Metabolism-Related Genes SMOX and SUCLG2 as Immunological and Prognostic Biomarkers in Colorectal Cancer: A Pan-Cancer Analysis
Abstract
1. Introduction
2. Materials and Methods
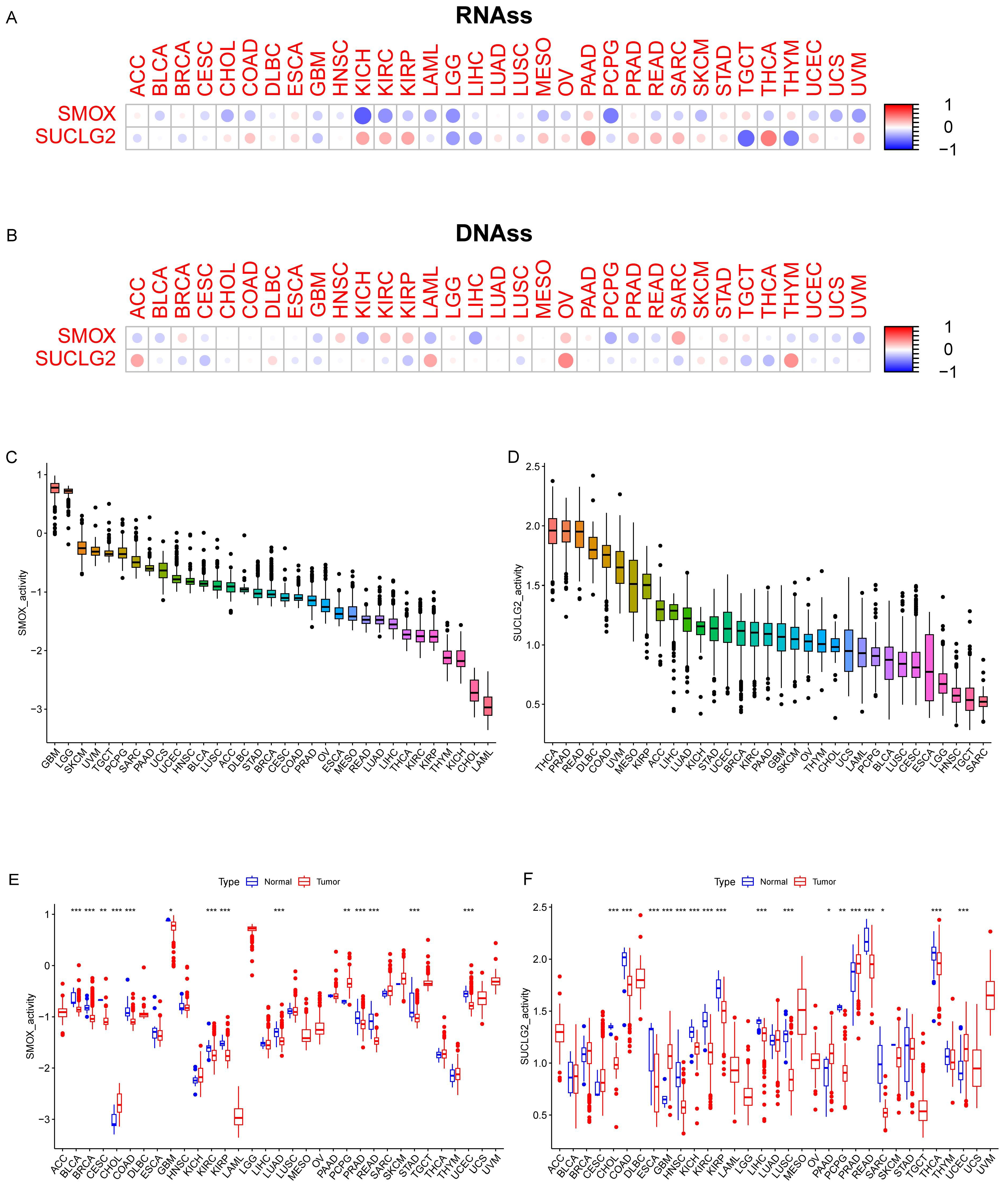
2.1. Pan-Cancer Data and Expression and Correlation Analyses
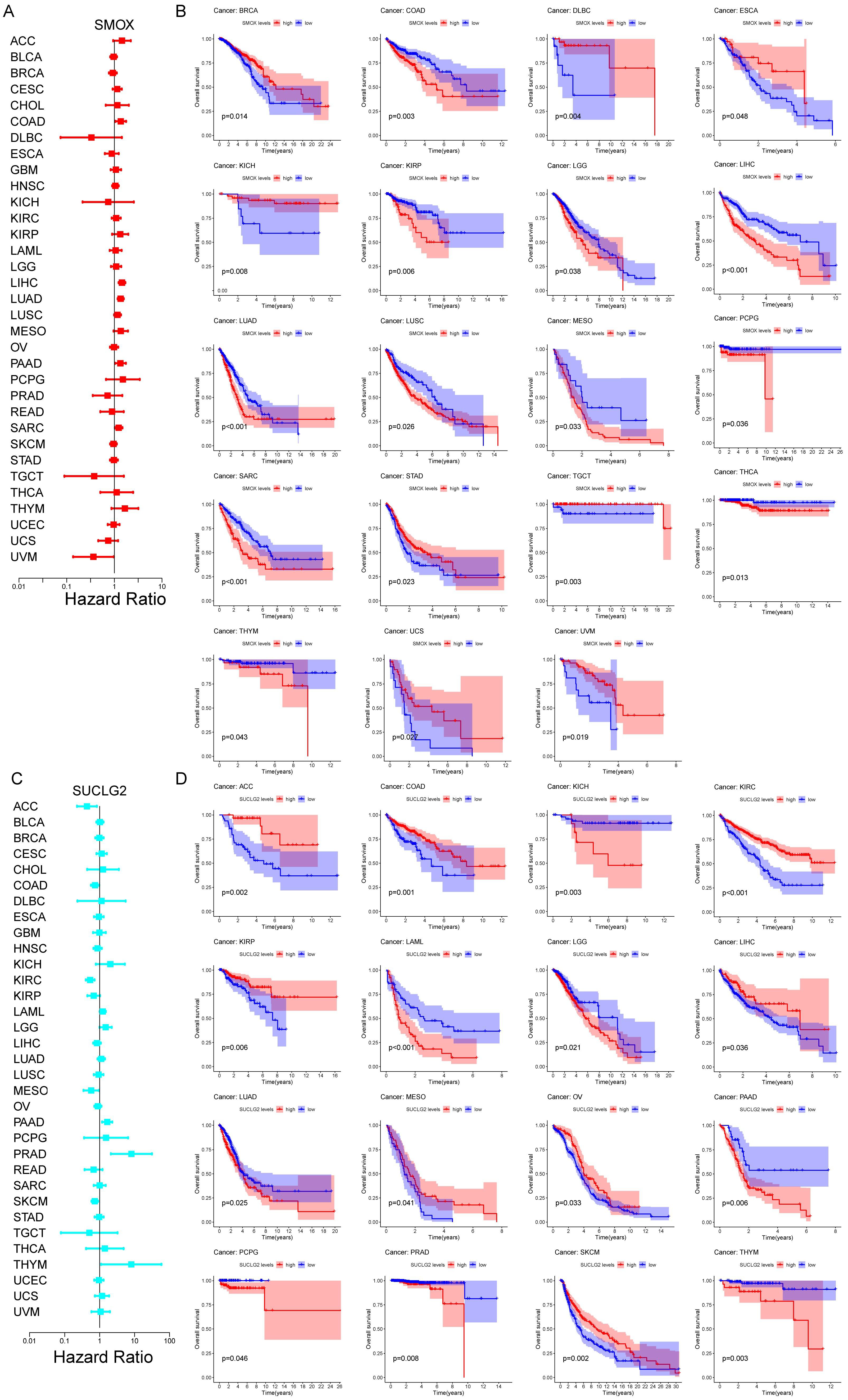
2.2. Analyzing the Association of SMOX and SUCLG2 with Pan-Cancer Prognosis
2.3. Analyzing the Association of SMOX and SUCLG2 with Pan-Cancer Cancer Immunity
2.4. Analyzing the Association of SMOX and SUCLG2 with the Characteristics of Pan-Cancer Cells
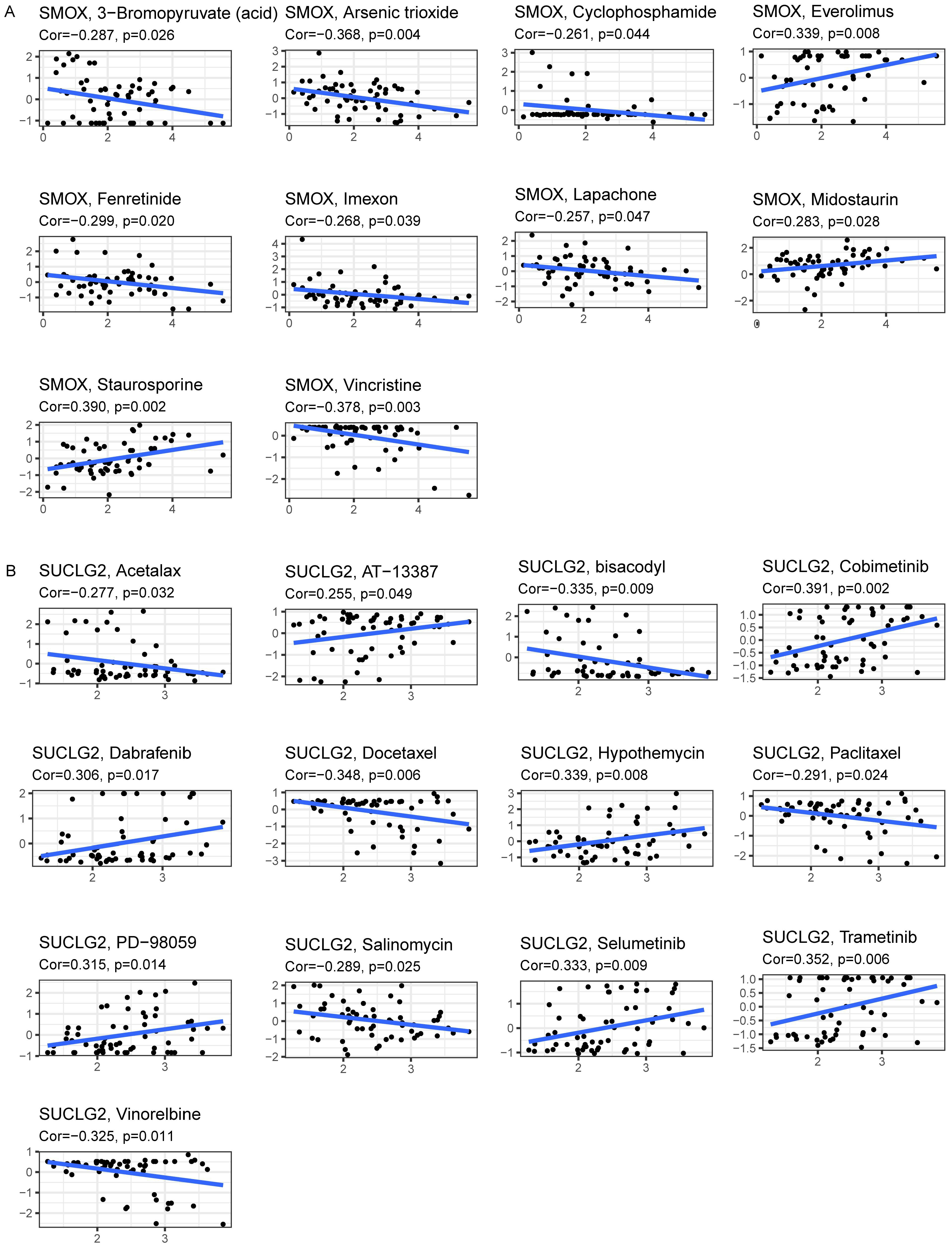
2.5. Drug Sensitivity Analysis
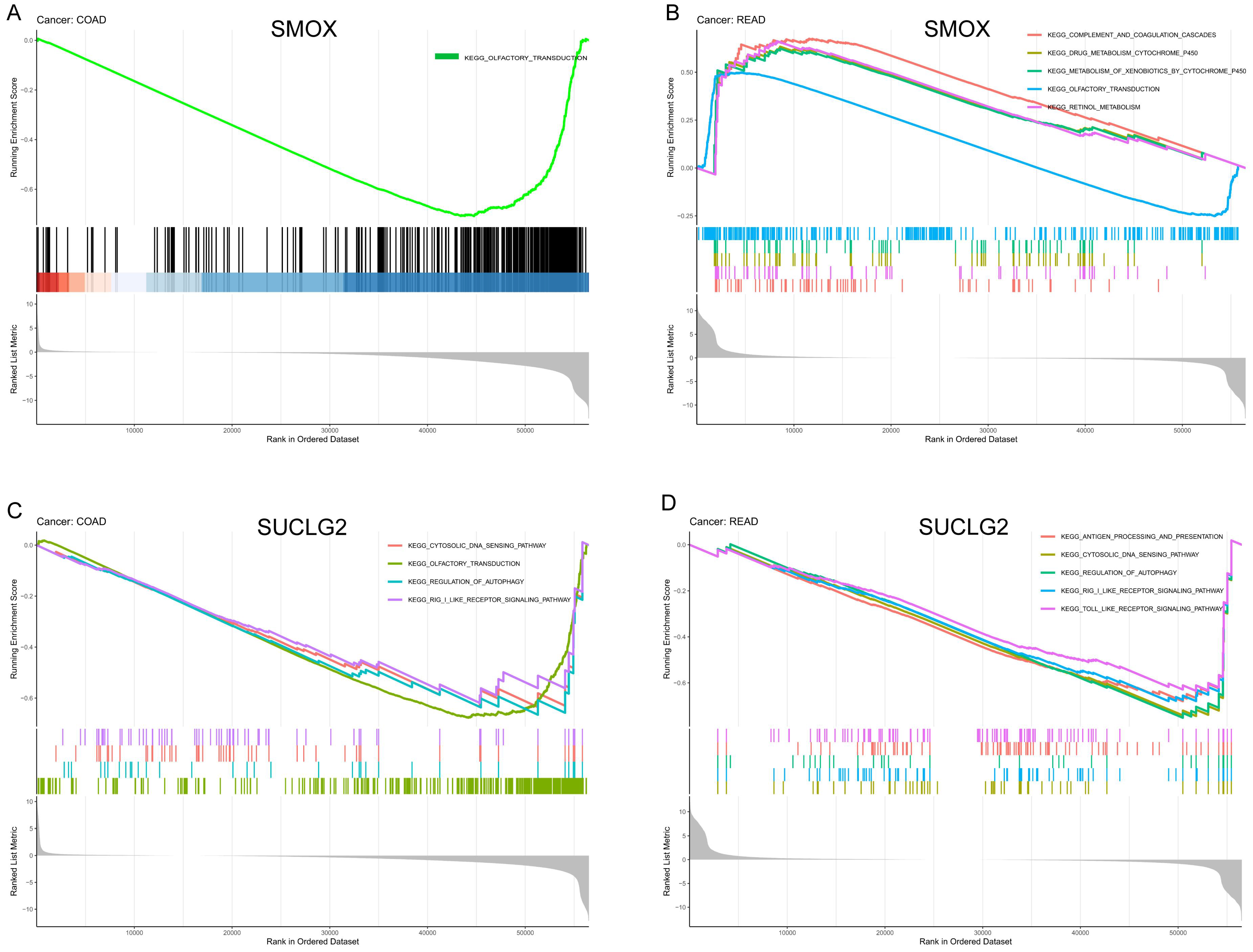
2.6. Functional Enrichment Analysis
2.7. Association with CRC, External Validation, and ceRNA Network Construction
2.8. qPCR
2.9. Cell Culture, Transfection and CCK-8 of SUCLG2
2.10. Statistical Analysis
3. Results
3.1. Differential Expression Analysis and Correlation Between SMOX and SUCLG2 Across Different Cancers
3.2. Prognostic Significance of SMOX and SUCLG2 in Pan-Cancer
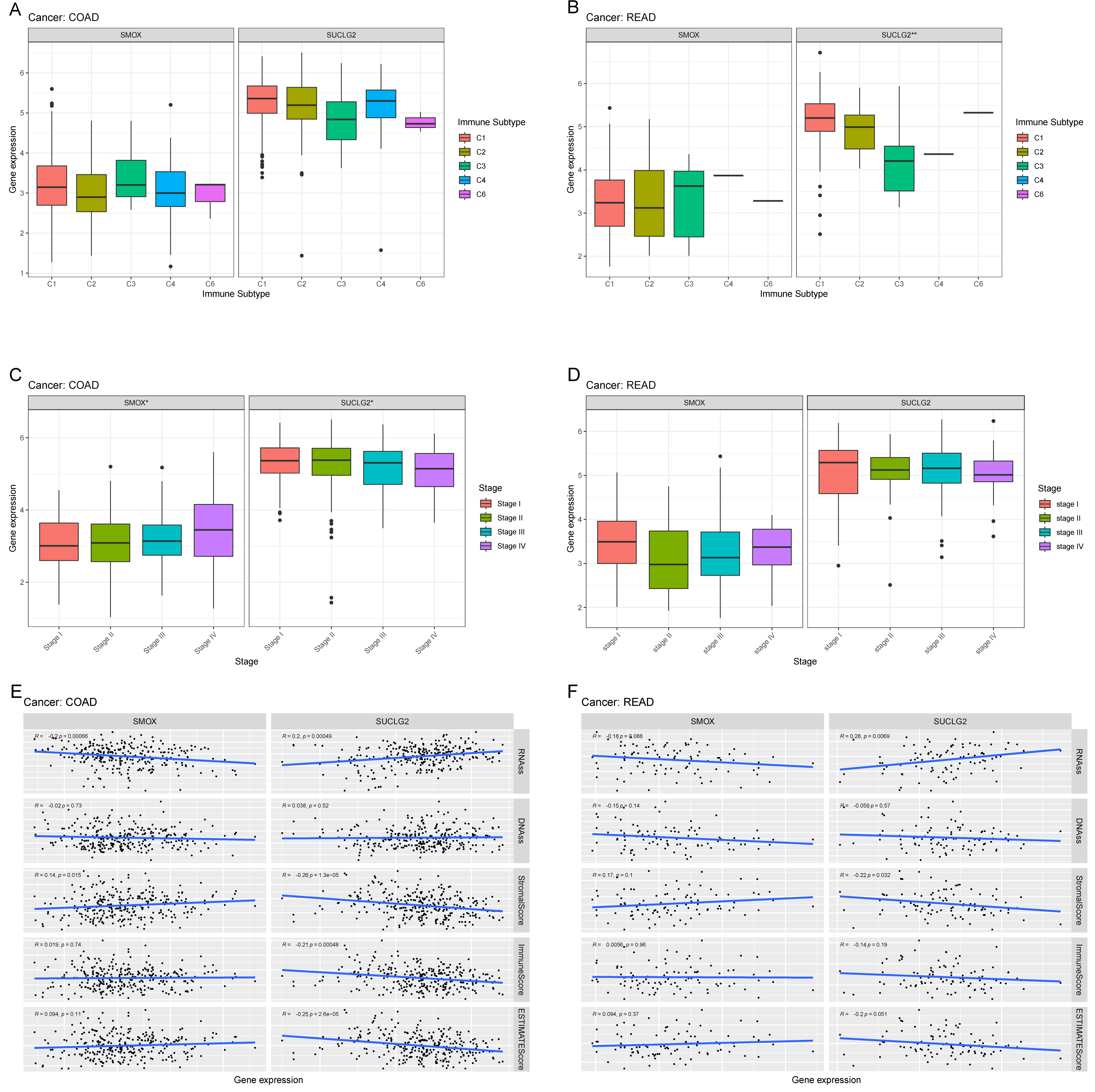
3.3. Relationship Between SMOX and SUCLG and Pan-Cancer Cancer Immunity
3.4. Relationship Between SMOX and SUCLG2 and Pan-Cancer Cancer Characteristics
3.5. Association of SMOX and SUCLG2 with Drug Sensitivity
3.6. Functional Enrichment Analysis
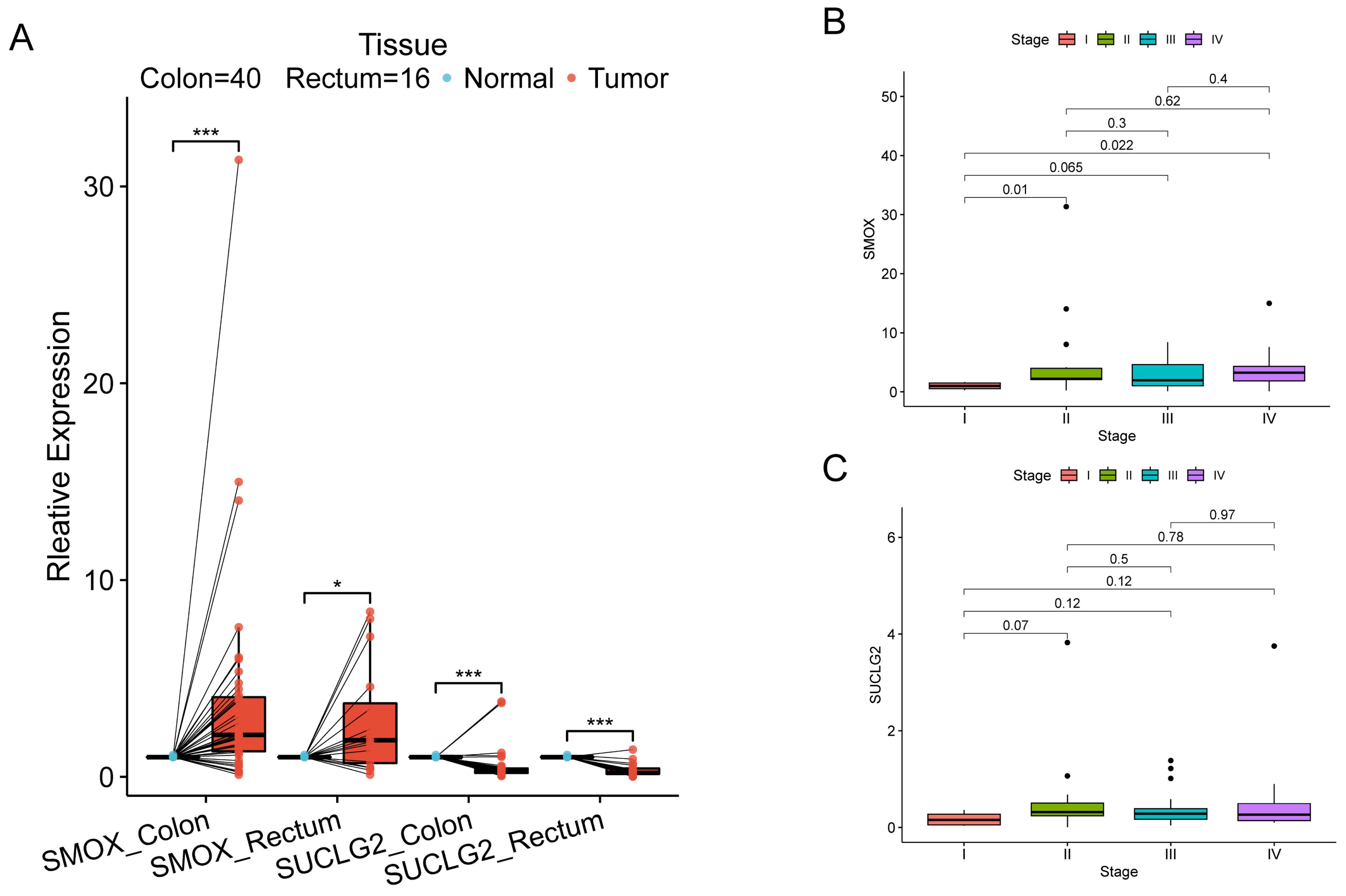
3.7. Association of SMOX and SUCLG2 with CRC
3.8. External Validation of SMOX and SUCLG2 in CRC with Gene Expression Omnibus and TCGA Datasets
3.9. CeRNA Network Establishment
3.10. Validation by HPA and qPCR
3.11. SUCLG2 Inhibited the Proliferation of CRC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder Urothelial Carcinoma |
| BRCA | Breast invasive carcinoma |
| CCK-8 | Cell Counting Kit-8 |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| CRC | Colorectal cancer |
| DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma |
| ESCA | Esophageal carcinoma |
| GBM | Glioblastoma multiforme |
| GEO | Gene Expression Omnibus |
| HNSC | Head and Neck squamous cell carcinoma |
| KICH | Kidney Chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LAML | Acute Myeloid Leukemia |
| LGG | Brain Lower Grade Glioma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| MESO | Mesothelioma |
| MSI | Microsatellite instability |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and Paraganglioma |
| PRAD | Prostate adenocarcinoma |
| READ | Rectum adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin Cutaneous Melanoma |
| STAD | Stomach adenocarcinoma |
| TCGA | The Cancer Genome Atlas |
| TGCT | Testicular Germ Cell Tumors |
| THCA | Thyroid carcinoma |
| THYM | Thymoma |
| TMB | Tumor mutational burden |
| UCEC | Uterine Corpus Endometrial Carcinoma |
| UCS | Uterine Carcinosarcoma |
| UCSC | University of California Santa Cruz |
| UVM | Uveal Melanoma |
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Huang, X.; Chen, Z.; Xiang, X.; Liu, Y.; Long, X.; Li, K.; Qin, M.; Long, C.; Mo, X.; Tang, W.; et al. Comprehensive multi-omics analysis of the m7G in pan-cancer from the perspective of predictive, preventive, and personalized medicine. EPMA J. 2022, 13, 671–697. [Google Scholar] [CrossRef]
- Rodríguez, C.; Puente-Moncada, N.; Reiter, R.J.; Sánchez-Sánchez, A.M.; Herrera, F.; Rodríguez-Blanco, J.; Duarte-Olivenza, C.; Turos-Cabal, M.; Antolín, I.; Martín, V. Regulation of cancer cell glucose metabolism is determinant for cancer cell fate after melatonin administration. J. Cell. Physiol. 2021, 236, 27–40. [Google Scholar] [CrossRef]
- Boroughs, L.K.; DeBerardinis, R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biology. 2015, 17, 351–359. [Google Scholar] [CrossRef]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 2021, 20, 28. [Google Scholar] [CrossRef]
- Callao, V.; Montoya, E. Toxohormone-like factor from microorganisms with impaired respiration. Science 1961, 134, 2041–2042. [Google Scholar] [CrossRef] [PubMed]
- Guerra, L.; Bonetti, L.; Brenner, D. Metabolic Modulation of Immunity: A New Concept in Cancer Immunotherapy. Cell Rep. 2020, 32, 107848. [Google Scholar] [CrossRef] [PubMed]
- Cronin, S.J.F.; Seehus, C.; Weidinger, A.; Talbot, S.; Reissig, S.; Seifert, M.; Pierson, Y.; McNeill, E.; Longhi, M.S.; Turnes, B.L.; et al. The metabolite BH4 controls T cell proliferation in autoimmunity and cancer. Nature 2018, 563, 564–568. [Google Scholar] [CrossRef]
- Wang, Y.; Devereux, W.; Woster, P.M.; Stewart, T.M.; Hacker, A.; A Casero, R. Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001, 61, 5370–5373. [Google Scholar]
- Babbar, N.; Murray-Stewart, T.; Casero, R. Inflammation and polyamine catabolism: The good, the bad and the ugly. Biochem. Soc. Trans. 2007, 35, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Tepper, A.; Chu, G.; Klaren, V.N.A.; Kalin, J.H.; Molina-Ortiz, P.; Impagliazzo, A. Development and characterization of rabbit monoclonal antibodies that recognize human spermine oxidase and application to immunohistochemistry of human cancer tissues. PLoS ONE 2022, 17, e0267046. [Google Scholar] [CrossRef]
- Sierra, J.C.; Piazuelo, M.B.; Luis, P.B.; Barry, D.P.; Allaman, M.M.; Asim, M.; Sebrell, T.A.; Finley, J.L.; Rose, K.L.; Hill, S.; et al. Spermine oxidase mediates Helicobacter pylori-induced gastric inflammation, DNA damage, and carcinogenic signaling. Oncogene 2020, 39, 4465–4474. [Google Scholar] [CrossRef] [PubMed]
- Fratini, E.; Cervelli, M.; Mariottini, P.; Kanamori, Y.; Amendola, R.; Agostinelli, E. Link between spermine oxidase and apoptosis antagonizing transcription factor: A new pathway in neuroblastoma. Int. J. Oncol. 2019, 55, 1149–1156. [Google Scholar] [CrossRef]
- Johnson, J.D.; Mehus, J.G.; Tews, K.; Milavetz, B.I.; Lambeth, D.O. Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes. J. Biol. Chem. 1998, 273, 27580–27586. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Lin, S.R.; Wen, Y.C.; Yeh, H.L.; Jiang, K.C.; Chen, W.H.; Mokgautsi, N.; Huang, J.; Chen, W.-Y.; Liu, Y.-N. EGFR-upregulated LIFR promotes SUCLG2-dependent castration resistance and neuroendocrine differentiation of prostate cancer. Oncogene 2020, 39, 6757–6775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, M.; Liu, M.; Xu, Y.; Wu, G. Glycolysis-Related Genes Serve as Potential Prognostic Biomarkers in Clear Cell Renal Cell Carcinoma. Oxid. Med. Cell. Longev. 2021, 2021, 6699808. [Google Scholar] [CrossRef]
- Chen, F.; Fan, Y.; Cao, P.; Liu, B.; Hou, J.; Zhang, B.; Tan, K. Pan-Cancer Analysis of the Prognostic and Immunological Role of HSF1: A Potential Target for Survival and Immunotherapy. Oxid. Med. Cell. Longev. 2021, 2021, 5551036. [Google Scholar] [CrossRef]
- Shi, Z.-Z.; Tao, H.; Fan, Z.-W.; Song, S.-J.; Bai, J. Prognostic and Immunological Role of Key Genes of Ferroptosis in Pan-Cancer. Front. Cell Dev. Biol. 2021, 9, 748925. [Google Scholar] [CrossRef]
- Ma, B.; Wang, K.; Liang, Y.; Meng, Q.; Li, Y. Molecular Characteristics, Oncogenic Roles, and Relevant Immune and Pharmacogenomic Features of EVA1B in Colorectal Cancer. Front. Immunol. 2022, 13, 809837. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lin, Y.-C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Bi, G.; Bian, Y.; Liang, J.; Yin, J.; Li, R.; Zhao, M.; Huang, Y.; Lu, T.; Zhan, C.; Fan, H.; et al. Pan-cancer characterization of metabolism-related biomarkers identifies potential therapeutic targets. J. Transl. Med. 2021, 19, 219. [Google Scholar] [CrossRef]
- Deng, L.; Jiang, A.; Zeng, H.; Peng, X.; Song, L. Comprehensive analyses of PDHA1 that serves as a predictive biomarker for immunotherapy response in cancer. Front. Pharmacol. 2022, 13, 947372. [Google Scholar] [CrossRef]
- Sung, J.-Y.; Cheong, J.-H. Pan-Cancer Analysis Reveals Distinct Metabolic Reprogramming in Different Epithelial-Mesenchymal Transition Activity States. Cancers 2021, 13, 1778. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Roh, S.; Hong, I.; Kim, H.; Ahn, T.S.; Kang, D.H.; Lee, M.S.; Baek, M.J.; Kwak, K.J.; et al. Expression of Spermine Oxidase Is Associated with Colorectal Carcinogenesis and Prognosis of Patients. Biomedicines 2022, 10, 626. [Google Scholar] [CrossRef]
- Hu, T.; Sun, D.; Zhang, J.; Xue, R.; Janssen, H.L.; Tang, W.; Dong, L. Spermine oxidase is upregulated and promotes tumor growth in hepatocellular carcinoma. Hepatol. Res. 2018, 48, 967–977. [Google Scholar] [CrossRef]
- Chen, S.; Duan, Y.; Wu, Y.; Yang, D.; An, J. A Novel Integrated Metabolism-Immunity Gene Expression Model Predicts the Prognosis of Lung Adenocarcinoma Patients. Front. Pharmacol. 2021, 12, 728368. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.C.; Kenny, H.A.; Grassl, N.; Watters, K.M.; Litchfield, L.M.; Coscia, F.; Blaženović, I.; Ploetzky, L.; Fiehn, O.; Mann, M.; et al. Mesothelial Cell HIF1α Expression Is Metabolically Downregulated by Metformin to Prevent Oncogenic Tumor-Stromal Crosstalk. Cell Rep. 2019, 29, 4086–4098.e6. [Google Scholar] [CrossRef] [PubMed]
- Ghesquière, B.; Wong, B.W.; Kuchnio, A.; Carmeliet, P. Metabolism of stromal and immune cells in health and disease. Nature 2014, 511, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Sukumar, M.; Roychoudhuri, R.; Restifo, N.P. Nutrient Competition: A New Axis of Tumor Immunosuppression. Cell 2015, 162, 1206–1208. [Google Scholar] [CrossRef]
- Qiu, J.; Villa, M.; Sanin, D.E.; Buck, M.D.; O’sullivan, D.; Ching, R.; Matsushita, M.; Grzes, K.M.; Winkler, F.; Chang, C.-H.; et al. Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep. 2019, 27, 2063–2074.e5. [Google Scholar] [CrossRef]
- Huang, Z.; Gan, J.; Long, Z.; Guo, G.; Shi, X.; Wang, C.; Zang, Y.; Ding, Z.; Chen, J.; Zhang, J.; et al. Targeted delivery of let-7b to reprogramme tumor-associated macrophages and tumor infiltrating dendritic cells for tumor rejection. Biomaterials 2016, 90, 72–84. [Google Scholar] [CrossRef]
- Lin, A.; Zhang, J.; Luo, P. Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer. Front. Immunol. 2020, 11, 2039. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaiser, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [PubMed]
- Shimasaki, N.; Jain, A.; Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020, 19, 200–218. [Google Scholar] [CrossRef] [PubMed]
- Niogret, J.; Berger, H.; Rebe, C.; Mary, R.; Ballot, E.; Truntzer, C.; Thibaudin, M.; Derangère, V.; Hibos, C.; Hampe, L.; et al. Follicular helper-T cells restore CD8+-dependent antitumor immunity and anti-PD-L1/PD-1 efficacy. J. Immunother. Cancer 2021, 9, e002157. [Google Scholar] [CrossRef]
- Buchan, S.L.; Fallatah, M.; Thirdborough, S.M.; Taraban, V.Y.; Rogel, A.; Thomas, L.J.; Penfold, C.A.; He, Z.-L.; Curran, M.A.; Keler, T.; et al. PD-1 Blockade and CD27 Stimulation Activate Distinct Transcriptional Programs That Synergize for CD8+ T-Cell-Driven Antitumor Immunity. Clin. Cancer Res. 2018, 24, 2383–2394. [Google Scholar] [CrossRef]
- Chen, B.-J.; Zhao, J.-W.; Zhang, D.-H.; Zheng, A.-H.; Wu, G.-Q. Immunotherapy of Cancer by Targeting Regulatory T cells. Int. Immunopharmacol. 2022, 104, 108469. [Google Scholar] [CrossRef]
- Kim, H.M.; Koo, J.S. Clinicopathologic Characteristics of Breast Cancer According to the Infiltrating Immune Cell Subtypes. Int. J. Mol. Sci. 2020, 21, 4438. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, X.; Jiang, Y.; Cheng, W. Development of an immune gene prognostic classifier for survival prediction and respond to immunocheckpoint inhibitor therapy/chemotherapy in endometrial cancer. Int. Immunopharmacol. 2020, 86, 106735. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, Z.; Li, L.; Ma, M.; Long, F.; Wu, R.; Huang, L.; Chou, J.; Yang, K.; Zhang, Y.; et al. Identification and Validation of Ferroptosis-Related LncRNA Signatures as a Novel Prognostic Model for Colon Cancer. Front. Immunol. 2021, 12, 783362. [Google Scholar] [CrossRef]
- Ugel, S.; Canè, S.; De Sanctis, F.; Bronte, V. Monocytes in the Tumor Microenvironment. Annu. Rev. Pathol. 2021, 16, 93–122. [Google Scholar] [CrossRef]
- Gobert, A.P.; Latour, Y.L.; Asim, M.; Barry, D.P.; Allaman, M.M.; Finley, J.L.; Smith, T.M.; McNamara, K.M.; Singh, K.; Sierra, J.C.; et al. Protective Role of Spermidine in Colitis and Colon Carcinogenesis. Gastroenterology 2022, 162, 813–827.e8. [Google Scholar] [CrossRef]
- Snezhkina, A.V.; Krasnov, G.S.; Lipatova, A.V.; Sadritdinova, A.F.; Kardymon, O.L.; Fedorova, M.S.; Melnikova, N.V.; Stepanov, O.A.; Zaretsky, A.R.; Kaprin, A.D.; et al. The Dysregulation of Polyamine Metabolism in Colorectal Cancer Is Associated with Overexpression of c-Myc and C/EBPβ rather than Enterotoxigenic Bacteroides fragilis Infection. Oxid. Med. Cell. Longev. 2016, 2016, 2353560. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Song, N.; Choi, J.-Y.; Shin, A. Effect of Citric Acid Cycle Genetic Variants and Their Interactions with Obesity, Physical Activity and Energy Intake on the Risk of Colorectal Cancer: Results from a Nested Case-Control Study in the UK Biobank. Cancers 2020, 12, 2939. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Zhang, Y.; Song, F.; Han, Y.; Ding, Y. A newly identified small molecular compound acts as a protein kinase inhibitor to suppress metastasis of colorectal cancer. Bioorg. Chem. 2021, 107, 104625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lin, L.; Xia, L.; Cai, W.; Dai, W.; Zou, C.; Yin, L.; Tang, D.; Xu, Y.; Dai, Y. Multi-omics analyses of human colorectal cancer revealed three mitochondrial genes potentially associated with poor outcomes of patients. J. Transl. Med. 2021, 19, 273. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Z.; Lin, Y.; Yang, Y.; Li, W.; Huang, W.; Zhang, S. Metabolism-Related Genes SMOX and SUCLG2 as Immunological and Prognostic Biomarkers in Colorectal Cancer: A Pan-Cancer Analysis. Curr. Issues Mol. Biol. 2025, 47, 465. https://doi.org/10.3390/cimb47060465
Xiong Z, Lin Y, Yang Y, Li W, Huang W, Zhang S. Metabolism-Related Genes SMOX and SUCLG2 as Immunological and Prognostic Biomarkers in Colorectal Cancer: A Pan-Cancer Analysis. Current Issues in Molecular Biology. 2025; 47(6):465. https://doi.org/10.3390/cimb47060465
Chicago/Turabian StyleXiong, Zuming, Yirong Lin, Yongjun Yang, Wenxin Li, Wei Huang, and Sen Zhang. 2025. "Metabolism-Related Genes SMOX and SUCLG2 as Immunological and Prognostic Biomarkers in Colorectal Cancer: A Pan-Cancer Analysis" Current Issues in Molecular Biology 47, no. 6: 465. https://doi.org/10.3390/cimb47060465
APA StyleXiong, Z., Lin, Y., Yang, Y., Li, W., Huang, W., & Zhang, S. (2025). Metabolism-Related Genes SMOX and SUCLG2 as Immunological and Prognostic Biomarkers in Colorectal Cancer: A Pan-Cancer Analysis. Current Issues in Molecular Biology, 47(6), 465. https://doi.org/10.3390/cimb47060465




