Selection and Validation of Reliable Reference Genes for Liquidambar formosana Leaves with Different Leaf Colors
Abstract
1. Introduction
2. Materials and Methods
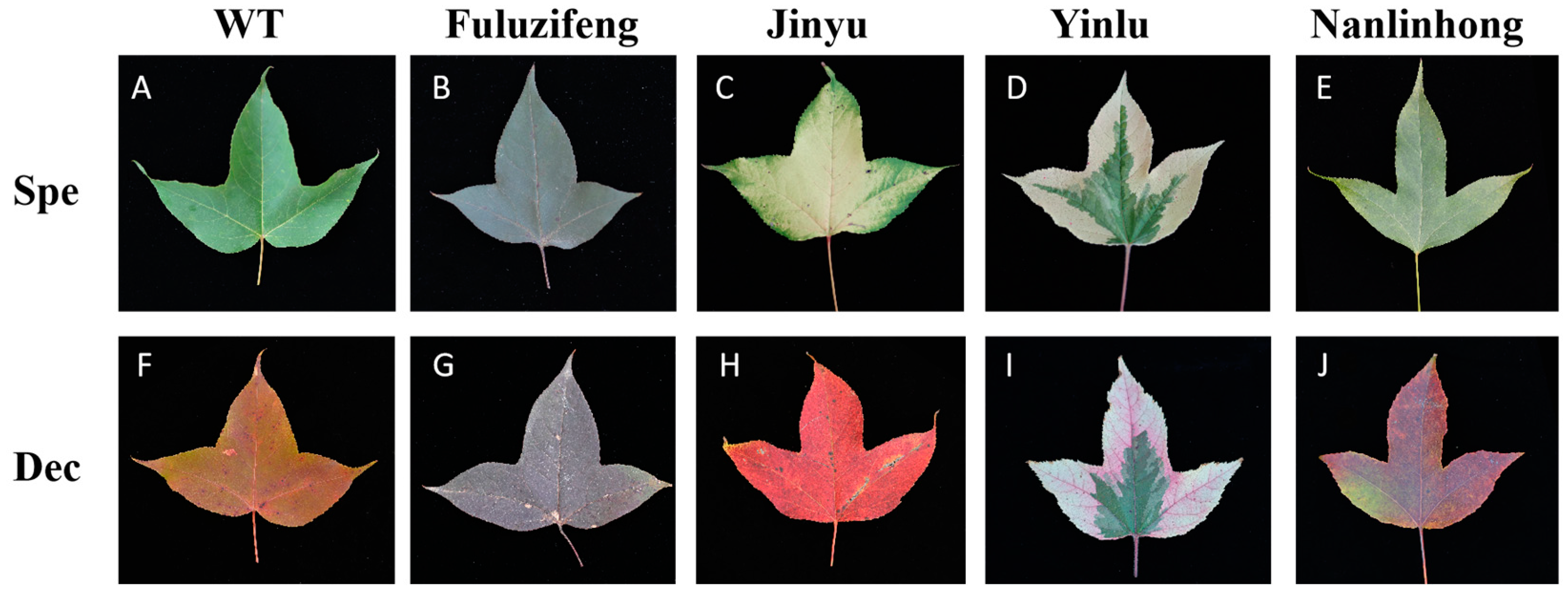
2.1. Plant Materials
2.2. Total RNA Extraction and cDNA First-Strand Synthesis
2.3. Primer Design and Specific Detection
2.4. Real-Time Fluorescence-Quantitative PCR of Candidate Reference Genes
2.5. Establishment of Reference Gene Primer Standard Curves
2.6. Data Analysis
2.7. Stability Verification of Reference Genes
3. Results
3.1. RNA Quality Testing and Primer Specificity Validation
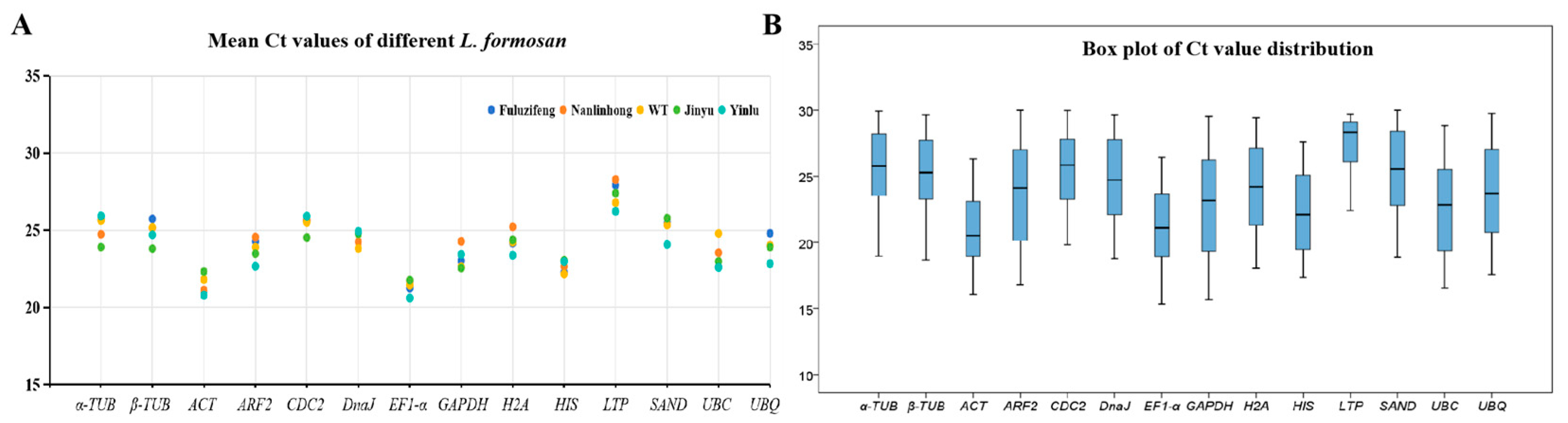
3.2. Reference Primer Amplification Ct Value Analysis
3.3. Analysis of Gene Expression Stability
3.3.1. ΔCt Analysis
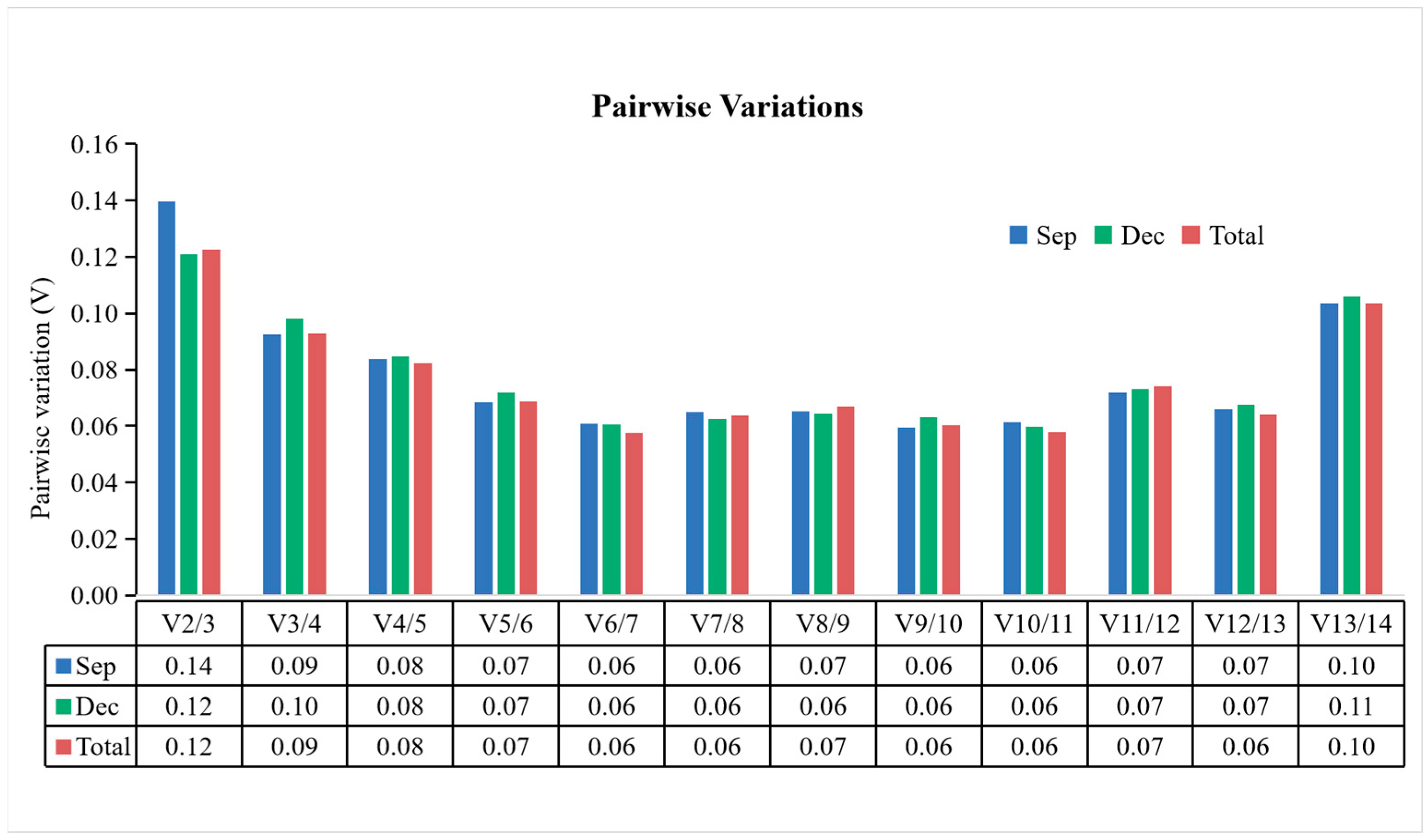
3.3.2. geNorm Analysis
3.3.3. BestKeeper Analysis
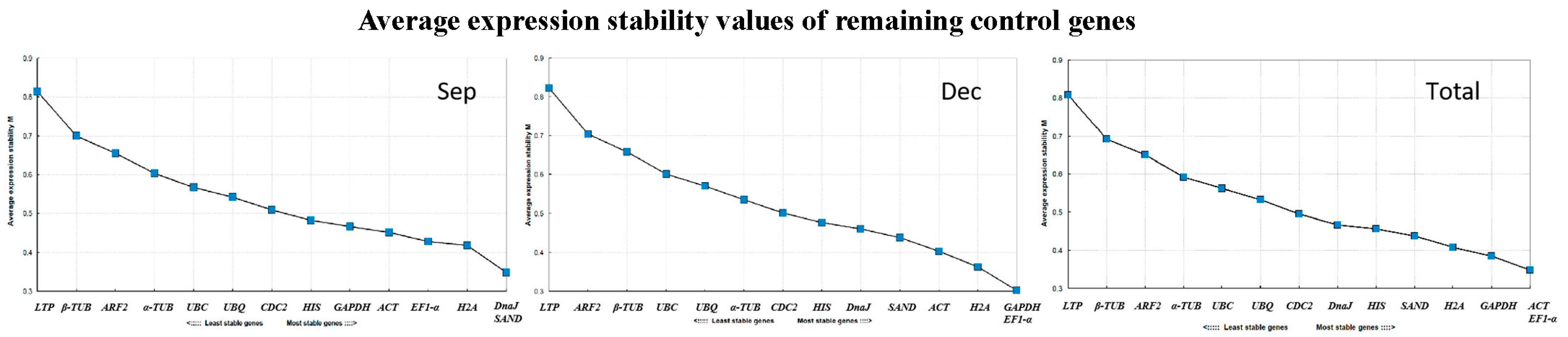
3.3.4. NormFinder Analysis
3.3.5. RefFinder Analysis
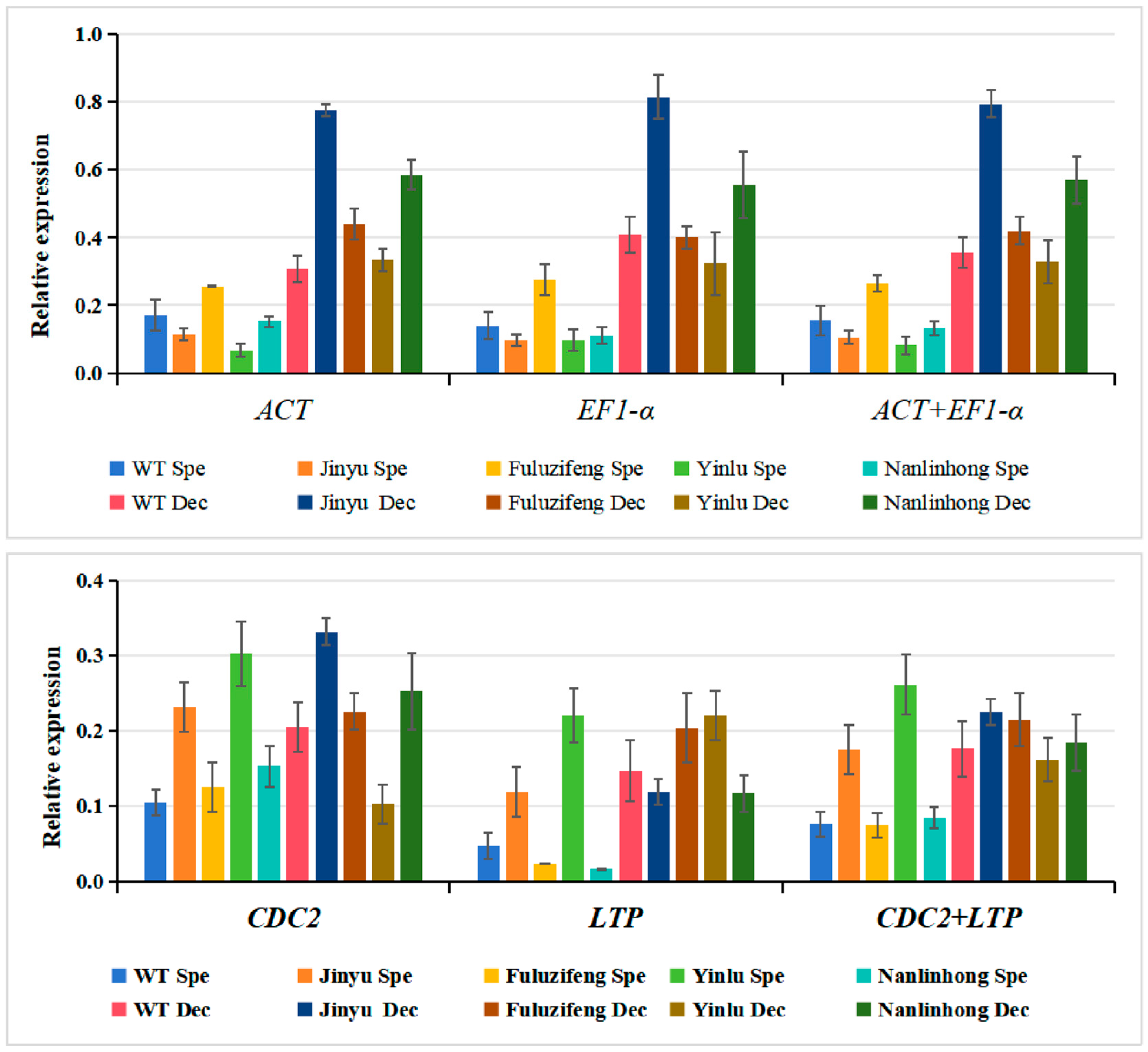
3.4. Reference Gene Stability Verification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, Z.; Guan, H.D.; Zhang, X.P.; Liu, N. Photosynthetic capacity of senescent leaves for a subtropical broadleaf deciduous tree species Liquidambar formosana Hance. Sci. Rep. 2017, 7, 6323. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.P.; Wang, Y.; Xiao, Y.F.; Yang, J.S.; Wang, R.J.; Jiang, Y.; Huang, R.L.; Liu, X.S.; Jiang, Y. Relationships between leaf color changes, pigment levels, enzyme activity, photosynthetic fluorescence characteristics and chloroplast ultrastructure of Liquidambar formosana Hance. J. For. Res. 2022, 33, 1559–1572. [Google Scholar] [CrossRef]
- Lai, J.X.; Lin, F.R.; Huang, P.; Guo, W.Y.; Han, L.B.; Zheng, Y.Q. Seasonal variation of flavonoid metabolites in leaves of Liquidambar formosana ‘Nanlinhong’ trees revealed by UPLC-ESI–MS/MS. Rev. Bras. Bot. 2021, 44, 575–586. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Gil, N.H. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Schaberg, P.; Murakami, P.; Turner, M.; Heitz, H.; Hawley, G. Association of red coloration with senescence of sugar maple leaves in autumn. Trees–Struct. Funct. 2008, 22, 573–578. [Google Scholar] [CrossRef]
- Archetti, M. Phylogenetic analysis reveals a scattered distribution of autumn colours. Ann. Bot. 2009, 103, 703–713. [Google Scholar] [CrossRef][Green Version]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.X.; Lin, F.R.; Huang, P.; Zheng, Y.Q. Moderate genetic diversity and genetic differentiation in the relict tree Liquidambar formosana Hance revealed by genic simple sequence repeat markers. Front. Plant Sci. 2016, 7, 1411. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, Y.; Chen, H.; Chen, C.; Liu, Z.; Han, C.; Wu, Q.; Yu, F. Transcriptomic analyses reveal key genes involved in pigment biosynthesis related to leaf color change of Liquidambar formosana Hance. Molecules 2022, 27, 5433. [Google Scholar] [CrossRef]
- Xu, W.Q.; Ren, C.Q.; Zhang, X.Y.; Comes, H.P.; Liu, X.H.; Li, Y.G.; Kettle, C.J.; Jalonen, R.; Gaisberger, H.; Ma, Y.Z.; et al. Genome sequences and population genomics reveal climatic adaptation and genomic divergence between two closely related sweetgum species. Plant J. 2024, 118, 1372–1387. [Google Scholar] [CrossRef]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Klein, D. Quantifification using real-time PCR technology: Applications and limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, X.; Chen, S.; Zheng, L.; He, X.; Liu, M.; Qiao, G.; Wang, Y.; Zhou, R. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci. Rep. 2017, 7, 40290. [Google Scholar] [CrossRef]
- Ambroise, V.; Legay, S.; Guerriero, G.; Hausman, J.F.; Cuypers, A.; Sergeant, K. Selection of appropriate reference genes for gene expression analysis under abiotic stresses in Salix viminalis. Int. J. Mol. Sci. 2019, 20, 4210. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.C.; Cai, Y.M.; Zhao, B.X.; Fu, C.Q.; Yang, L.Y. Screening of RT-qPCR reference genes in different varieties and tissues of Zantedeschia hybrida. Mol. Plant Breed 2020, 18, 3971–3979. [Google Scholar]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Lu, M.; Chen, Y.; Zhan, Z.; Cui, Q.; Wang, Y. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS ONE 2012, 7, e43084. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Pfaffl, M.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Zhang, Q.; Yuan, L.; Zhou, X. Reference gene selection for transcriptional profiling in Cryptocercus punctulatus, an evolutionary link between Isoptera and Blattodea. Sci. Rep. 2020, 10, 22169. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Wei, W.; Fan, Z.Q.; Chen, J.Y.; You, Y.L.; Huang, W.D.; Zhan, J.C. VabHLH137 promotes proanthocyanidin and anthocyanin biosynthesis and enhances resistance to Colletotrichum gloeosporioides in grapevine. Hortic. Res. 2022, 10, uhac261. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, F.; Chen, Y.; Wu, H.; Yin, T. Genome-Wide comparative analysis of R2R3 MYB gene family in Populus and Salix and identification of male flower bud development-related genes. Front. Plant Sci. 2021, 12, 721558. [Google Scholar] [CrossRef]
- Li, J.; Bogle, A.L.; Klein, A.S. Interspecific relationships and genetic divergence of the disjunct genus Liquidambar (Hamamelidaceae) inferred from DNA sequences of plastid gene matK. Rhodora 1997, 99, 229–240. [Google Scholar]
- Wen, C.H.; Chu, F.H. A R2R3-MYB gene LfMYB113 is responsible for autumn leaf coloration in Formosan sweet gum (Liquidambar formosana Hance). Plant Cell Physiol. 2017, 58, 508–521. [Google Scholar]
- Niu, X.P.; Qi, J.M.; Zhang, G.Y.; Xu, J.T.; Tao, A.F.; Fang, P.P.; Su, J.G. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 2015, 6, 848. [Google Scholar] [CrossRef]
- Kanakachari, M.; Solanke, A.U.; Prabhakaran, N.; Ahmad, I.; Dhandapani, G.; Jayabalan, N.; Kumar, P.A. Evaluation of suitable reference genes for normalization of qPCR gene expression studies in Brinjal (Solanum melongena L.) during fruit developmental stages. Appl. Biochem. Biotechnol. 2016, 178, 433–450. [Google Scholar] [CrossRef]
- Qi, S.; Yang, L.; Wen, X.; Hong, Y.; Song, X.; Zhang, M.; Dai, S. Reference Gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 2016, 7, 287. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.B.; Lukaszewicz, G.; Lamattina, L.; Cassia, R. Selection and optimization of reference genes for RT-qPCR normalization: A case study in Solanum lycopersicum exposed to UV-B. Plant Physiol. Biochem. 2021, 160, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.W.; Chen, Y.N.; Wu, H.T.; Yin, T.M. A Selection of Reliable Reference Genes for Gene Expression Analysis in the Female and Male Flowers of Salix suchowensis. Plants 2022, 11, 647. [Google Scholar] [CrossRef] [PubMed]
| Gene | Gene Description | Primer Sequence F/R (5′-3′) | Product Size (bp) | Efficiency (%) | R2 |
|---|---|---|---|---|---|
| α-TUB1 | Alpha-tubulin 1 | AGCAACTCATCAGCGGCAAGG | 264 | 101.21 | 0.9801 |
| TGGCTCCACAACAGAGGTAGAAA | |||||
| β-TUB | Tubulin | ACGAGGCTCTTTATGATATTTG | 264 | 107.22 | 0.9960 |
| GTTGGGTGAGTTCAGGGACA | |||||
| ACT | Actin | ATGTTCCCTGGCATTGCAGAC | 181 | 102.45 | 0.9916 |
| ACTCATCATATTCACCCTTCG | |||||
| ARF2 | ADP-ribosylation factor 2 | GTTGAATGAGGATGAGTTGAGGGAA | 177 | 105.14 | 0.9813 |
| CGTTGACGGAGCGAGTGAAGA | |||||
| CDC2 | Cyclin-dependent kinase-putative | TGGCATTGCTTATTGTCATTC | 123 | 106.40 | 0.9875 |
| GGTGCTCTGTACCACAGGGTC | |||||
| DnaJ | Chaperone protein DnaJ 49 | TGGGTCAGATGAGCCCGTTTA | 135 | 105.27 | 0.9923 |
| AATTGGGTCGGTTGCATTCCT | |||||
| EF1-α | Elongation factor 1-alpha | GATTGGTGGCATTGGAACTGT | 124 | 109.53 | 0.9903 |
| GTGGTGCATCTCAACGGACTT | |||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | GGTGGATTTGGCACATTTGGT | 108 | 105.90 | 0.9944 |
| CACTCCTCATCAGCAGGGTTT | |||||
| H2A | Histone H2A | GCTTGAGTTGGCGGGAAATGC | 168 | 103.12 | 0.9961 |
| TTCGGAAGGAGAAGGTTGTGAATGT | |||||
| HIS | Histone superfamily protein H3 | GAAGAAGCCTCACAGATACCG | 135 | 105.02 | 0.9950 |
| GTCTTGAAGTCCTGGGCAAT | |||||
| LTP | Seed storage/lipid transfer protein | CATCAGCAGCACAAGATTCAA | 102 | 117.49 | 0.9765 |
| AAAGCATAACAGCACAGAGGC | |||||
| SAND | SAND family protein | GGCTTCAGAGTTTCCATCACC | 157 | 107.50 | 0.9843 |
| GACCCAGCAGAGTAGAACATAG | |||||
| UBC | Ubiquitin conjugating enzyme | AGCCCTGCCCTTACCATTTCC | 141 | 108.51 | 0.9920 |
| GCTCCTTGCGGTTGTTTCATA | |||||
| UBQ | Ubiquitin family | AGGAGTGCCCTAATGCCGAGTG | 105 | 99.31 | 0.9947 |
| CAGCCTTCTGATAAACGTAAGTCAA | |||||
| Target gene | |||||
| bHLH137 | Basic helix-loop-helix | AAGAAGCTCCAACAGGGTAC | 200 | 101.77 | 0.9972 |
| TTGTAGGGACTGGACATAGTT | |||||
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spe | Gene | EF1-α | DnaJ | SAND | H2A | CDC2 | α-TUB | UBQ | HIS | UBC | β-TUB | ACT | GAPDH | ARF2 | LTP |
| Stability | 0.63 | 0.65 | 0.67 | 0.68 | 0.69 | 0.74 | 0.75 | 0.76 | 0.78 | 0.8 | 0.92 | 1.02 | 1.1 | 1.45 | |
| Dec | Gene | DnaJ | EF1-α | SAND | HIS | β-TUB | H2A | CDC2 | UBQ | α-TUB | UBC | ACT | ARF2 | GAPDH | LTP |
| Stability | 0.6 | 0.64 | 0.64 | 0.65 | 0.66 | 0.66 | 0.69 | 0.75 | 0.77 | 0.79 | 0.82 | 0.96 | 1.03 | 1.6 | |
| Total | Gene | EF1-α | ACT | HIS | UBQ | SAND | H2A | DnaJ | β-TUB | GAPDH | α-TUB | UBC | ARF2 | CDC2 | LTP |
| Stability | 0.41 | 0.42 | 0.44 | 0.46 | 0.51 | 0.58 | 0.69 | 0.87 | 0.93 | 1.04 | 1.25 | 1.68 | 2.58 | 2.67 | |
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spe | Gene | EF1-α | ACT | β-TUB | DnaJ | UBQ | HIS | LTP | SAND | α-TUB | H2A | CDC2 | UBC | GAPDH | ARF2 |
| SD | 0.69 | 0.76 | 0.78 | 0.86 | 0.91 | 0.92 | 1.08 | 1.19 | 1.27 | 1.34 | 1.51 | 1.57 | 1.71 | 1.93 | |
| CV | 2.07 | 2.36 | 2.52 | 3.21 | 3.41 | 3.47 | 3.88 | 4.87 | 4.98 | 5.37 | 5.62 | 6.02 | 7.03 | 8.41 | |
| Dec | Gene | EF1-α | ACT | DnaJ | β-TUB | α-TUB | UBQ | SAND | HIS | CDC2 | H2A | LTP | UBC | ARF2 | GAPDH |
| SD | 0.61 | 0.64 | 0.72 | 0.81 | 2.47 | 2.55 | 2.58 | 2.64 | 2.76 | 2.89 | 3.05 | 3.09 | 3.32 | 3.33 | |
| CV | 5.81 | 8.55 | 8.86 | 9.43 | 11.49 | 11.69 | 9.94 | 11.65 | 10.88 | 11.61 | 12.42 | 13.29 | 13.61 | 14.12 | |
| Total | Gene | EF1-α | SAND | ACT | β-TUB | DnaJ | HIS | UBQ | H2A | LTP | α-TUB | GAPDH | UBC | CDC2 | ARF2 |
| SD | 0.70 | 0.77 | 0.80 | 0.82 | 0.91 | 0.94 | 1.16 | 1.21 | 1.48 | 1.62 | 1.94 | 1.96 | 2.21 | 2.48 | |
| CV | 2.38 | 2.55 | 2.76 | 3.15 | 3.21 | 3.48 | 4.16 | 4.85 | 5.41 | 6.21 | 8.65 | 8.98 | 9.85 | 10.81 | |
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spe | Gene | EF1-α | SAND | DnaJ | CDC2 | H2A | α-TUB | HIS | UBQ | UBC | β-TUB | ACT | GAPDH | ARF2 | LTP |
| Stability | 0.158 | 0.249 | 0.267 | 0.34 | 0.34 | 0.425 | 0.467 | 0.496 | 0.537 | 0.544 | 0.74 | 0.899 | 0.988 | 1.381 | |
| Dec | Gene | DnaJ | SAND | EF1-α | HIS | β-TUB | H2A | CDC2 | α-TUB | UBQ | UBC | ACT | ARF2 | GAPDH | LTP |
| Stability | 0.114 | 0.242 | 0.264 | 0.285 | 0.292 | 0.329 | 0.387 | 0.525 | 0.527 | 0.583 | 0.588 | 0.84 | 0.941 | 1.560 | |
| Total | Gene | ACT | EF1-α | SAND | UBQ | HIS | H2A | DnaJ | β-TUB | GAPDH | α-TUB | UBC | ARF2 | CDC2 | LTP |
| Stability | 0.176 | 0.263 | 0.274 | 0.378 | 0.411 | 0.467 | 0.482 | 0.497 | 0.896 | 1.063 | 1.250 | 1.453 | 1.533 | 1.874 | |
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ranking order of leaves in September (better average) | ||||||||||||||
| Delta Ct | EF1-α | DnaJ | SAND | H2A | CDC2 | α-TUB | UBQ | HIS | UBC | β-TUB | ACT | GAPDH | ARF2 | LTP |
| geNorm | DnaJ | SAND | H2A | EF1-α | ACT | GAPDH | HIS | CDC2 | UBQ | UBC | α-TUB | ARF2 | β-TUB | LTP |
| BestKeeper | EF1-α | ACT | β-TUB | DnaJ | UBQ | HIS | LTP | SAND | α-TUB | H2A | CDC2 | UBC | GAPDH | ARF2 |
| NormFinder | EF1-α | SAND | DnaJ | CDC2 | H2A | α-TUB | HIS | UBQ | UBC | β-TUB | ACT | GAPDH | ARF2 | LTP |
| Comprehensive | EF1-α | SAND | DnaJ | H2A | CDC2 | α-TUB | HIS | UBQ | UBC | β-TUB | ACT | GAPDH | ARF2 | LTP |
| Ranking order of leaves in December (better average) | ||||||||||||||
| Delta Ct | DnaJ | EF1-α | SAND | HIS | β-TUB | H2A | CDC2 | UBQ | α-TUB | UBC | ACT | ARF2 | GAPDH | LTP |
| geNorm | GAPDH | EF1-α | H2A | ACT | SAND | DnaJ | HIS | CDC2 | α-TUB | UBQ | UBC | β-TUB | ARF2 | LTP |
| BestKeeper | EF1-α | ACT | DnaJ | β-TUB | α-TUB | UBQ | SAND | HIS | CDC2 | H2A | LTP | UBC | ARF2 | GAPDH |
| NormFinder | DnaJ | SAND | EF1-α | HIS | β-TUB | H2A | CDC2 | α-TUB | UBQ | UBC | ACT | ARF2 | GAPDH | LTP |
| Comprehensive | DnaJ | EF1-α | ACT | SAND | β-TUB | H2A | CDC2 | HIS | α-TUB | UBQ | UBC | ARF2 | GAPDH | LTP |
| Ranking order under total samples (better average) | ||||||||||||||
| Delta Ct | EF1-α | ACT | HIS | UBQ | SAND | H2A | DnaJ | β-TUB | GAPDH | α-TUB | UBC | ARF2 | CDC2 | LTP |
| geNorm | ACT | EF1-α | GAPDH | H2A | SAND | HIS | DnaJ | CDC2 | UBQ | UBC | α-TUB | ARF2 | β-TUB | LTP |
| BestKeeper | EF1-α | SAND | ACT | β-TUB | DnaJ | HIS | UBQ | H2A | LTP | α-TUB | GAPDH | UBC | CDC2 | ARF2 |
| NormFinder | ACT | EF1-α | SAND | UBQ | HIS | H2A | DnaJ | β-TUB | GAPDH | α-TUB | UBC | ARF2 | CDC2 | LTP |
| Comprehensive | EF1-α | ACT | SAND | UBQ | HIS | H2A | DnaJ | β-TUB | GAPDH | α-TUB | UBC | ARF2 | CDC2 | LTP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, F.; Xu, L.; Shi, C.; Yang, S.; Chen, Y. Selection and Validation of Reliable Reference Genes for Liquidambar formosana Leaves with Different Leaf Colors. Curr. Issues Mol. Biol. 2024, 46, 9449-9462. https://doi.org/10.3390/cimb46090560
Zhou F, Xu L, Shi C, Yang S, Chen Y. Selection and Validation of Reliable Reference Genes for Liquidambar formosana Leaves with Different Leaf Colors. Current Issues in Molecular Biology. 2024; 46(9):9449-9462. https://doi.org/10.3390/cimb46090560
Chicago/Turabian StyleZhou, Fangwei, Liang Xu, Congguang Shi, Shaozong Yang, and Yahui Chen. 2024. "Selection and Validation of Reliable Reference Genes for Liquidambar formosana Leaves with Different Leaf Colors" Current Issues in Molecular Biology 46, no. 9: 9449-9462. https://doi.org/10.3390/cimb46090560
APA StyleZhou, F., Xu, L., Shi, C., Yang, S., & Chen, Y. (2024). Selection and Validation of Reliable Reference Genes for Liquidambar formosana Leaves with Different Leaf Colors. Current Issues in Molecular Biology, 46(9), 9449-9462. https://doi.org/10.3390/cimb46090560





