1. Introduction
Plants are threatened by a diversity of pathogen species living in complex communities, including also a variety of other non-pathogenic microorganisms. A plant’s pathobiomes can be defined as a collection of coexisting phytopathogens that affect each other and the plant [
1]. They are formed by pathogens inhabiting the same ecological niche and either cooperating or competing for the same plant resources. Two or more phytopathogens on the same host can result in significantly different disease outcomes compared to single infections [
2]. The communities of natural microorganisms inhabiting the plant phyllosphere, the aboveground portions of the plant’s habitat, or the rhizosphere, the roots’ surrounding habitat, also include non-pathogenic members that can have protective effects against pathogens. For example, it has been shown lately that maize (
Zea mays) grains are populated with a diversity of fungi and bacteria [
3], some of which are known phytopathogens such as
Alternaria alternata and
Fusarium proliferatum, which can be opportunists, causing disease when the conditions are favorable. Other members of the maize grains’ microflora are beneficial bioprotective species, including the fungal species
T. asperellum,
Penicillium citrinum,
Chaetomium cochliodes, and the bacteria
Bacillus subtilis.
The interactions between microorganisms occur in the soil of commercial fields before sowing, and their outcome can drastically affect disease severity [
4]. In cotton (
Gossypium hirsutum), interactions between
Fusarium oxysporum, a wilt agent, and
Magnaporthiopsis maydis, a root-surface inhabitant and assumedly harmless endophyte, were shown to have an intriguing result [
5]. These interspecies relationships are associated with an appreciable decrease in the severity of cotton wilt disease. The infection reduction was maximum when
M. maydis preceded
F. oxysporum in the soil compared to when they were inoculated simultaneously. Co-inoculation of the plants with both fungi resulted in some protection compared to
F. oxysporum alone. In contrast, when
M. maydis was added to the ground after
F. oxysporum, it exerted little or no protective effect [
5].
In a previous study, we reported similar antagonistic relationships between
M. maydis and
Macrophomina phaseolina, the charcoal rot disease (CRD) agent in cotton plants [
2,
6]. The fungus
M. maydis (synonyms are
Cephalosporium maydis [
7] and
Harpophora maydis [
8]) is known as the maize late wilt disease causal agent [
6,
9]. This disease severely affects sensitive maize plants at the maturity stage [
10,
11,
12].
M. maydis was recently identified in several secondary hosts, including cotton (Pima or Goliath cv.) [
13]. The fungus is associated with other plant pathogenic fungi such as
Fusarium verticillioides, the stalk rot agent, and
M. phaseolina, causing maize post-flowering stalk rot diseases [
14].
In cotton plants,
M. maydis infection caused increased growth in lateral roots and the presence of shallow cracks and dark red lesions on young roots up to 45 days after sowing (DAS) [
5]. Still, no such aboveground effects were observed throughout the growth of cotton plants up to maturity. Additionally,
M. maydis was not recovered from these symptomatic cotton plants [
5]. It was recently reported that
M. maydis infection reduced wet biomass and phenological development in young cotton plants in a growth chamber [
13]. However, tracking
M. maydis infection on cotton plants throughout the whole season in a two-year study shows that the fungi did not affect cotton growth parameters or yield [
2]. While this result was obtained in a regular irrigation regime, under drought stress,
M. maydis infection led to decreased weight and height of the aboveground parts of cotton plants. Taking these lines of evidence together, it should be considered that some opportunistic behavior of this pathogen could exist.
The common pathogen
M. phaseolina (Tassi) Goidanich affects a wide range of cultivated and wild species in temperate, warm, and tropical regions worldwide [
15]. The fungus has multiple host plants and is a threat to more than 500 plant species [
16], including maize and cotton. Like
M. maydis,
M. phaseolina is a primarily soil-borne and seed-borne pathogen, but unlike
M. maydis, it has a highly competitive saprophytic capability [
17]. Both fungi have a similar pathogenic action mode and cause similar growth reduction and wilt symptoms. These species are a challenge to control because they can survive for long periods in the ground. The fungi penetrate the sprouts’ roots, causing damping-off, root necrosis, and sprout development restriction [
16,
18].
M. phaseolina and
M. maydis may spread upwards inside the plant’s xylem. Typically, the symptoms develop later in the season near the flowering stage and are enhanced under drought conditions [
19,
20]. When the growing season progresses, the pathogens block the water supply and lead to dehydration and yield reduction [
11,
21]. Both
M. phaseolina and
M. maydis culture filtrate negatively affect maize seed development [
22].
M. phaseolina can produce phytotoxins [
16], while
M. maydis-secreted phytotoxins have not yet been identified.
The interactions between
M. phaseolina and
M. maydis are particularly interesting since both pathogens can be found abundantly in Israeli commercial fields that share the two summer crop hosts, cotton and maize [
2,
13]. These crops are cultivated alternately in two-year rotations in Israel and Egypt [
5]. Even though neither of the pathogens was superior in a growth plate confrontation assay, their co-inoculum significantly influenced the plants under field conditions. Compared to inoculation with each fungus alone, when the soil was co-inoculated by both, their antagonistic interactions were significantly reflected [
2]. Their coexistence within the plants led to growth promotion, yield improvement, and a decrease in disease symptoms. Interestingly, the dual infection led to decreased
M. maydis DNA and increased
M. phaseolina DNA levels in the plants’ stem tissue.
The current study explores these pathogen–pathogen interactions and their influence on cotton plants during the sprout growth phase. For this purpose, sprouts were inoculated with M. phaseolina and M. maydis, together or one after the other, before, during, and after sowing. The plants’ aboveground emergence percentages, phenological progression, and health status indices were used to evaluate the outcome of these pathogenic interactions. Furthermore, the molecular tracking of both fungi DNA inside the host plant’s roots using quantitative real-time PCR (qPCR) enables a close evaluation of their co-influence inside their host habitat. A deeper understanding of such interactions could uncover hidden data layers regarding controlling the pathogens, their virulence, and the plant’s immunity and assist in developing new plant protection strategies.
2. Materials and Methods
2.1. Fungal Species and Growth Conditions
The
M. maydis isolate
Hm-2 (CBS 133165, CBS-KNAW Fungal Biodiversity Center, Utrecht, The Netherlands) was isolated from diseased maize plants collected from Sde Nehemia (Hula Valley, Upper Galilee, northern Israel) in 2001 and identified. Like the other isolates, which were also deposited in CBS-KNAW in the same collection, this representative
M. maydis strain was previously characterized by its pathogenicity, physiology, colony morphology, and microscopic traits [
23,
24]. The morphological and microscopic characteristics of the pathogen were similar to strains described earlier in Egypt and India [
7,
25]. Using PCR-based DNA analysis [
24,
26], a final confirmation of those strains was achieved.
The
M. phaseolina isolate
Mp-1 was recovered from wilted cotton plants in 2017 (Roni Cohen’s lab, Newe Ya’ar Research Center, Ramat Yishay, northern Israel) and identified using pathogenicity, physiology, colony morphology, microscopic characteristics, and molecular traits [
2]. The fungi were grown on solid, rich potato dextrose agar (PDA) (Difco, Detroit, MI, USA) medium in the dark at 28 ± 1 °C for 4–7 days.
2.2. Cotton Cultivar Selected for This Study
Pima cotton, Goliath cv. (extra-long staple (ELS) cotton), was cultivated routinely in different regions of Israel (supplied by Israel Seeds, Kibbutz Shefaim, Israel). This cotton cultivar was also grown on late wilt-contaminated fields during crop rotation and was recently reported to be
M. maydis-vulnerable at the early growth stages [
13]. In a survey conducted by Roni Cohen across Israel (Newe Ya’ar Research Center, northern Israel), Pima cotton, Goliath cv., was found to be sensitive to
M. phaseolina charcoal rot disease [
27].
2.3. Seedlings Experiment in a Growth Chamber under Controlled Conditions
The pot experiment was performed twice using the same experimental design. The results of both experiments are presented. The first repeat also included acute CRD cotton sprouts induced by puncturing them with infected wooden toothpicks, as will be described in
Section 2.5.
Two-liter pots were filled with 70% commercial, non-sterilized garden soil and 30% Perlite No. 4 for aeration. The commercial garden soil mixture (Garden Mix, Deshanit, Be’er Yaakov, Israel) comprised fibers, coconut peat, a relatively low amount of tuff, and Osmocote (ScottsMiracle-Gro, Marysville, OH, USA), a 3–4 month slow-release fertilizer. The Pima cotton cv. seeds were soaked for about 15 min in distilled water and sown in pots to a depth of 5 cm (five seeds per pot). The pots were kept in a growth room with a photoperiod of 8 h of light and 16 h of darkness. All the plants were grown under the same conditions of constant humidity of 45% and a temperature of 25 ± 2 °C. Immediately after sowing, the pots were irrigated to initiate germination. Irrigation was carried out after the sprouts’ first emergence, 100 mL once every two days, using a computerized irrigation system. Throughout the experiment, treatments against various pests and fertilization treatments were performed according to instructions by the Israel Ministry of Agriculture Consultation Service (SAHAM). The experiments were conducted in 6 repetitions per treatment, whereby each repetition is a pot containing five sprouts.
2.4. The Preparation of Infected Sterilized Wheat Grains
Infected sterilized wheat grains were used to spread the fungi in the soil, as previously reported [
12]. This experimental setting, using sterilized wheat grains substrate, is a common method to induce maximum inoculation pressure under controlled conditions. This is especially needed when pathogens with a limited saprophytic ability (such as
M. maydis) are involved.
Wheat grains were soaked in tap water overnight. The grains were then dried for about four hours in a fume hood on paper towels and autoclave-sterilized for 30 min at a temperature of 120 °C. Disinfected plastic 0.5 L boxes were used to inoculate 100 g sterilized wheat grains with 10 mycelial discs of each fungal species,
M. maydis or
M. phaseolina (separately). Mycelia disks (6 mm in diameter) were taken from the margins of a 4–6-day-old fungal colony grown as described in
Section 2.1. The boxes were sealed with a lid that was tightened to the box using Saran wrap, covered with aluminum foil to guarantee dark conditions, and incubated at 28 ± 1 °C in the dark for 10 days.
2.5. The Inoculation Method in the Potted Plants’ Experiments
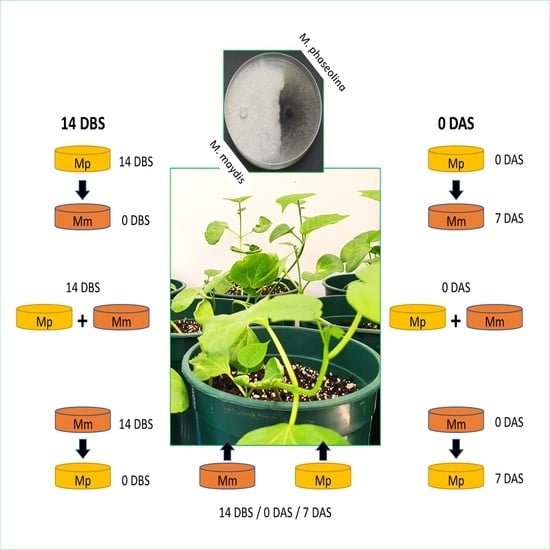
The experiment comprised the following treatments: M. maydis inoculation, M. phaseolina inoculation, and dual fungi inoculation. Each infection treatment was implemented on three dates: 14 days before sowing (DBS), at sowing, or 7 days after planting (DAS). In addition, the infection order—one fungus after the other—was examined. This setting aimed at investigating possible advantages for each pathogen when it preceded the other in the soil. In both seedling experiment repetitions, we also included a non-infected negative control.
In the pre-sowing treatment, the pots were inoculated two weeks before planting with infected sterilized wheat grains, prepared as described in
Section 2.4. In each pot, the soil’s top layer up to a depth of 10 cm root area was mixed with 12 g of those grains enriched with the selected fungus. In the other treatments, where inoculation was done with seeding or one-week post sowing, three fungal discs 6 mm in diameter, taken from the margins of a 4–6-day-old fungal colony grown as described in
Section 2.1, were added to each seed/seedling. This procedure is conducted by making a small hole in the soil adjacent to the plant, inserting the fungal discs to a depth of 5 cm, and sealing it with soil.
Another treatment group was set up to induce acute CRD in the cotton sprouts. This procedure was conducted by puncturing the sprouts, as previously reported [
28]. To this end, infected wooden toothpicks were prepared by adding sterilized toothpicks to the surface of PDA plates seeded with
M. maydis or
M. phaseolina. The plates were kept at 28 ± 1 °C in the dark for about one week until the fungal hyphae covered the toothpicks (
Figure 1). At 14 DAS (when the plants were mature enough to perform this procedure), each plant was stabbed in its lower stem part near the ground with
M. maydis- and/or
M. phaseolina-infected wooden toothpicks. Each stalk was inoculated once by puncturing and leaving the infected wooden toothpick stuck in the stem. In the
M. maydis and
M. phaseolina combined puncturing inoculation, the stalk was inoculated by stabbing with two toothpicks, one for each fungus.
2.6. Data Collection during and at the End of the Growth Period
The emergence degree of the plants revealing the tip of the coleoptile that grew aboveground was determined 10 DAS. At the end of the experiment (day 40), all seedlings were gently removed from the ground, thoroughly rinsed with running tap water, and dried using paper towels. The plants’ phenological development estimation included wet biomass and the number of leaves. In addition, the survival rate was evaluated, and a 0.7 g root sample was taken from each pot for DNA extraction.
2.7. DNA Extraction
For DNA purification, the plants’ roots were washed twice in sterile DDW for 30 s. Tissues were sampled by removing a cross-section of approximately 2 cm in length from each plant’s root and near-surface hypocotyl under a sterile biosafety hood. Samples from the five plants of each plant pot were combined and considered as one repeat. The total weight was adjusted to 0.7 g and considered one repeat. All tissue samples were inserted into universal extraction bags (Bioreba, Switzerland) and 4 mL of CTAB buffer (0.7 M NaC1, 1% cetyltriammonium bromide (CTAB), 50 mM Tris-HC1 pH 8.8, 10 mM EDTA, and 1% 2-mercaptoethanol) was added. The tissues were ground with a tissue homogenizer (Bioreba, Switzerland) for five minutes until they were completely homogenous. The DNA samples were suspended in 100 microliters of ultra-pure quality water and stored in a freezer at −20 °C until used in the qPCR reaction.
The homogenized samples were treated for DNA purification according to the Murray and Thompson procedure [
29] with slight modifications [
10]. First, 1.2 mL of each DNA sample in CTAB buffer was incubated for 20 min at 65 °C. Then, the DNA samples were centrifuged at 14,000 rpm for 5 min at room temperature (24 °C). The upper phase of the lysate (usually 700 µL) was then extracted with an equal volume of chloroform/isoamyl-alcohol (24:1). After mixing by vortex, the blend was centrifuged again at 14,000 rpm for 5 min at room temperature. This stage of chloroform/isoamyl alcohol extraction was repeated twice. The supernatant, usually 300 µL, was then separated into a new Eppendorf tube and mixed with cold isopropanol (2:3). The DNA solution was mixed gently by inverting the tube several times, kept at −20 °C for 20–60 min, and centrifuged (14,000 rpm at 4 °C for 20 min). The precipitate DNA was isolated and resuspended in 0.5 mL of 70% ethanol. After another centrifugation (14,000 rpm at 4 °C for 10 min), the precipitate DNA was isolated and left to dry in a sterile hood overnight. Finally, the DNA was suspended in 100 µL HPLC-grade water and kept at −20 °C until use.
2.8. Real-Time PCR-Based Molecular Test
The qPCR reactions were performed as previously described [
10], using the ABI PRISM
® 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) for 384-well plates. The molecular method is based on a standard qPCR protocol used to detect mRNA (converted to cDNA) [
30]. Instead, it was modified to detect the DNA of the pathogen
M. maydis using species-specific primers [
31,
32]. The A200a primers set was used for
M. maydis detection (200 bp species-specific fragment) [
24], while the MpK FI RI primers were used for
M. phaseolina detection (300–400 bp species-specific fragment) [
33]. The primers’ sequences are listed in
Table 1.
The housekeeping
COX gene encoding the enzyme cytochrome c oxidase—the mitochondria’s last enzyme in the cellular respiratory electron transport chain—was aimed at normalizing the
M. maydis pathogen DNA [
35]. The
COX gene is used as a reference “housekeeping” gene representing the samples’ total plant and fungal DNA. This gene was designated to normalize the amount of
M. maydis DNA and was proven sufficiently stable in many similar works (see, for example, refs. [
10,
12,
28,
36,
37]). The use of the
COX gene provides a steady, strong, and easily detectable qPCR reading value after 22–25 cycles with minor variations and values that rarely exceed this range. The COX amplification was done using the COX F/R primer set [
34,
35] (
Table 1). The calculation of relative gene abundance was made according to the ΔCt model [
38]. Similar efficacy was assumed for all samples. All amplifications were performed in triplicate.
The qPCR conditions were as follows: 5 µL total reaction volume was used per sample well—2 µL of DNA sample extract, 2.5 µL of iTaq™ Universal SYBR Green Supermix (Bio-Rad Laboratories Ltd., Hercules, CA, USA), and 0.25 µL of each of the forward and reverse primers (10 µM from each primer per well). The qPCR cycle program was as follows: pre-cycle activation phase (1 min at 95 °C), denaturation (15 s at 95 °C) for 40 cycles, and annealing and extension (30 s at 60 °C). A melting curve was used to analyze the results.
2.9. Statistical Processing
All statistical processing was done using JMP software, 15th edition (SAS Institute Inc., Cary, NC, USA). To test the significance of the results, we used a one-way analysis of variance (ANOVA) and a post hoc Student-based t-test for each pair of averages, without correction based on multiple tests. The significance threshold was p < 0.05. The random effect on the results of the biological repeats, the fungus-alone compared to the co-inoculation, and pre-inoculation compared to post-inoculation was insignificant. The experiment’s biological repeats 1 and 2 are presented separately in the Results section and analyzed comparatively to each other.
3. Results
The relationships between M. phaseolina and M. maydis and their impact on cotton plants at their early growth stage under controlled conditions were studied. Sprouts were inoculated with M. phaseolina and M. maydis, together or one after the other, before, during, and after sowing. The pot experiment was performed twice with the exact experimental design, and the results of both experiments’ repeats are presented. The two biological repeats obtained some similar results but also revealed an inquisitive perspective of the role of M. maydis in the sprouts’ CRD outcome. This pathogen caused significant disease symptoms when applied alone before the seedling. However, it acted differently in the presence of M. phaseolina in the experiment’s two biological repeats. In the first repetition, M. maydis pre- or combined infection shielded the plants against the CRD agent’s destructive impact. In contrast, in the second repeat, M. maydis was virulent, causing harsh disease symptoms in all combinations tested. These pathogenic variations will be described and analyzed in detail below.
3.1. The Experiment’s First Biological Repeat
3.1.1. Sprouts’ Aboveground Emergence
The combined effect of the two soil pathogens,
M. phaseolina and
M. maydis, was tested at the plant’s early aboveground emergence stage (
Figure 2). The aboveground emergence estimation performed 10 days after sowing (DAS) shows that the two pathogens harmed the sprouts when applied separately 14 days before sowing (DBS) compared to soil inoculation with sowing. In those early soil infection treatments, the emergence percentages dropped to 33% compared to the non-infected control, in which 96% of the plants emerged above the ground surface. In contrast, inoculating the soil along with sowing resulted in a lesser emergence inhibition effect. The difference between the two infection dates (14 DBS and 0 DAS) was an emergence improvement of 25% in
M. phaseolina treatment (without a statistical difference) and 28% in
M. maydis (
p < 0.05) treatment.
Combining the two pathogens 14 DBS or on the sowing day caused mutual suppression and higher emergence indices (58% compared to 33% when the plants were treated with pathogens separately 14 DBS). This difference, dual vs. single infection, was statistically significant regarding M. maydis. A noticeable emergence improvement was found when M. maydis inoculation preceded M. phaseolina. The emergence was 64% and 58% in the M. maydis pre-treatment applied 14 or 0 DBS, respectively, compared to 47% and 44% in the M. phaseolina pre-treatment, respectively, on the same date.
3.1.2. Plants Development
At age 40 DAS (
Table 2), the plants’ growth indices show trends similar to the emergence values. The early infection of the soil 14 DBS with each of the pathogens had a significant (
p < 0.05) inhibitory effect on growth in terms of the plants’ wet weight and the number of leaves. This adverse effect was reduced when the inoculation was performed on the sowing day, when both pathogens were applied together, or when
M. maydis preceded
M. phaseolina in the soil, 14 DBS for the former fungus and 0 DAS for the latter.
Indeed, there is a close similarity to the non-infected control of the
M. phaseolina treatment alone at sowing (
Mp 0 DAS) and the
Mm 14 DBS +
Mp 0 DAS treatment regarding all measures in
Table 2. This evidence suggests that the addition of
M. phaseolina alone with the sowing has a weak impact on the plants’ growth indices and survival. Still, it shields them against the devastating effect of the 14 DBS soil pre-inoculation with
M. maydis alone.
3.1.3. Sprouts’ Survival Rate
The treatments had a prominent effect on the plants’ survival rate. In the control group, 96% of the plants survived, while the sole infection of both pathogens 14 days post-seeding led to significant (
p < 0.05) sprout death (only 33–43% were vivid). A slight improvement (50% survival rate) was achieved when
M. maydis was added to the soil after
M. phaseolina, regardless of the soil infection day. Better survival percentages (63–70%) were recorded in the 14 and 0 DBS combined infection. Those values were still significantly lower than the control. The best survival rates (70–73%) were achieved when the fungi were added separately to the soil on the sowing day or when
M. maydis had inhabited the soil 14 DBS before
M. phaseolina (
Table 2).
3.1.4. The Pathogens’ DNA in Plant Roots
Real-time PCR-based molecular monitoring of the pathogens
M. maydis and
M. phaseolina DNA in plant roots on day 40 of growth (
Figure 3) indicates, in most cases, a correlation between the presence of pathogens and the severity of symptoms (emergence values,
Figure 2 and growth indices,
Table 2). In analyzing the qPCR results, no statistical significance was found due to high natural variations in the fungi pathogenesis. The highest DNA amount of
M. maydis (4.3 × 10
−4) was recorded when the infection was done on the sowing day alone or at sowing, before infection with
M. phaseolina was performed one week later. The
M. maydis DNA level was especially low (7.2 × 10
−5) when the soil had been previously inoculated with
M. phaseolina.
Changes in
M. phaseolina DNA amounts in those plants were also noticeable (
Figure 3). In contrast to
M. maydis, the cotton CRD agent (
M. phaseolina) DNA levels were higher (4.2 × 10
−3) when the pathogen was added to the soil 14 DBS compared to on the sowing day (1.4 × 10
−3). Simultaneous inoculation in both pathogens or pre-treatment in
M. maydis 14 DBS or on the sowing day dramatically reduced the amount of DNA of the cotton pathogen (ca. 10-fold reduction). However, pre-inoculating the soil with
M. phaseolina led to high levels of this pathogens’ DNA levels, in accordance with the growth reduction results.
3.1.5. The Puncturing Inoculation Effect
The addition of puncturing inoculation with wooden toothpicks in the lower stem section (
Figure 1) produced mechanical damage to the plants, which could induce acute infection. This procedure was conducted two weeks after sowing when the plants were mature enough, causing a drastic reduction in growth indices, as expected. The outcome of this intervention on the growth indices was especially noticed in the combined infection by both fungi or when
M. phaseolina preceded
M. maydis (
p < 0.05,
Table 2). These results were similar to the severe disease symptoms recorded in the second experiment’s repeat, detailed in
Section 3.2.
Infecting the sprouts with
M. maydis at sowing and infecting them again by puncturing with
M. phaseolina two weeks later led to an 8.5-fold increase in
M. maydis DNA compared to the reverse procedure (
Mp 0 DAS +
Mm 14 DAS St.) (
Figure 4). This procedure of
M. maydis first followed by
M. phaseolina stabbing inoculation caused
M. phaseolina DNA values to drop to near-zero levels (ca. 100-fold lower levels). These tendencies are similar to those obtained without stabbing (
Figure 3).
3.2. The Experiment’s Second Biological Repeat
In the first experiment, M. maydis infection shielded the plants from the full impact of CRD when applied before or with M. phaseolina. However, in the second repeat, the picture was different. M. maydis enhanced the disease symptoms rather than eliminating them. This pathogenic behavior change led to harsh disease symptoms in almost all of the treatments.
3.2.1. Sprouts’ Aboveground Emergence
Similar to the first repetition, here, too, the two pathogens harmed the sprouts when applied separately 14 DBS (
Table 3). This infection procedure caused the emergence values to drop from 96% emergence in the non-infected control to 64–68% (
p < 0.05). Compared to the 14 DBS results, the separate infection on the sowing day had a lesser effect in the
M. phaseolina treatment (84% emergence) but a stronger impact in the
M. maydis treatment (55% emergence). The combined infection caused close to 60% aboveground sprouting, with minor differences between the dates 14 and 0 DBS.
Interestingly, it appears that the addition of M. phaseolina to the soil 14 DBS shielded the plants from the devastating effect of M. maydis, which was added afterward. This 14 DBS pre-inoculation with M. phaseolina caused drastic emergence recovery (72%). Yet, applying this sequence inoculation on sowing days (Mp 0 DAS + Mm 7 DAS) failed to protect the plants, and only 32% of them emerged above the ground surface. In contrast, sequenced soil inoculation with M. maydis before M. phaseolina caused acute emergence suppression on both dates applied (48% and 36%).
3.2.2. Plants Development
Forty days after sowing (
Table 4), the plants’ growth indices had the same tendencies as the emergence parameters. Early soil infection 14 DBS with
M. maydis or
M. phaseolina had an inhibitory effect on the plants’ wet weight and the number of leaves—6–16% and 31–33% reduction, respectively (statistical significance measured in the number of leaves). As in the first biological repeat, soil inoculation with
M. phaseolina at sowing had a lesser impact on the leaves number (weight value was slightly reduced). However, unlike the first repeat,
M. maydis infection with sowing caused more severe growth inhibition (44% and 54% wet weight and leaves number reduction).
When M. maydis combined or preceded M. phaseolina in the soil 14 DBS, the growth depression was more severe than the single infection (38% fresh weight and 40–52% leaves number reduction). Applying the pathogens in a sequence with M. phaseolina first at 14 DBS had some protective effect against the destructive activity of M. maydis. Yet, sequence infection starting from sowing, regardless of the pathogens’ order, resulted in poor growth indices (59–69% biomass and 62–71% leaves number reduction).
3.2.3. Sprouts’ Survival Rate
Regarding the plants’ survival percentages at the experiment end, the second repetition (
Table 4) also revealed quite a different picture from the first repeat. Here,
M. maydis or
M. phaseolina sole infection 14 DBS caused a minor survival decrease (70% compared to 96% in the control treatment). However, decreased survival rates were caused when
M. maydis was applied alone with the sowing (52%) or with
M. phaseolina simultaneously (60–63%), or even more in a sequence where
M. maydis is the first 14 DBS or at sowing (43–53%). At the same time, the addition of
M. phaseolina before
M. maydis 14 DBS rescued these values (70% survival rate). This survival’s protective impact was not observed when the inoculation sequence
M. phaseolina–
M. maydis was started at the seeding (
Mp 0 DAS +
Mm 7 DAS, 32% survival rate).
3.2.4. The Pathogens’ DNA in Plant Roots
In the second experiment’s repeat, the highest DNA values of each of the pathogens were recorded in the sole infection (
Figure 5). A peak of 0.5 relative DNA was recorded when
M. maydis was applied alone in the soil 14 DBS, a nearly 1000 times higher value than the same treatment in the first experiment’s repeat. In the same procedure on the sowing day, the
M. maydis DNA levels dropped by half to 0.25. Still, those values were higher than the
M. phaseolina pre-treatment. Moreover, they were higher by more than two orders of magnitude than the combined infection and
M. maydis pre-inoculation treatments. Similar to fluctuations in the growth parameters, combined infection on 14 DBS resulted in higher
M. maydis DNA levels than combined infection with seeding. Moreover, pre-treatment with this pathogen in a sequence with
M. phaseolina starting from 14 DBS was less infectious than at sowing.
Changes in the DNA amounts of the
M. phaseolina pathogen in those plants were similar to those of
M. maydis, except for the ordered inoculation when
M. maydis was applied first (
Figure 5H). Soil sequence inoculation with
M. maydis first, starting 14 DBS, was more potent (ca. 10-fold higher CRD pathogen DNA levels) than implementing this sequence from the sowing day. This is in contrast with the higher growth and survival indices measured in the 14 DBS sequence treatment.
3.3. Similarities and Differences between the Two Experiments’ Repeats
The two experiment’s biological repeats had some similar results and some differences, as summarized in
Figure 6. Similarities in both repetitions’ results include a strong impact on the sprouts’ development and survival (86%) when each pathogen is applied separately 14 DBS. A similar severe result (75–86%) was the sequence inoculation when
M. phaseolina was added to the soil before
M. maydis 14 or 0 DBS. In contrast,
M. phaseolina sole infection on the sowing days had a minor or no effect on the sprouts.
All other treatments varied between the two experiment’s repeats. The main difference was the strong growth depression influence of the treatments in the second repeat. Indeed, the total disease severity estimation based on combining all symptoms measured was 40% in the first experiment and 80% in the second. The cause of this result is presumed to be the high virulence of M. maydis in this repeat since sole infection with this pathogen with seeding led to drastic growth repression.
4. Discussion
In Israel,
M. maydis and
M. phaseolina can be found in commercial cotton and maize fields [
13]. While the former is considered a major threat to sensitive maize cultivars [
37], it has an endophytic or opportunistic lifestyle in cotton plants [
2,
5].
M. phaseolina is the primary causal agent of cotton charcoal rot disease (CRD). This disease has become of increasing concern to Israel’s cotton production over the past decade. Since both pathogens can be found in diseased cotton plants, they may contribute to the disease’s outbreaks and spread [
13,
39]. However, when the two fungi had been previously inoculated simultaneously in the soil, CRD symptoms were prominently reduced while the plants’ growth and yield improved [
2]. It was concluded that competitive interactions between the two fungal species are the cause of this outcome. This antagonistic co-influence resulted in restricting the spread of
M. maydis in maize plants and of
M. phaseolina in cotton.
The current study aimed at deepening our understanding of these intriguing relationships found in cotton sprouts. Indeed, the results from two independent growth room trials indicate that the picture is more complex than a simple two-directional antagonism. Variations in the CRD severity or the pathogens’ virulence probably exist. M. maydis can act as a bioprotective endophyte in the presence of M. phaseolina. Supporting this are the first experiment’s repetition results. Simultaneous infection by both pathogens or pre-treating the soil with M. maydis 14 DBS or on the sowing day dramatically reduced the amount of CRD agent (M. phaseolina) DNA in the plants’ roots and improved the sprouts’ growth indices and survival. This result was achieved when sole inoculation of M. maydis on the sowing day had a mild influence on the experiment’s first repeat.
Where
M. maydis inoculation with sowing had an aggressive impact in the second experiment’s repetition, it had a prominent effect on the plants’ growth and survival. In this scenario, the pre-inoculation of the soil 14 DBS with
M. phaseolina protected the plants. Thus, mutual inhibition between
M. phaseolina and
M. maydis exists, and each of the partners restricts the other fungal population from developing uncontrollably. One possible explanation is that antagonism is formed in situ since both fungi occupy the same space. Therefore, the fungi interfere less with plant growth. An alternative explanation is that the antagonism directly results from inhibitory metabolites secreted by both fungi [
2].
The qPCR results raise an interesting question—why are there different tendencies between 14 and 0 DBS when the pathogens are applied alone? It appears that M. maydis could successfully inoculate the plants when added at sowing. In contrast, M. phaseolina is best established in the plants when added to the soil at 14 DBS. This is emphasized even more when M. maydis is inoculated at 0 DAS and M. phaseolina at 7 DAS.
Tracking the effect of
M. maydis soil inoculation on cotton plants under field conditions throughout the whole season in a two-year study shows that this fungus did not affect cotton growth parameters or yield [
2]. These results imply that the maize pathogen maintained an endophytic lifestyle in cotton plants. Yet, under drought stress,
M. maydis infection led to the decreased growth of the cotton plants, without any measurable effect on yield production [
2]. So, some opportunistic behavior of this pathogen in cotton could exist. The current study’s results are in line with this conclusion. The second experiment’s repeat conditions were probably favorable for the pathogenic behavior of
M. maydis and evoked its aggressive impact. This assumption and the nature of these interesting relationships and their causes over a full growth period will require further work to be resolved.
Similar antagonistic interactions have been previously reported in phytoparasitic fungi pairs of other plants. One example is the suppression of
Cochliobolus sativus by the
Fusarium (
roseum) species [
40]. In this case,
C. sativus and
F. pseudograminearum consistently and significantly reduced one another under field conditions. Another example is the interactions of
F. oxysporum and
F. solani on pea roots [
41]. While these pathogens inhibit one another, co-infection by other pathogens may result in a different outcome—increased disease severity. For example, roots infected with
Aphanomyces euteiches were more susceptible to
Fusarium root rot than those exposed only to
Fusarium spp. [
42]. This observation was confirmed by qPCR, which revealed significant changes in colonization rates when multiple species were present. Yet, the underlying mechanism in the
M. maydis and
M. phaseolina co-infection seems somewhat different. According to former studies,
M. maydis does not cause actual disease in cotton. It is a root and stem inhabitant [
2,
5,
13] that may provide some defense against other intruding soil pathogens. For example, Sabet et al. [
5] demonstrated that the interaction between the cotton pathogen
F. oxysporum and
M. maydis on the roots induces a decrease in the severity of
Fusarium cotton wilt.
It is curious why M. maydis failed to protect the plants when it was added to the soil after M. phaseolina. It is possible that the inhibitory metabolites produced by M. maydis did not reach an effective dose before the fungus had grown for some time. Thus, the action of M. maydis against infection with the CRD fungus is considerably reduced when inoculated into the soil after M. phaseolina and is enhanced when inoculated before it. While this assumption should be based on future studies, it should also be considered that other partners in the soil microbiome may be involved. Since the soil and roots microflora include many different microorganisms, the fabric of the relationship between them most probably affects the interactions between M. maydis and M. phaseolina as well. Yet, this study’s results provide an essential opening stage for subsequent studies and implications for future control strategies development. The findings suggest a decreased risk of yield loss in regions where M. maydis and M. phaseolina co-occur, especially when the former precedes the latter.
The role of
M. maydis as an endophyte in cotton is still unclear. This is also true for other fungal endophytes such as pathogens and harmless species. Until lately, studies of fungi in cultivated cotton have focused primarily on monitoring and controlling plant pathogens [
43]. Today, there is increasing interest in asymptomatic fungal endophytes, and attempts are being made to unravel their potential as biological control agents. Still, surveys are needed to better characterize their distribution patterns, diversity, and possible implementation in integrated pest management. In such a survey, the pathogenicity of 42 isolates was tested on cultivated cotton in Australia [
44]. All isolates caused a localized discoloration in stem tissue when infection was performed with the stem-stabbing method. Still, none of them could induce any foliar symptoms during the experiment’s five-week period, suggesting that the endophytic fungi of native
Gossypium species are unlikely sources of cotton pathogens. In cotton cultivated in the United States, members of the genera
Alternaria,
Colletotrichum, and
Phomopsis, as well as the species
Drechslerella dactyloides (formerly
Arthrobotrys dactyloides) and
Exserohilum rostratum, were isolated and identified as endophytes [
43]. In addition, many latent pathogens were found. Some of those latent species are known to be antagonists against plant pathogens. Interestingly, no differences were found in endophyte species diversity among different cotton varieties or richness. Yet, the researchers did detect differences in plant tissues and during the growth period [
43].
The above-mentioned data and other scientific studies together suggest that pathogenic soil biota, particularly soil-borne fungi, play an essential role in regulating plant productivity, i.e., biomass [
45]. This hypothesis holds that plant species accumulate species-specific pathogens that reduce their host’s performance, especially when the plant diversity is low, i.e., in monocultures where the relative abundance of the host is 100%, such as in commercial fields. This claim relies on crop yield reductions from pests, which typically increase with repeated crop cultivation in the same area and can be reduced by crop rotation [
46]. Moreover, fungicide treatment or sterilization of the soil enhanced plant biomass production, even at low plant species richness [
47,
48].
Bioassays that inoculate plant species with isolates of one or two fungi may reveal host-specific pathogenic effects and provide proof of principle that these belowground organisms can act as pathogens and affect various plant species differently. However, such single-fungus single-plant bioassays are relatively simple [
45]. The next step would be to determine the effects of multiple fungal interactions on plant growth since fungal co-infection is the rule rather than the exception. Combining next-generation sequencing and complex culture-based approaches will reveal the most influential fungal pathogenic actors and their roles in such biodiversity-ecosystem functioning.
5. Conclusions
Crop rotation has long been known as an effective means to reduce plant diseases. Even if late wilt disease-resistant cultivars are used, maize cultivation enhances the abundance of the disease agent Magnaporthiopsis maydis. The current study revealed that the presence of this pathogen in the ground might drastically affect the ability of the soil fungus Macrophomina phaseolina, the charcoal rot disease (CRD) agent, to harm cotton sprouts. Infection of the soil 14 days before sowing with each pathogen caused the severe inhibition of the sprouts’ emergence, development, and survival, accompanied by high DNA levels of each pathogen in the sprouts’ roots. Similar results were obtained when M. phaseolina soil infection was performed before M. maydis. However, as demonstrated here, M. maydis may act as a beneficial endophyte, protecting the plants from CRD or as a dangerous pathogen. In some circumstances, M. maydis showed strong virulence potential, proving its true nature as an opportunist in cotton. Interestingly we demonstrated that in this situation, soil inoculation with M. phaseolina 14 days before sowing protected the plants from M. maydis. Thus, the two pathogens restrict each other, and this equilibrium may lead to a moderate disease burst. The results obtained in this study improve our understanding of the relationship between M. phaseolina, the CRD agent, and M. maydis. They suggest that the coexistence of two pathogens is an important balancing factor that may result from maize-cotton crop rotation. Any intervention control strategy—agrotechnical, chemical, or biological—in these two-year-rotation cultivated fields should consider this knowledge since an attempt to restrict one of the partners could unintentionally contribute to the thriving of the other.