145. [Google Scholar] [CrossRef]
- Sies, H.; Mailloux, R.J.; Jakob, U. Fundamentals of redox regulation in biology. Nat. Rev. Mol. Cell Biol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.J.; Shah, Z.H.; Allison, S.J.; Ahmed, S.U.; Ford, J.; Warnock, L.J.; Li, H.; Serrano, M.; Milner, J. SIRT1 undergoes alternative splicing in a novel auto-regulatory loop with p53. PLoS ONE 2010, 5, e13502. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative splicing and related RNA binding proteins in human health and disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.Y.; Al-Salami, H.; Dass, C.R. C2C12 cell model: Its role in understanding of insulin resistance at the molecular level and pharmaceutical development at the preclinical stage. J. Pharm. Pharmacol. 2020, 72, 1667–1693. [Google Scholar] [CrossRef] [PubMed]
- Ren, N.S.X.; Ji, M.; Tokar, E.J.; Busch, E.L.; Xu, X.; Lewis, D.; Li, X.; Jin, A.; Zhang, Y.; Wu, W.K.K.; et al. Haploinsufficiency of SIRT1 Enhances Glutamine Metabolism and Promotes Cancer Development. Curr. Biol. 2017, 27, 483–494. [Google Scholar] [CrossRef]
- Patyal, P.; Nguyen, B.; Zhang, X.; Azhar, G.; Ameer, F.S.; Verma, A.; Crane, J.; Kc, G.; Che, Y.; Wei, J.Y. Rho/SRF Inhibitor Modulates Mitochondrial Functions. Int. J. Mol. Sci. 2022, 23, 11536. [Google Scholar] [CrossRef]
- Zhang, X.; Ameer, F.S.; Azhar, G.; Wei, J.Y. Alternative Splicing Increases Sirtuin Gene Family Diversity and Modulates Their Subcellular Localization and Function. Int. J. Mol. Sci. 2021, 22, 473. [Google Scholar] [CrossRef]
- Zhang, X.; Williams, E.D.; Azhar, G.; Rogers, S.C.; Wei, J.Y. Does p49/STRAP, a SRF-binding protein (SRFBP1), modulate cardiac mitochondrial function in aging? Exp. Gerontol. 2016, 82, 150–159. [Google Scholar] [CrossRef]
- Clutton, G.; Mollan, K.; Hudgens, M.; Goonetilleke, N. A Reproducible, Objective Method Using MitoTracker Fluorescent Dyes to Assess Mitochondrial Mass in T Cells by Flow Cytometry. Cytom. Part A 2019, 95, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Peng, Y.; Fan, W.; Han, S.; Peng, Q.; Xue, C.; Qin, X.; Liu, Y.; Ding, Z. Obesity-induced oxidative stress and mitochondrial dysfunction negatively affect sperm quality. FEBS Open Bio 2023, 13, 763–778. [Google Scholar] [CrossRef]
- Desai, S.; Grefte, S.; van de Westerlo, E.; Lauwen, S.; Paters, A.; Prehn, J.H.; Gan, Z.; Keijer, J.; Adjobo-Hermans, M.J.; Koopman, W.J. Performance of TMRM and Mitotrackers in mitochondrial morphofunctional analysis of primary human skin fibroblasts. Biochim. Biophys. Acta (BBA) Bioenerg. 2024, 1865, 149027. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Azhar, G.; Zhang, X.; Patyal, P.; Kc, G.; Sharma, S.; Che, Y.; Wei, J.Y. P. gingivalis-LPS Induces Mitochondrial Dysfunction Mediated by Neuroinflammation through Oxidative Stress. Int. J. Mol. Sci. 2023, 24, 950. [Google Scholar] [CrossRef]
- Zhang, X.; Azhar, G.; Wei, J.Y. The expression of microRNA and microRNA clusters in the aging heart. PLoS ONE 2012, 7, e34688. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Patyal, P.; Zhang, X.; Verma, A.; Azhar, G.; Wei, J.Y. Inhibitors of Rho/MRTF/SRF Transcription Pathway Regulate Mitochondrial Function. Cells 2024, 13, 392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Azhar, G.; Helms, S.; Zhong, Y.; Wei, J.Y. Identification of a subunit of NADH-dehydrogenase as a p49/STRAP-binding protein. BMC Cell Biol. 2008, 9, 8. [Google Scholar] [CrossRef]
- Filippi, M.D.; Ghaffari, S. Mitochondria in the maintenance of hematopoietic stem cells: New perspectives and opportunities. Blood 2019, 133, 1943–1952. [Google Scholar] [CrossRef] [PubMed]
- Letts, J.A.; Fiedorczuk, K.; Sazanov, L.A. The architecture of respiratory supercomplexes. Nature 2016, 537, 644–648. [Google Scholar] [CrossRef]
- Zhou, Y.; Zou, J.; Xu, J.; Cen, X.; Zhao, Y. Recent advances of mitochondrial complex I inhibitors for cancer therapy: Current status and future perspectives. Eur. J. Med. Chem. 2023, 251, 115219. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.S.; Fernand, V.; Zhang, Y.; Shin, J.; Jun, H.J.; Joshi, Y.; Gettys, T.W. NT-PGC-1alpha protein is sufficient to link beta3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis. J. Biol. Chem. 2012, 287, 9100–9111. [Google Scholar] [CrossRef] [PubMed]
- Torres-Mendez, A.; Pop, S.; Bonnal, S.; Almudi, I.; Avola, A.; Roberts, R.J.V.; Paolantoni, C.; Alcaina-Caro, A.; Martin-Anduaga, A.; Haussmann, I.U.; et al. Parallel evolution of a splicing program controlling neuronal excitability in flies and mammals. Sci. Adv. 2022, 8, eabk0445. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M.; Pozo, F.; di Domenico, T.; Vazquez, J.; Tress, M.L. An analysis of tissue-specific alternative splicing at the protein level. PLoS Comput. Biol. 2020, 16, e1008287. [Google Scholar] [CrossRef]
- Shah, Z.H.; Ahmed, S.U.; Ford, J.R.; Allison, S.J.; Knight, J.R.; Milner, J. A deacetylase-deficient SIRT1 variant opposes full-length SIRT1 in regulating tumor suppressor p53 and governs expression of cancer-related genes. Mol. Cell. Biol. 2012, 32, 704–716. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanders, B.D.; Jackson, B.; Marmorstein, R. Structural basis for sirtuin function: What we know and what we don’t. Biochim. Biophys. Acta 2010, 1804, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Tanno, M.; Sakamoto, J.; Miura, T.; Shimamoto, K.; Horio, Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007, 282, 6823–6832. [Google Scholar] [CrossRef]
- Hallows, W.C.; Lee, S.; Denu, J.M. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 2006, 103, 10230–10235. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Dong, H.; Yong, S.; Li, X.; Olashaw, N.; Kruk, P.A.; Cheng, J.Q.; Bai, W.; Chen, J.; et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene 2009, 28, 445–460. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar] [CrossRef]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Li, P.; Ge, J.; Li, H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 2020, 17, 96–115. [Google Scholar] [CrossRef]
- Ye, J.; Ai, X.; Eugeni, E.E.; Zhang, L.; Carpenter, L.R.; Jelinek, M.A.; Freitas, M.A.; Parthun, M.R. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell 2005, 18, 123–130. [Google Scholar] [CrossRef]
- Dujardin, G.; Lafaille, C.; de la Mata, M.; Marasco, L.E.; Munoz, M.J.; Le Jossic-Corcos, C.; Corcos, L.; Kornblihtt, A.R. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 2014, 54, 683–690. [Google Scholar] [CrossRef]
- Rahhal, R.; Seto, E. Emerging roles of histone modifications and HDACs in RNA splicing. Nucleic Acids Res. 2019, 47, 4911–4926. [Google Scholar] [CrossRef]
- Chemello, F.; Chai, A.C.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Atmanli, A.; Mireault, A.A.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021, 7, eabg4910. [Google Scholar] [CrossRef]
- Nikom, D.; Zheng, S. Alternative splicing in neurodegenerative disease and the promise of RNA therapies. Nat. Rev. Neurosci. 2023, 24, 457–473. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patyal, P.; Ameer, F.S.; Verma, A.; Zhang, X.; Azhar, G.; Shrivastava, J.; Sharma, S.; Zhang, R.; Wei, J.Y. The Role of Sirtuin-1 Isoforms in Regulating Mitochondrial Function. Curr. Issues Mol. Biol. 2024, 46, 8835-8851. https://doi.org/10.3390/cimb46080522
Patyal P, Ameer FS, Verma A, Zhang X, Azhar G, Shrivastava J, Sharma S, Zhang R, Wei JY. The Role of Sirtuin-1 Isoforms in Regulating Mitochondrial Function. Current Issues in Molecular Biology. 2024; 46(8):8835-8851. https://doi.org/10.3390/cimb46080522
Chicago/Turabian StylePatyal, Pankaj, Fathima S. Ameer, Ambika Verma, Xiaomin Zhang, Gohar Azhar, Jyotsna Shrivastava, Shakshi Sharma, Rachel Zhang, and Jeanne Y. Wei. 2024. "The Role of Sirtuin-1 Isoforms in Regulating Mitochondrial Function" Current Issues in Molecular Biology 46, no. 8: 8835-8851. https://doi.org/10.3390/cimb46080522
APA StylePatyal, P., Ameer, F. S., Verma, A., Zhang, X., Azhar, G., Shrivastava, J., Sharma, S., Zhang, R., & Wei, J. Y. (2024). The Role of Sirtuin-1 Isoforms in Regulating Mitochondrial Function. Current Issues in Molecular Biology, 46(8), 8835-8851. https://doi.org/10.3390/cimb46080522





