NOD1 and NOD2: Essential Monitoring Partners in the Innate Immune System
Abstract
:1. Introduction
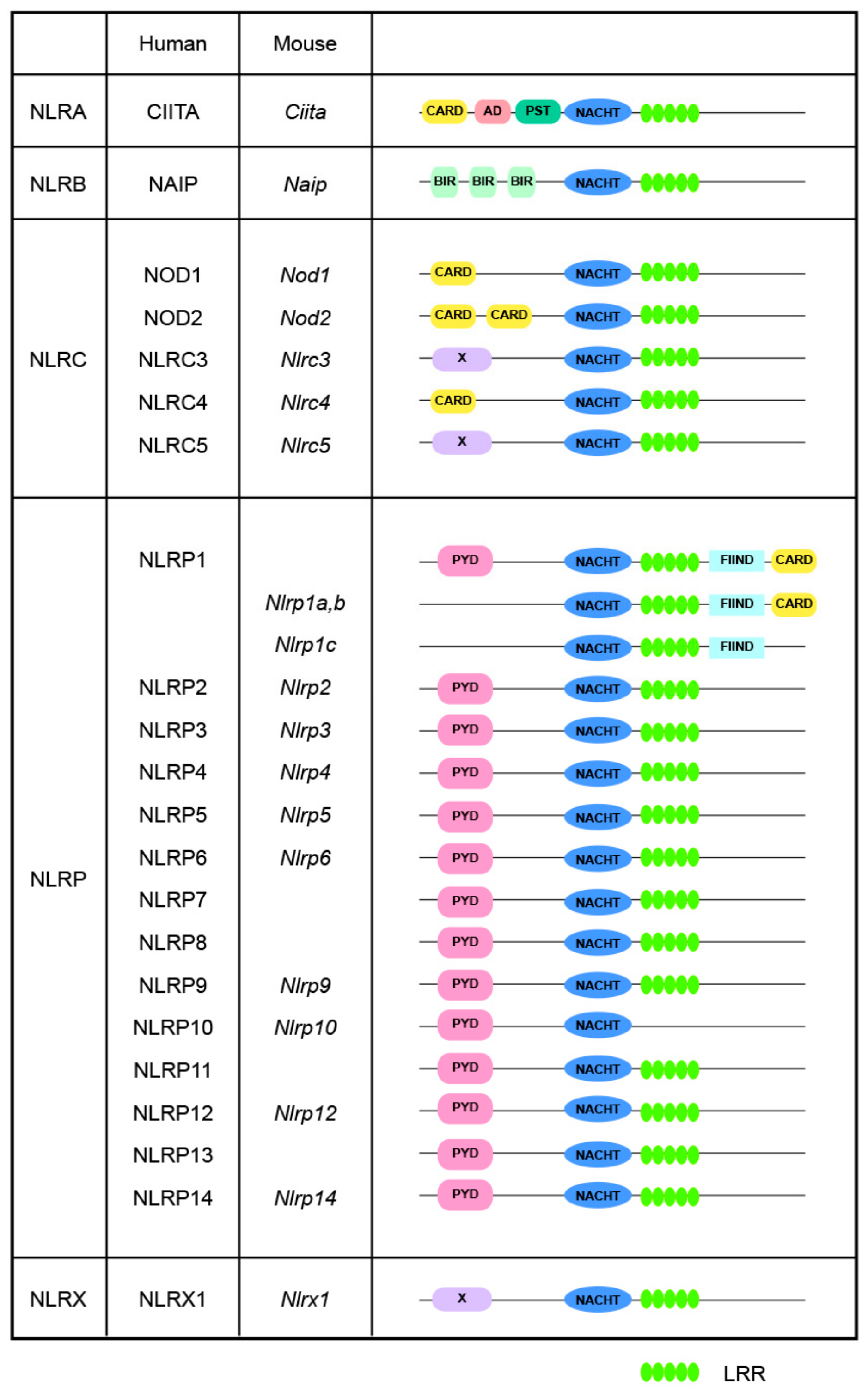
2. Basic Structure
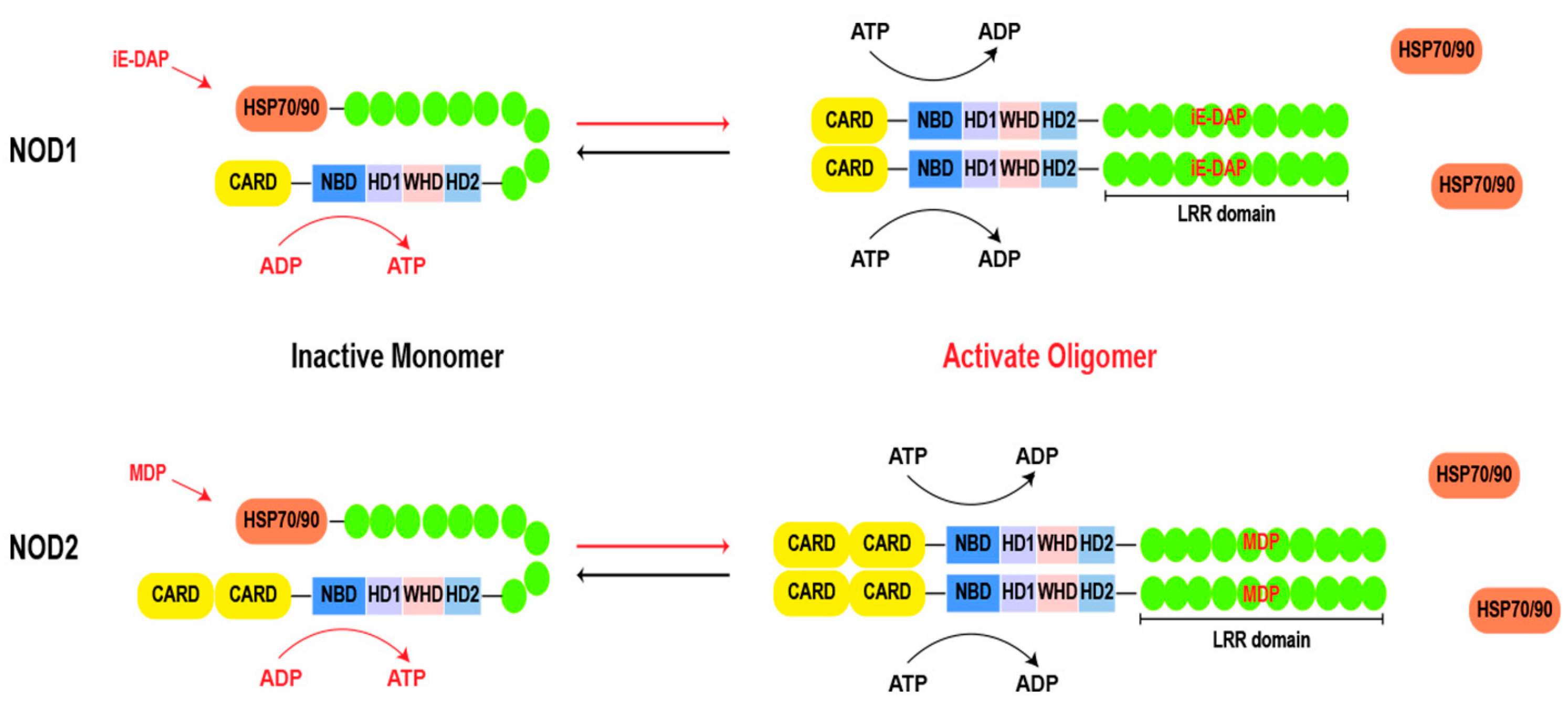
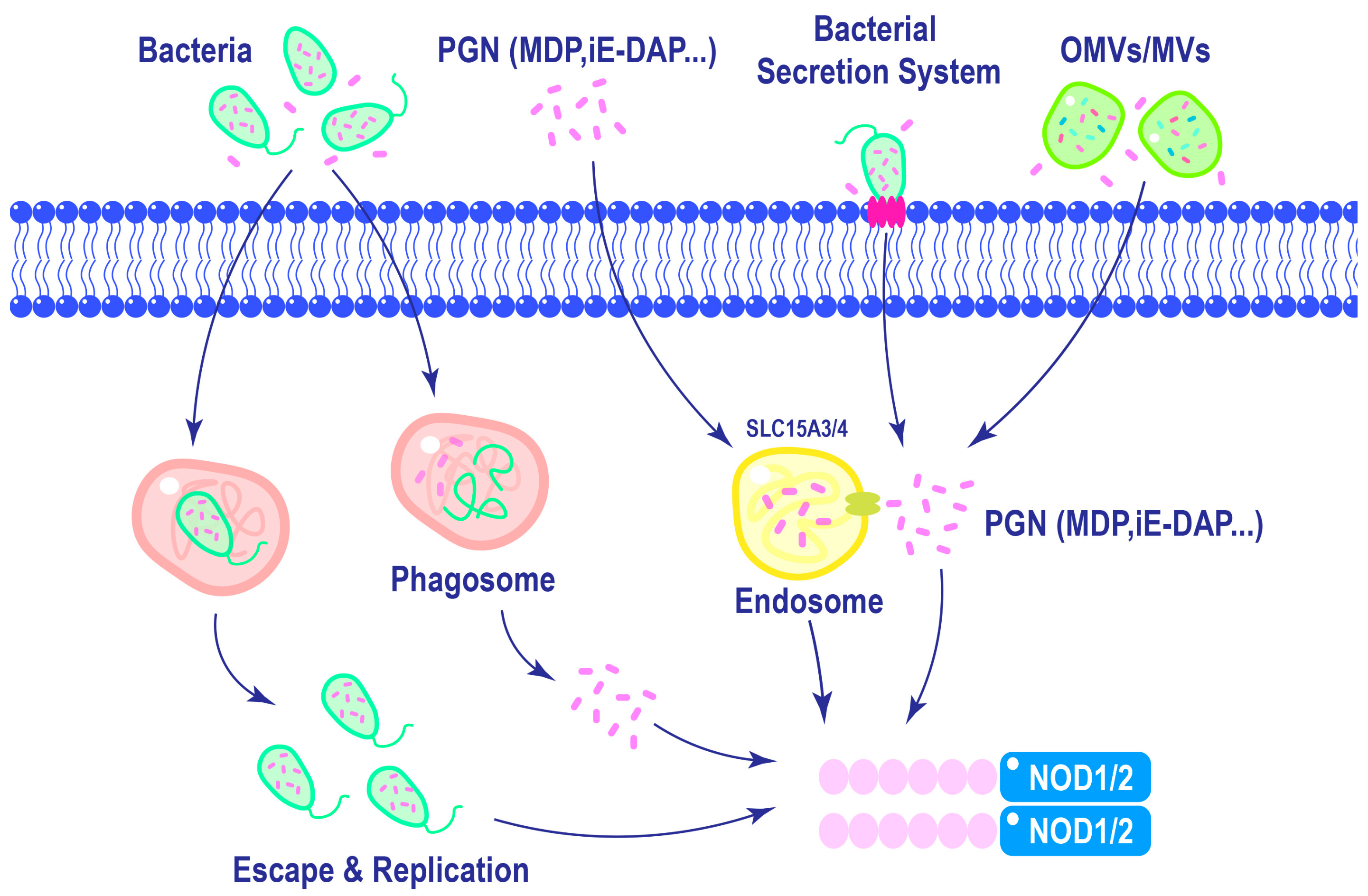
3. Activation of NOD1 and NOD2
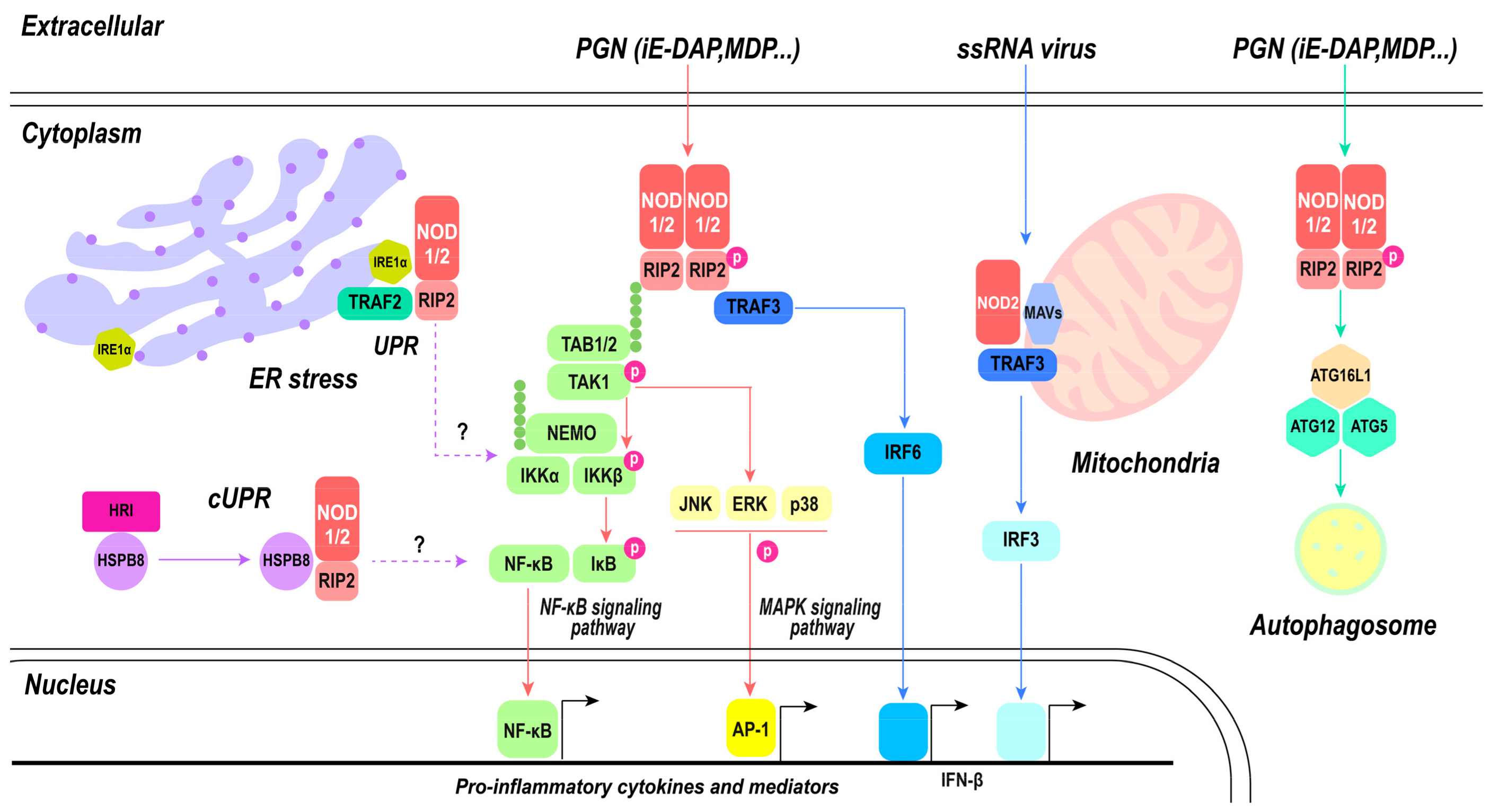
4. Signaling Pathways of NOD1 of NOD2
5. Position of NOD1and NOD2 in Innate Immunity
6. NOD1 and NOD2 in Cancer
7. NOD1 and NOD2 in Metabolic Diseases
8. NOD1 and NOD2 as Potential Therapeutic Targets
9. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Inohara, N.; Nunez, G. NODs: Intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Kibby, E.M.; Conte, A.N.; Burroughs, A.M.; Nagy, T.A.; Vargas, J.A.; Whalen, L.A.; Aravind, L.; Whiteley, A.T. Bacterial NLR-related proteins protect against phage. Cell 2023, 186, 2410–2424.e2418. [Google Scholar] [CrossRef] [PubMed]
- Philpott, D.J.; Sorbara, M.T.; Robertson, S.J.; Croitoru, K.; Girardin, S.E. NOD proteins: Regulators of inflammation in health and disease. Nat. Rev. Immunol. 2014, 14, 9–23. [Google Scholar] [CrossRef]
- Caruso, R.; Warner, N.; Inohara, N.; Nunez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef]
- Sundaram, B.; Tweedell, R.E.; Prasanth Kumar, S.; Kanneganti, T.D. The NLR family of innate immune and cell death sensors. Immunity 2024, 57, 674–699. [Google Scholar] [CrossRef]
- Howe, K.; Schiffer, P.H.; Zielinski, J.; Wiehe, T.; Laird, G.K.; Marioni, J.C.; Soylemez, O.; Kondrashov, F.; Leptin, M. Structure and evolutionary history of a large family of NLR proteins in the zebrafish. Open Biol. 2016, 6, 160009. [Google Scholar] [CrossRef]
- Chou, W.C.; Jha, S.; Linhoff, M.W.; Ting, J.P. The NLR gene family: From discovery to present day. Nat. Rev. Immunol. 2023, 23, 635–654. [Google Scholar] [CrossRef]
- Tattoli, I.; Carneiro, L.A.; Jehanno, M.; Magalhaes, J.G.; Shu, Y.; Philpott, D.J.; Arnoult, D.; Girardin, S.E. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008, 9, 293–300. [Google Scholar] [CrossRef]
- Motta, V.; Soares, F.; Sun, T.; Philpott, D.J. NOD-like receptors: Versatile cytosolic sentinels. Physiol. Rev. 2015, 95, 149–178. [Google Scholar] [CrossRef]
- Velloso, F.J.; Trombetta-Lima, M.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like receptors: Major players (and targets) in the interface between innate immunity and cancer. Biosci. Rep. 2019, 39, BSR20181709. [Google Scholar] [CrossRef] [PubMed]
- Keestra-Gounder, A.M.; Tsolis, R.M. NOD1 and NOD2: Beyond Peptidoglycan Sensing. Trends Immunol. 2017, 38, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, T.; Hovingh, E.S.; Foerster, E.G.; Abdel-Nour, M.; Philpott, D.J.; Girardin, S.E. NOD1 and NOD2 in inflammation, immunity and disease. Arch. Biochem. Biophys. 2019, 670, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Parkhouse, R.; Boyle, J.P.; Mayle, S.; Sawmynaden, K.; Rittinger, K.; Monie, T.P. Interaction between NOD2 and CARD9 involves the NOD2 NACHT and the linker region between the NOD2 CARDs and NACHT domain. FEBS Lett. 2014, 588, 2830–2836. [Google Scholar] [CrossRef]
- Zurek, B.; Proell, M.; Wagner, R.N.; Schwarzenbacher, R.; Kufer, T.A. Mutational analysis of human NOD1 and NOD2 NACHT domains reveals different modes of activation. Innate Immun. 2012, 18, 100–111. [Google Scholar] [CrossRef]
- Alipoor, S.D.; Mirsaeidi, M. Inborn Errors in the LRR Domain of Nod2 and Their Potential Consequences on the Function of the Receptor. Cells 2021, 10, 31. [Google Scholar] [CrossRef]
- Hugot, J.P. CARD15/NOD2 mutations in Crohn’s disease. Ann. N. Y. Acad. Sci. 2006, 1072, 9–18. [Google Scholar] [CrossRef]
- Hahn, J.S. Regulation of Nod1 by Hsp90 chaperone complex. FEBS Lett. 2005, 579, 4513–4519. [Google Scholar] [CrossRef]
- Lee, K.H.; Biswas, A.; Liu, Y.J.; Kobayashi, K.S. Proteasomal degradation of Nod2 protein mediates tolerance to bacterial cell wall components. J. Biol. Chem. 2012, 287, 39800–39811. [Google Scholar] [CrossRef]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef]
- Marino, N.D.; Brodsky, I.E. Immunology: NACHT domain proteins get a prokaryotic origin story. Curr. Biol. 2023, 33, R875–R878. [Google Scholar] [CrossRef]
- Wu, X.; Yang, Z.H.; Wu, J.; Han, J. Ribosome-rescuer PELO catalyzes the oligomeric assembly of NOD-like receptor family proteins via activating their ATPase enzymatic activity. Immunity 2023, 56, 926–943.e927. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.I.; Whiteheart, S.W. AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 2005, 6, 519–529. [Google Scholar] [CrossRef]
- Maharana, J.; Sahoo, B.R.; Bej, A.; Jena, I.; Parida, A.; Sahoo, J.R.; Dehury, B.; Patra, M.C.; Martha, S.R.; Balabantray, S.; et al. Structural models of zebrafish (Danio rerio) NOD1 and NOD2 NACHT domains suggest differential ATP binding orientations: Insights from computational modeling, docking and molecular dynamics simulations. PLoS ONE 2015, 10, e0121415. [Google Scholar] [CrossRef]
- Parkhouse, R.; Boyle, J.P.; Monie, T.P. Blau syndrome polymorphisms in NOD2 identify nucleotide hydrolysis and helical domain 1 as signalling regulators. FEBS Lett. 2014, 588, 3382–3389. [Google Scholar] [CrossRef]
- Mo, J.; Boyle, J.P.; Howard, C.B.; Monie, T.P.; Davis, B.K.; Duncan, J.A. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 2012, 287, 23057–23067. [Google Scholar] [CrossRef]
- Maekawa, S.; Ohto, U.; Shibata, T.; Miyake, K.; Shimizu, T. Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 2016, 7, 11813. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, T.; Chamaillard, M.; Ogura, Y.; Zhu, L.; Qiu, S.; Masumoto, J.; Ghosh, P.; Moran, A.; Predergast, M.M.; Tromp, G.; et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004, 23, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Vijayrajratnam, S.; Pushkaran, A.C.; Balakrishnan, A.; Vasudevan, A.K.; Biswas, R.; Mohan, C.G. Understanding the molecular differential recognition of muramyl peptide ligands by LRR domains of human NOD receptors. Biochem. J. 2017, 474, 2691–2711. [Google Scholar] [CrossRef]
- Meunier, E.; Broz, P. Evolutionary Convergence and Divergence in NLR Function and Structure. Trends Immunol. 2017, 38, 744–757. [Google Scholar] [CrossRef]
- Laing, K.J.; Purcell, M.K.; Winton, J.R.; Hansen, J.D. A genomic view of the NOD-like receptor family in teleost fish: Identification of a novel NLR subfamily in zebrafish. BMC Evol. Biol. 2008, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Chuphal, B.; Rai, U.; Roy, B. Teleost NOD-like receptors and their downstream signaling pathways: A brief review. Fish. Shellfish. Immunol. Rep. 2022, 3, 100056. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, S.; Till, A.; Sina, C.; Arlt, A.; Grasberger, H.; Schreiber, S.; Rosenstiel, P. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J. Cell Sci. 2009, 122 Pt 19, 3522–3530. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.P.; Mayle, S.; Parkhouse, R.; Monie, T.P. Comparative Genomic and Sequence Analysis Provides Insight into the Molecular Functionality of NOD1 and NOD2. Front. Immunol. 2013, 4, 317. [Google Scholar] [CrossRef]
- Sabbah, A.; Chang, T.H.; Harnack, R.; Frohlich, V.; Tominaga, K.; Dube, P.H.; Xiang, Y.; Bose, S. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 2009, 10, 1073–1080. [Google Scholar] [CrossRef]
- Trindade, B.C.; Chen, G.Y. NOD1 and NOD2 in inflammatory and infectious diseases. Immunol. Rev. 2020, 297, 139–161. [Google Scholar] [CrossRef]
- Dixon, C.L.; Wu, A.; Fairn, G.D. Multifaceted roles and regulation of nucleotide-binding oligomerization domain containing proteins. Front. Immunol. 2023, 14, 1242659. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, Q.; Cao, P.; Shi, C.; Zhang, L.; Wang, L.; Gong, Z. NOD2-mediated HDAC6/NF-kappab signalling pathway regulates ferroptosis induced by extracellular histone H3 in acute liver failure. J. Cell Mol. Med. 2022, 26, 5528–5538. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, R.; Kong, Y.; Feng, Z.; Xu, Q. METTL3 regulates LPS-induced inflammatory response via the NOD1 signaling pathway. Cell Signal 2022, 93, 110283. [Google Scholar] [CrossRef]
- Nakamura, N.; Lill, J.R.; Phung, Q.; Jiang, Z.; Bakalarski, C.; de Maziere, A.; Klumperman, J.; Schlatter, M.; Delamarre, L.; Mellman, I. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 2014, 509, 240–244. [Google Scholar] [CrossRef]
- Kufer, T.A.; Kremmer, E.; Adam, A.C.; Philpott, D.J.; Sansonetti, P.J. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2008, 10, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Woodley, K.T.; Collins, M.O. Regulation and function of the palmitoyl-acyltransferase ZDHHC5. FEBS J. 2021, 288, 6623–6634. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zheng, Y.; Coyaud, É.; Zhang, C.; Selvabaskaran, A.; Yu, Y.; Xu, Z.; Weng, X.; Chen, J.S.; Meng, Y.; et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science 2019, 366, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Stevens, C.; Henderson, P.; Nimmo, E.R.; Soares, D.C.; Dogan, B.; Simpson, K.W.; Barrett, J.C.; Wilson, D.C.; Satsangi, J.; International Inflammatory Bowel Disease Genetics Consortium. The intermediate filament protein, vimentin, is a regulator of NOD2 activity. Gut 2013, 62, 695–707. [Google Scholar] [CrossRef]
- Haglund, C.M.; Welch, M.D. Pathogens and polymers: Microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 2011, 195, 7–17. [Google Scholar] [CrossRef]
- Dorflinger, B.; Badr, M.T.; Haimovici, A.; Fischer, L.; Vier, J.; Metz, A.; Eisele, B.; Bronsert, P.; Aumann, K.; Hoppner, J.; et al. Mitochondria supply sub-lethal signals for cytokine secretion and DNA-damage in H. pylori infection. Cell Death Differ. 2022, 29, 2218–2232. [Google Scholar] [CrossRef]
- Keestra, A.M.; Winter, M.G.; Auburger, J.J.; Frassle, S.P.; Xavier, M.N.; Winter, S.E.; Kim, A.; Poon, V.; Ravesloot, M.M.; Waldenmaier, J.F.; et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature 2013, 496, 233–237. [Google Scholar] [CrossRef]
- Friebel, A.; Ilchmann, H.; Aepfelbacher, M.; Ehrbar, K.; Machleidt, W.; Hardt, W.D. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J. Biol. Chem. 2001, 276, 34035–34040. [Google Scholar] [CrossRef]
- Legrand-Poels, S.; Kustermans, G.; Bex, F.; Kremmer, E.; Kufer, T.A.; Piette, J. Modulation of Nod2-dependent NF-kappaB signaling by the actin cytoskeleton. J. Cell Sci. 2007, 120 Pt 7, 1299–1310. [Google Scholar] [CrossRef]
- Eitel, J.; Krull, M.; Hocke, A.C.; N’Guessan, P.D.; Zahlten, J.; Schmeck, B.; Slevogt, H.; Hippenstiel, S.; Suttorp, N.; Opitz, B. Beta-PIX and Rac1 GTPase mediate trafficking and negative regulation of NOD2. J. Immunol. 2008, 181, 2664–2671. [Google Scholar] [CrossRef]
- De Marzi, M.C.; Todone, M.; Ganem, M.B.; Wang, Q.; Mariuzza, R.A.; Fernández, M.M.; Malchiodi, E.L. Peptidoglycan recognition protein–peptidoglycan complexes increase monocyte/macrophage activation and enhance the inflammatory response. Immunology 2015, 145, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Park, B.G.; Wolf, A.J.; Brikos, C.; Goodridge, H.S.; Becker, C.A.; Reyes, C.N.; Miao, E.A.; Aderem, A.; Gotz, F.; et al. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe 2010, 7, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Bitto, N.J.; Cheng, L.; Johnston, E.L.; Pathirana, R.; Phan, T.K.; Poon, I.K.H.; O’Brien-Simpson, N.M.; Hill, A.F.; Stinear, T.P.; Kaparakis-Liaskos, M. Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. J. Extracell. Vesicles 2021, 10, e12080. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, Y.G.; McDonald, C.; Kanneganti, T.D.; Hasegawa, M.; Body-Malapel, M.; Inohara, N.; Nunez, G. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 2007, 178, 2380–2386. [Google Scholar] [CrossRef]
- Pellegrini, E.; Desfosses, A.; Wallmann, A.; Schulze, W.M.; Rehbein, K.; Mas, P.; Signor, L.; Gaudon, S.; Zenkeviciute, G.; Hons, M.; et al. RIP2 filament formation is required for NOD2 dependent NF-kappaB signalling. Nat. Commun. 2018, 9, 4043. [Google Scholar] [CrossRef]
- Rivoal, M.; Dubuquoy, L.; Millet, R.; Leleu-Chavain, N. Receptor Interacting Ser/Thr-Protein Kinase 2 as a New Therapeutic Target. J. Med. Chem. 2023, 66, 14391–14410. [Google Scholar] [CrossRef]
- Gong, Q.; Long, Z.; Zhong, F.L.; Teo, D.E.T.; Jin, Y.; Yin, Z.; Boo, Z.Z.; Zhang, Y.; Zhang, J.; Yang, R.; et al. Structural basis of RIP2 activation and signaling. Nat. Commun. 2018, 9, 4993. [Google Scholar] [CrossRef]
- Goncharov, T.; Hedayati, S.; Mulvihill, M.M.; Izrael-Tomasevic, A.; Zobel, K.; Jeet, S.; Fedorova, A.V.; Eidenschenk, C.; deVoss, J.; Yu, K.; et al. Disruption of XIAP-RIP2 Association Blocks NOD2-Mediated Inflammatory Signaling. Mol. Cell 2018, 69, 551–565.e557. [Google Scholar] [CrossRef]
- Strober, W.; Murray, P.J.; Kitani, A.; Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006, 6, 9–20. [Google Scholar] [CrossRef]
- Travassos, L.H.; Carneiro, L.A.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhaes, J.G.; Yuan, L.; Soares, F.; Chea, E.; Le Bourhis, L.; et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010, 11, 55–62. [Google Scholar] [CrossRef]
- Cooney, R.; Baker, J.; Brain, O.; Danis, B.; Pichulik, T.; Allan, P.; Ferguson, D.J.; Campbell, B.J.; Jewell, D.; Simmons, A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010, 16, 90–97. [Google Scholar] [CrossRef]
- Anand, P.K.; Tait, S.W.; Lamkanfi, M.; Amer, A.O.; Nunez, G.; Pages, G.; Pouyssegur, J.; McGargill, M.A.; Green, D.R.; Kanneganti, T.D. TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J. Biol. Chem. 2011, 286, 42981–42991. [Google Scholar] [CrossRef] [PubMed]
- Homer, C.R.; Richmond, A.L.; Rebert, N.A.; Achkar, J.P.; McDonald, C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology 2010, 139, 1630–1641.e2. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Ellison, L.K.; Ramjeet, M.; Travassos, L.H.; Jones, N.L.; Girardin, S.E.; Philpott, D.J. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 2013, 39, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Chipurupalli, S.; Samavedam, U.; Robinson, N. Crosstalk Between ER Stress, Autophagy and Inflammation. Front. Med. 2021, 8, 758311. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef] [PubMed]
- Celli, J.; Tsolis, R.M. Bacteria, the endoplasmic reticulum and the unfolded protein response: Friends or foes? Nat. Rev. Microbiol. 2015, 13, 71–82. [Google Scholar] [CrossRef]
- Keestra-Gounder, A.M.; Byndloss, M.X.; Seyffert, N.; Young, B.M.; Chavez-Arroyo, A.; Tsai, A.Y.; Cevallos, S.A.; Winter, M.G.; Pham, O.H.; Tiffany, C.R.; et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature 2016, 532, 394–397. [Google Scholar] [CrossRef]
- Baeyens, A.; Bracero, S.; Chaluvadi, V.S.; Khodadadi-Jamayran, A.; Cammer, M.; Schwab, S.R. Monocyte-derived S1P in the lymph node regulates immune responses. Nature 2021, 592, 290–295. [Google Scholar] [CrossRef]
- Lu, Y.; Neculai, D. Sphingosine-1-phosphate: The missing link between NOD1/2 and ER stress. EMBO J. 2021, 40, e108812. [Google Scholar] [CrossRef]
- Tsai, H.C.; Han, M.H. Sphingosine-1-Phosphate (S1P) and S1P Signaling Pathway: Therapeutic Targets in Autoimmunity and Inflammation. Drugs 2016, 76, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Godkowicz, M.; Druszczynska, M. NOD1, NOD2, and NLRC5 Receptors in Antiviral and Antimycobacterial Immunity. Vaccines 2022, 10, 1487. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef]
- Limonta, D.; Dyna-Dagman, L.; Branton, W.; Mancinelli, V.; Makio, T.; Wozniak, R.W.; Power, C.; Hobman, T.C. Nodosome Inhibition as a Novel Broad-Spectrum Antiviral Strategy against Arboviruses, Enteroviruses, and SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65, e0049121. [Google Scholar] [CrossRef]
- Kim, J.; Yang, Y.L.; Jang, Y.S. Human beta-defensin 2 is involved in CCR2-mediated Nod2 signal transduction, leading to activation of the innate immune response in macrophages. Immunobiology 2019, 224, 502–510. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Z.; Xue, Q.; Yang, F.; Cao, W.; Zhang, K.; Liu, X.; Zheng, H. Foot-and-Mouth Disease Virus Antagonizes NOD2-Mediated Antiviral Effects by Inhibiting NOD2 Protein Expression. J. Virol. 2019, 93, e00124-19. [Google Scholar] [CrossRef] [PubMed]
- Vegna, S.; Gregoire, D.; Moreau, M.; Lassus, P.; Durantel, D.; Assenat, E.; Hibner, U.; Simonin, Y. NOD1 Participates in the Innate Immune Response Triggered by Hepatitis C Virus Polymerase. J. Virol. 2016, 90, 6022–6035. [Google Scholar] [CrossRef]
- Wu, X.M.; Zhang, J.; Li, P.W.; Hu, Y.W.; Cao, L.; Ouyang, S.; Bi, Y.H.; Nie, P.; Chang, M.X. NOD1 Promotes Antiviral Signaling by Binding Viral RNA and Regulating the Interaction of MDA5 and MAVS. J. Immunol. 2020, 204, 2216–2231. [Google Scholar] [CrossRef]
- Roczkowsky, A.; Limonta, D.; Fernandes, J.P.; Branton, W.G.; Clarke, M.; Hlavay, B.; Noyce, R.S.; Joseph, J.T.; Ogando, N.S.; Das, S.K.; et al. COVID-19 Induces Neuroinflammation and Suppresses Peroxisomes in the Brain. Ann. Neurol. 2023, 94, 531–546. [Google Scholar] [CrossRef]
- Carreto-Binaghi, L.E.; Herrera, M.T.; Guzman-Beltran, S.; Juarez, E.; Sarabia, C.; Salgado-Cantu, M.G.; Juarez-Carmona, D.; Guadarrama-Perez, C.; Gonzalez, Y. Reduced IL-8 Secretion by NOD-like and Toll-like Receptors in Blood Cells from COVID-19 Patients. Biomedicines 2023, 11, 1078. [Google Scholar] [CrossRef]
- Rivera, E.G.; Patnaik, A.; Salvemini, J.; Jain, S.; Lee, K.; Lozeau, D.; Yao, Q. SARS-CoV-2/COVID-19 and its relationship with NOD2 and ubiquitination. Clin. Immunol. 2022, 238, 109027. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Ying, L.; D’Costa, K.; Wray-McCann, G.; Kerr, G.; Le, L.; Allison, C.C.; Ferrand, J.; Chaudhry, H.; Emery, J.; et al. NOD1 mediates interleukin-18 processing in epithelial cells responding to Helicobacter pylori infection in mice. Nat. Commun. 2023, 14, 3804. [Google Scholar] [CrossRef]
- Kim, J.G.; Lee, S.J.; Kagnoff, M.F. Nod1 is an essential signal transducer in intestinal epithelial cells infected with bacteria that avoid recognition by toll-like receptors. Infect. Immun. 2004, 72, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- van der Vorst, E.P.C.; Döring, Y.; Weber, C. Chemokines. Arterioscler. Thromb. Vasc. Biol. 2015, 35, e52–e56. [Google Scholar] [CrossRef]
- Chan, L.; Karimi, N.; Morovati, S.; Alizadeh, K.; Kakish, J.E.; Vanderkamp, S.; Fazel, F.; Napoleoni, C.; Alizadeh, K.; Mehrani, Y.; et al. The Roles of Neutrophils in Cytokine Storms. Viruses 2021, 13, 2318. [Google Scholar] [CrossRef] [PubMed]
- Hedl, M.; Abraham, C. Secretory mediators regulate Nod2-induced tolerance in human macrophages. Gastroenterology 2011, 140, 231–241. [Google Scholar] [CrossRef]
- Nayar, S.; Morrison, J.K.; Giri, M.; Gettler, K.; Chuang, L.-s.; Walker, L.A.; Ko, H.M.; Kenigsberg, E.; Kugathasan, S.; Merad, M.; et al. A myeloid–stromal niche and gp130 rescue in NOD2-driven Crohn’s disease. Nature 2021, 593, 275–281. [Google Scholar] [CrossRef]
- Pei, G.; Dorhoi, A. NOD-Like Receptors: Guards of Cellular Homeostasis Perturbation during Infection. Int. J. Mol. Sci. 2021, 22, 6714. [Google Scholar] [CrossRef]
- Ryu, J.K.; Kim, S.J.; Rah, S.H.; Kang, J.I.; Jung, H.E.; Lee, D.; Lee, H.K.; Lee, J.O.; Park, B.S.; Yoon, T.Y.; et al. Reconstruction of LPS Transfer Cascade Reveals Structural Determinants within LBP, CD14, and TLR4-MD2 for Efficient LPS Recognition and Transfer. Immunity 2017, 46, 38–50. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Tsai, W.H.; Huang, D.Y.; Yu, Y.H.; Chen, C.Y.; Lin, W.W. Dual roles of NOD2 in TLR4-mediated signal transduction and -induced inflammatory gene expression in macrophages. Cell Microbiol. 2011, 13, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Budikhina, A.S.; Murugina, N.E.; Maximchik, P.V.; Dagil, Y.A.; Nikolaeva, A.M.; Balyasova, L.S.; Murugin, V.V.; Selezneva, E.M.; Pashchenkova, Y.G.; Chkadua, G.Z.; et al. Interplay between NOD1 and TLR4 Receptors in Macrophages: Nonsynergistic Activation of Signaling Pathways Results in Synergistic Induction of Proinflammatory Gene Expression. J. Immunol. 2021, 206, 2206–2220. [Google Scholar] [CrossRef]
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef]
- Zhou, H.; Coveney, A.P.; Wu, M.; Huang, J.; Blankson, S.; Zhao, H.; O’Leary, D.P.; Bai, Z.; Li, Y.; Redmond, H.P.; et al. Activation of Both TLR and NOD Signaling Confers Host Innate Immunity-Mediated Protection Against Microbial Infection. Front. Immunol. 2018, 9, 3082. [Google Scholar] [CrossRef]
- Santecchia, I.; Ferrer, M.F.; Vieira, M.L.; Gomez, R.M.; Werts, C. Phagocyte Escape of Leptospira: The Role of TLRs and NLRs. Front. Immunol. 2020, 11, 571816. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, R. Innate Receptor Activation Patterns Involving TLR and NLR Synergisms in COVID-19, ALI/ARDS and Sepsis Cytokine Storms: A Review and Model Making Novel Predictions and Therapeutic Suggestions. Int. J. Mol. Sci. 2021, 22, 2108. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef] [PubMed]
- da Silva Correia, J.; Miranda, Y.; Austin-Brown, N.; Hsu, J.; Mathison, J.; Xiang, R.; Zhou, H.; Li, Q.; Han, J.; Ulevitch, R.J. Nod1-dependent control of tumor growth. Proc. Natl. Acad. Sci. USA. 2006, 103, 1840–1845. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y. Activation of RIPK2-mediated NOD1 signaling promotes proliferation and invasion of ovarian cancer cells via NF-kappaB pathway. Histochem. Cell Biol. 2022, 157, 173–182. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Yuan, G.; Yao, H.; Zhang, D.; Li, N.; Zhang, G.; Sun, Y.; Wang, W.; Zeng, J.; et al. Upregulation of NOD1 and NOD2 contribute to cancer progression through the positive regulation of tumorigenicity and metastasis in human squamous cervical cancer. BMC Med. 2022, 20, 55. [Google Scholar] [CrossRef]
- Ma, X.; Qiu, Y.; Zhu, L.; Zhao, Y.; Lin, Y.; Ma, D.; Qin, Z.; Sun, C.; Shen, X.; Li, T.; et al. NOD1 inhibits proliferation and enhances response to chemotherapy via suppressing SRC-MAPK pathway in hepatocellular carcinoma. J. Mol. Med. 2020, 98, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Gurses, S.A.; Banskar, S.; Stewart, C.; Trimoski, B.; Dziarski, R.; Gupta, D. Nod2 protects mice from inflammation and obesity-dependent liver cancer. Sci. Rep. 2020, 10, 20519. [Google Scholar] [CrossRef]
- Zangara, M.T.; Johnston, I.; Johnson, E.E.; McDonald, C. Mediators of Metabolism: An Unconventional Role for NOD1 and NOD2. Int. J. Mol. Sci. 2021, 22, 1156. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Shiny, A.; Regin, B.; Balachandar, V.; Gokulakrishnan, K.; Mohan, V.; Babu, S.; Balasubramanyam, M. Convergence of innate immunity and insulin resistance as evidenced by increased nucleotide oligomerization domain (NOD) expression and signaling in monocytes from patients with type 2 diabetes. Cytokine 2013, 64, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Li, L.; Li, M.; Wang, W.; Yin, W.; Liu, W.; Hu, Y. Silencing of NOD2 protects against diabetic cardiomyopathy in a murine diabetes model. Int. J. Mol. Med. 2018, 42, 3017–3026. [Google Scholar] [CrossRef]
- Cuda, C.; Badawi, A.; Karmali, M.; El-Sohemy, A. Effects of polymorphisms in nucleotide-binding oligomerization domains 1 and 2 on biomarkers of the metabolic syndrome and type II diabetes. Genes. Nutr. 2012, 7, 427–435. [Google Scholar] [CrossRef]
- Carlos, D.; Perez, M.M.; Leite, J.A.; Rocha, F.A.; Martins, L.M.S.; Pereira, C.A.; Fraga-Silva, T.F.C.; Pucci, T.A.; Ramos, S.G.; Camara, N.O.S.; et al. NOD2 Deficiency Promotes Intestinal CD4+ T Lymphocyte Imbalance, Metainflammation, and Aggravates Type 2 Diabetes in Murine Model. Front. Immunol. 2020, 11, 1265. [Google Scholar] [CrossRef]
- Johansson, M.E.; Zhang, X.Y.; Edfeldt, K.; Lundberg, A.M.; Levin, M.C.; Borén, J.; Li, W.; Yuan, X.-M.; Folkersen, L.; Eriksson, P.; et al. Innate immune receptor NOD2 promotes vascular inflammation and formation of lipid-rich necrotic cores in hypercholesterolemic mice. Eur. J. Immunol. 2014, 44, 3081–3092. [Google Scholar] [CrossRef]
- Vlacil, A.K.; Schuett, J.; Ruppert, V.; Soufi, M.; Oberoi, R.; Shahin, K.; Wachter, C.; Tschernig, T.; Lei, Y.; Liu, F.; et al. Deficiency of Nucleotide-binding oligomerization domain-containing proteins (NOD) 1 and 2 reduces atherosclerosis. Basic. Res. Cardiol. 2020, 115, 47. [Google Scholar] [CrossRef]
- Gresnigt, M.S.; Cunha, C.; Jaeger, M.; Goncalves, S.M.; Malireddi, R.K.S.; Ammerdorffer, A.; Lubbers, R.; Oosting, M.; Rasid, O.; Jouvion, G.; et al. Genetic deficiency of NOD2 confers resistance to invasive aspergillosis. Nat. Commun. 2018, 9, 2636. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, N.; Murch, O.; McMaster, S.K.; Paul-Clark, M.J.; van Heel, D.A.; Ryffel, B.; Quesniaux, V.F.; Evans, T.W.; Thiemermann, C.; Mitchell, J.A. Selective NOD1 agonists cause shock and organ injury/dysfunction in vivo. Am. J. Respir. Crit. Care Med. 2007, 175, 595–603. [Google Scholar] [CrossRef]
- Bonen, D.K.; Ogura, Y.; Nicolae, D.L.; Inohara, N.; Saab, L.; Tanabe, T.; Chen, F.F.; Foster, S.J.; Duerr, R.H.; Brant, S.R.; et al. Crohn’s disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology 2003, 124, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Voss, E.; Wehkamp, J.; Wehkamp, K.; Stange, E.F.; Schroder, J.M.; Harder, J. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J. Biol. Chem. 2006, 281, 2005–2011. [Google Scholar] [CrossRef]
- Girardelli, M.; Loganes, C.; Pin, A.; Stacul, E.; Decleva, E.; Vozzi, D.; Baj, G.; De Giacomo, C.; Tommasini, A.; Bianco, A.M. Novel NOD2 Mutation in Early-Onset Inflammatory Bowel Phenotype. Inflamm. Bowel Dis. 2018, 24, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Shang, D. NOD1 and NOD2: Essential Monitoring Partners in the Innate Immune System. Curr. Issues Mol. Biol. 2024, 46, 9463-9479. https://doi.org/10.3390/cimb46090561
Li Z, Shang D. NOD1 and NOD2: Essential Monitoring Partners in the Innate Immune System. Current Issues in Molecular Biology. 2024; 46(9):9463-9479. https://doi.org/10.3390/cimb46090561
Chicago/Turabian StyleLi, Zhenjia, and Dejing Shang. 2024. "NOD1 and NOD2: Essential Monitoring Partners in the Innate Immune System" Current Issues in Molecular Biology 46, no. 9: 9463-9479. https://doi.org/10.3390/cimb46090561
APA StyleLi, Z., & Shang, D. (2024). NOD1 and NOD2: Essential Monitoring Partners in the Innate Immune System. Current Issues in Molecular Biology, 46(9), 9463-9479. https://doi.org/10.3390/cimb46090561




