A Non-Invasive Neonatal Signature Predicts Later Development of Atopic Diseases
Abstract
:1. Introduction
2. Methods
2.1. Patients and Sampling
2.2. Ethics Statement
2.2.1. A-Immunological Analysis
Preparation of Fecal Samples
Total IgE, Tryptase, and EDN Determination
Calprotectin and Total Protein Determination
Immunoassays
2.2.2. A-Microbiological Analysis: Methanogenic Archaea by qPCR
DNA Extraction and PCR Assays
Statistical Analysis
3. Results
3.1. Demographic and Clinical Characteristics of Preterm Infants
3.2. Immune Profiling
3.2.1. Total Protein Determination
3.2.2. Immune Cell Markers and Cytokines
3.2.3. Correlation between Biomarkers
3.2.4. Meconium Samples
3.2.5. Samples at Two Weeks
3.2.6. Samples at Four Weeks
3.2.7. Samples at Six Weeks
3.3. Frequency of Detection of Methanogenic Archaea and Relationship with the Subsequent Development of Atopic Diseases
3.4. Unsupervised Analysis of Immunological Markers and Methanogenic Archaea Atthe Neonatal Period, and the Subsequent Occurrence of AD, Asthma and CMA during the First Year
4. Discussion
- (1)
- miniaturization and standardization, using small quantities of stool (1 g) and small volumes of extraction buffer (2 mL). The dilution of the samples was corrected by the freeze-drying process, as the lyophilizates were contained in 1 mL of buffer.
- (2)
- prevention, thanks to the use of protease inhibitors, of the risk of potential contamination of the handler.
- (3)
- suitability for a microarray platform yielding patterns of immune responses rather than individual measurements.
- (4)
- suitability for combined immune and microbiological assessment.
- (5)
- proof of concept of the immune profiling of fecal mediators in meconium and neonatal samples as predictors of later development of atopic disorders.
- (6)
- proof of concept for non-invasive investigation of the immune status of preterm neonates.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Crump, C.; Sundquist, J.; Winkleby, M.A.; Sundquist, K. Gestational age at birth and mortality from infancy into mid-adulthood: A national cohort study. Lancet Child Adolesc. Health 2019, 3, 408–417. [Google Scholar] [CrossRef]
- Chawanpaiboon, S.; Vogel, J.P.; Moller, A.B.; Lumbiganon, P.; Petzold, M.; Hogan, D.; Landoulsi, S.; Jampathong, N.; Kongwattanakul, K.; Laopaiboon, M.; et al. Global, regional, and national estimates of levels of preterm birth in 2014: A systematic review and modelling analysis. Lancet Glob. Health 2019, 7, e37–e46. [Google Scholar] [CrossRef]
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef]
- Schuster, J.; Uzun, A.; Stablia, J.; Schorl, C.; Mori, M.; Padbury, J.F. Effect of prematurity on genome wide methylation in the placenta. BMC Med. Genet. 2019, 20, 116. [Google Scholar] [CrossRef]
- Beck, S.; Wojdyla, D.; Say, L.; Betran, A.P.; Merialdi, M.; Requejo, J.H.; Rubens, C.; Menonf, M.; Look, V.F.P. The worldwide incidence of preterm birth: A systematic review of maternal mortality and morbidity. Bull. World Health Organ. 2010, 88, 31–38. [Google Scholar] [CrossRef]
- Gagneur, A.; Pinquier, D.; Quach, C. Immunization of preterm infants. Hum. Vaccines Immunother. 2015, 11, 2556–2563. [Google Scholar] [CrossRef]
- Marshall, H.; Clarke, M.; Rasiah, K.; Richmond, P.; Buttery, J.; Reynolds, G.; Andrews, R.; Nissen, M.; Wood, N.; McIntyre, P. Predictors of Disease Severity in Children Hospitalized for Pertussis During an Epidemic. Pediatr. Infect. Dis. J. 2015, 34, 339–345. [Google Scholar] [CrossRef]
- Gómez, M.; Moles, L.; Espinosa-Martos, I.; Bustos, G.; de Vos, W.; Fernández, L.; Rodríguez, M.J.; Fuentes, S.; Jiménez, E. Bacteriological and Immunological Profiling of Meconium and Fecal Samples from Preterm Infants: A Two-Year Follow-Up Study. Nutrients 2017, 9, 1293. [Google Scholar] [CrossRef]
- Helmo, F.R.; Alves, E.A.R.; Moreira, R.A.D.A.; Severino, V.O.; Rocha, L.P.; Monteiro, M.L.G.D.R.; Marlene Antônia dos Reis, D.A.M.; Etchebehere, M.R.; Machado, R.J.; Corrêa, M.R.R. Intrauterine infection, immune system and premature birth. J. Matern.-Fetal Neonatal Med. 2018, 31, 1227–1233. [Google Scholar] [CrossRef]
- Sonnenschein-van der Voort, A.M.; Arends, L.R.; de Jongste, J.C.; Annesi-Maesano, I.; Arshad, S.H.; Barros, H.; Basterrechea, M.; Bisgard, H.; Chatzi, L.; Corpeleijn, E.; et al. Preterm birth, infant weight gain, and childhood asthma risk: A meta-analysis of 147,000 European children. J. Allergy Clin. Immunol. 2014, 133, 1317–1329. [Google Scholar] [CrossRef]
- Wooldridge, A.L.; McMillan, M.; Kaur, M.; Giles, L.C.; Marshall, H.S.; Gatford, K.L. Relationship between birth weight or fetal growth rate and postnatal allergy: A systematic review. J. Allergy Clin. Immunol. 2019, 144, 1703–1713. [Google Scholar] [CrossRef]
- Mitselou, N.; Hallberg, J.; Stephansson, O.; Almqvist, C.; Melén, E.; Ludvigsson, J.F. Cesarean delivery, preterm birth, and risk of food allergy: Nationwide Swedish cohort study of more than 1 million children. J. Allergy Clin. Immunol. 2018, 142, 1510–1514. [Google Scholar] [CrossRef]
- Barbarot, S.; Gras-Leguen, C.; Colas, H.; Garrot, E.; Darmaun, D.; Larroque, B.; Roze, J.C.; Ancel, P.; Ancel. Lower risk of atopic dermatitis among infants born extremely preterm compared with higher gestational age. Br. J. Dermatol. 2013, 169, 1257–1264. [Google Scholar] [CrossRef]
- Al Nabhani, Z.; Eberl, G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. 2020, 13, 183–189. [Google Scholar] [CrossRef]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, M.A.; Kunøe, A.; Fink, R.N.; Chawes, C.B.; et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018, 9, 141. [Google Scholar] [CrossRef]
- Gomez Perdiguero, E.; Klapproth, K.; Schulz, C.; Busch, K.; Azzoni, E.; Crozet, L.; Garner, H.; Trouillet, C.; Bruijn, D.F.M.; Geissmann, F.; et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518, 547–551. [Google Scholar] [CrossRef]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; Horst, T.R.; Jansen, A.; Jacobs, L.; Bonder, J.M.; et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016, 167, 1897. [Google Scholar] [CrossRef]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 2018, 16, 457–470. [Google Scholar] [CrossRef]
- Sereme, Y.; Mezouar, S.; Grine, G.; Mege, J.L.; Drancourt, M.; Corbeau, P.; Vitte, J.L. Methanogenic Archaea: Emerging Partners in the Field of Allergic Diseases. Clin. Rev. Allergy Immunol. 2019, 57, 456–466. [Google Scholar] [CrossRef]
- Dridi, B.; Henry, M.; El Khéchine, A.; Raoult, D.; Drancourt, M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugère, J.F. Archaea and the human gut: New beginning of an old story. World J. Gastroenterol. 2014, 20, 16062–16078. [Google Scholar] [CrossRef]
- Bang, C.; Weidenbach, K.; Gutsmann, T.; Heine, H.; Schmitz, R.A. The Intestinal Archaea Methanosphaera stadtmanae and Methanobrevibacter smithii Activate Human Dendritic Cells. PLoS ONE 2014, 9, e99411. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Stein, K.; Heine, H. RNA is taking its Toll: Impact of RNA-specific Toll-like receptors on health and disease. Allergy 2019, 74, 223–235. [Google Scholar] [CrossRef]
- Bernatchez, E.; Gold, M.J.; Langlois, A.; Blais-Lecours, P.; Boucher, M.; Duchaine, C.; Marsolais, D.; McNagny, M.K.; Blanchet, R.M. Methanosphaera stadtmanae induces a type IV hypersensitivity response in a mouse model of airway inflammation. Physiol. Rep. 2017, 5, e13163. [Google Scholar] [CrossRef]
- Grine, G.; Boualam, M.A.; Drancourt, M. Methanobrevibacter smithii, a methanogen consistently colonising the newborn stomach. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2449–2455. [Google Scholar] [CrossRef]
- Sereme, Y.; Guindo, C.O.; Filleron, A.; Corbeau, P.; Tran, T.A.; Drancourt, M.; Vitte, J.; Grine, G. Meconial Methanobrevibacter smithii suggests intrauterine methanogen colonization in preterm neonates. Curr. Res. Microb. Sci. 2021, 2, 100034. [Google Scholar] [CrossRef]
- Baumann, R.; Untersmayr, E.; Zissler, U.M.; Eyerich, S.; Adcock, I.M.; Brockow, K.; Biedermann, T.; Ollert, M.; Chaker, M.A.; Pfaar, O.; et al. Noninvasive and minimally invasive techniques for the diagnosis and management of allergic diseases. Allergy 2021, 76, 1010–1023. [Google Scholar] [CrossRef]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Investig. 2021, 131, e141935. [Google Scholar] [CrossRef]
- Sereme, Y.; Zarza, S.M.; Medkour, H.; Amona, I.; Fenollar, F.; Akiana, J.; Mezouar, S.; Orain, N.; Vitte, J.; Davout, D.; et al. Stool Serology: Development of a Non-Invasive Immunological Method for the Detection of Enterovirus-Specific Antibodies in Congo Gorilla Faeces. Microorganisms 2021, 9, 810. [Google Scholar] [CrossRef]
- Lambert, C.; Sarrat, A.; Bienvenu, F.; Brabant, S.; Nicaise-Roland, P.; Alyanakian, M.-A.; Apoil, P.A.; Capron, C.; Couderc, R.; Evrard, B.; et al. The importance of EN ISO 15189 accreditation of allergen-specific IgE determination for reliable in vitro allergy diagnosis. Allergy 2015, 70, 180–186. [Google Scholar] [CrossRef]
- Drancourt, M.; Djemai, K.; Gouriet, F.; Grine, G.; Loukil, A.; Bedotto, M.; Levasseur, A.; Lépidi, H.; Khalil, B.J.; Khelaifia, S.; et al. Methanobrevibacter smithii archaemia in febrile patients with bacteremia, including those with endocarditis. Clin. Infect. Dis. 2020, 73, e2571–e2579. [Google Scholar] [CrossRef] [PubMed]
- Grine, G.; Terrer, E.; Boualam, M.A.; Aboudharam, G.; Chaudet, H.; Ruimy, R.; Drancourt, D. Tobacco-smoking-related prevalence of methanogens in the oral fluid microbiota. Sci. Rep. 2018, 8, 9197. [Google Scholar] [CrossRef] [PubMed]
- Nkamga, V.D.; Huynh, H.T.T.; Aboudharam, G.; Ruimy, R.; Drancourt, M. Diversity of human-associated Methanobrevibacter smithii isolates revealed by multispacer sequence typing. Curr. Microbiol. 2015, 70, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Guindo, C.O.; Davoust, B.; Drancourt, M.; Grine, G. Diversity of Methanogens in Animals’ Gut. Microorganisms 2020, 9, 13. [Google Scholar] [CrossRef]
- Muraro, A.; Werfel, T.; Hoffmann-Sommergruber, K.; Roberts, G.; Beyer, K.; Bindslev-Jensen, C.; Bindslev-Jensen, C.; Cardona, V.; Dubois, A.; duToit, G.; et al. EAACI food allergy and anaphylaxis guidelines: Diagnosis and management of food allergy. Allergy 2014, 69, 1008–1025. [Google Scholar] [CrossRef]
- Papadopoulos, N.G.; Arakawa, H.; Carlsen, K.H.; Custovic, A.; Gern, J.; Lemanske, R.; Le Souef, P.; Mäkelä, M.; Roberts, G.; Wong, G.; et al. International consensus on (ICON) pediatric asthma. Allergy 2012, 67, 976–997. [Google Scholar] [CrossRef]
- Lisowska-Myjak, B.; Skarżyńska, E.; Wojdan, K.; Nasierowska-Guttmejer, A. Protein and peptide profiles in neonatal meconium: Classification of meconium proteins. J. Obstet. Gynaecol. Res. 2019, 45, 556–564. [Google Scholar] [CrossRef]
- Kolmannskog, S.; Marhaug, G.; Haneberg, B. Fragments of IgE Antibodies in Human Feces. Int. Arch. Allergy Immunol. 1985, 78, 358–363. [Google Scholar] [CrossRef]
- Szépfalusi, Z.; Pichler, J.; Elsässer, S.; van Duren, K.; Ebner, C.; Bernaschek, G.; Urbanek, R. Transplacental priming of the human immune system with environmental allergens can occur early in gestation. J. Allergy Clin. Immunol. 2000, 106, 530–536. [Google Scholar] [CrossRef]
- Msallam, R.; Balla, J.; Rathore, A.P.S.; Kared, H.; Malleret, B.; Saron, W.A.A.; Liu, Z.; Hang, W.J.; Dutertre, A.C.; Larbi, A.; et al. Fetal mast cells mediate postnatal allergic responses dependent on maternal IgE. Science 2020, 370, 941–950. [Google Scholar] [CrossRef]
- Lyons, J.J.; Yi, T. Mast cell tryptases in allergic inflammation and immediate hypersensitivity. Curr. Opin. Immunol. 2021, 72, 94–106. [Google Scholar] [CrossRef]
- Edogawa, S.; Edwinson, A.L.; Peters, S.A.; Chikkamenahalli, L.L.; Sundt, W.; Graves, S.; Gurunathan, V.S.; Breen-Lyles, K.M.; Johnson, S.; Dyer, B.R.; et al. Serine proteases as luminal mediators of intestinal barrier dysfunction symptom severity in IBS. Gut 2020, 69, 62–73. [Google Scholar] [CrossRef]
- Raithel, M.; Winterkamp, S.; Pacurar, A.; Ulrich, P.; Hochberger, J.; Hahn, E.G. Release of mast cell tryptase from human colorectal mucosa in inflammatory bowel disease. Scand. J. Gastroenterol. 2001, 36, 174–179. [Google Scholar] [CrossRef]
- Peterson, C.G.B.; Hansson, T.; Skott, A.; Bengtsson, U.; Ahlstedt, S.; Magnussons, J. Detection of local mast-cell activity in patients with food hypersensitivity. J. Investig. Allergol. Clin. Immunol. 2007, 17, 314–320. [Google Scholar]
- Carroccio, A.; Brusca, I.; Mansueto, P.; Soresi, M.; D’Alcamo, A.; Ambrosiano, G.; Pepe, I.; Iacono, G.; Lospalluti, L.M.; Chiusa, L.M.S.; et al. Fecal assays detect hypersensitivity to cow’s milk protein and gluten in adults with irritable bowel syndrome. Clin. Gastroenterol. Hepatol. 2011, 9, 965–971. [Google Scholar] [CrossRef]
- Lettesjö, H.; Hansson, T.; Peterson, C.; Ung, K.-A.; Ringström, G.; Abrahamsson, H.; Simrén, M. Detection of inflammatory markers in stools from patients with irritable bowel syndrome and collagenous colitis. Scand. J. Gastroenterol. 2006, 41, 54–59. [Google Scholar] [CrossRef]
- Roca, M.; Rodriguez Varela, A.; Donat, E.; Cano, F.; Hervas, D.; Armisen, A.; Vaya, J.M.; Sjölander, A.; Ribes-Koninckx, C. Fecal Calprotectin and Eosinophil-derived Neurotoxin in Healthy Children Between 0 and 12 Years. J. Pediatr. Gastroenterol. Nutr. 2017, 65, 394–398. [Google Scholar] [CrossRef]
- Willers, M.; Ulas, T.; Völlger, L.; Vogl, T.; Heinemann, A.S.; Pirr, S.; Pagel, J.; Fehlhaber, B.; Halle, O.; Schöning, J.; et al. S100A8 and S100A9 Are Important for Postnatal Development of Gut Microbiota and Immune System in Mice and Infants. Gastroenterology 2020, 159, 2130–2145. [Google Scholar] [CrossRef]
- Olin, A.; Henckel, E.; Chen, Y.; Lakshmikanth, T.; Pou, C.; Mikes, J.; Gustafsson, A.; Bernhardsson, K.A.; Zhang, C.; Bohlin, K.; et al. Stereotypic Immune System Development in Newborn Children. Cell 2018, 174, 1277–1292. [Google Scholar] [CrossRef]
- Torow, N.; Marsland, B.J.; Hornef, M.W.; Gollwitzer, E.S. Neonatal mucosal immunology. Mucosal Immunol. 2017, 10, 5–17. [Google Scholar] [CrossRef]
- Goretzki, A.; Lin, Y.J.; Schülke, S. Immune metabolism in allergies, does it matter?—A review of immune metabolic basics and adaptations associated with the activation of innate immune cells in allergy. Allergy 2021, 76, 3314–3331. [Google Scholar] [CrossRef]
- Macpherson, A.J.; de Agüero, M.G.; Ganal-Vonarburg, S.C. How nutrition and the maternal microbiota shape the neonatal immune system. Nat. Rev. Immunol. 2017, 17, 508–517. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, D.P.; Manicassamy, S.; Munn, H.D.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Ando, M.; Kamada, N.; Nagano, Y.; Narushima, S.; Suda, W.; Imaoka, A.; Setoyama, H.; Nagamori, T.; et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 2015, 163, 367–380. [Google Scholar] [CrossRef]
- Vonk, M.M.; Blokhuis, B.R.J.; Diks, M.A.P.; Wagenaar, L.; Smit, J.J.; Pieters, R.H.H.; Garssen, J.; Knippels, J.M.L.; Esch, V.M.A.C.B. Butyrate Enhances Desensitization Induced by Oral Immunotherapy in Cow’s Milk Allergic Mice. Mediat. Inflamm. 2019, 2019, 9062537. [Google Scholar] [CrossRef]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, N.; et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Barnett, D.J.M.; Mommers, M.; Penders, J.; Arts, I.C.W.; Thijs, C. Intestinal archaea inversely associated with childhood asthma. J. Allergy Clin. Immunol. 2019, 143, 2305–2307. [Google Scholar] [CrossRef]
- Gomulka, K.; Liebhart, J.; Jaskula, E.; Lange, A.; Medrala, W. The –2549 –2567 del18 Polymorphism in VEGF and Irreversible Bronchoconstriction in Asthmatics. J. Investig. Allergol. Clin. Immunol. 2019, 29, 431–435. [Google Scholar] [CrossRef]



| Code | Meconium (M) | Two-Weeks (W2) | Four-Week (W4) | Six-Weeks (W6) | Peripartum Maternal Antibiotic Therapy | Mode of Delivery | Gestational Age | Weight | Size | Asthma or CMA | Atopic Dermatitis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | W2 | W4 | W6 | No | VD | 30 | 1275 | 37 | Yes | Yes |
| 2 | M | W2 | W4 | W6 | Yes | CS | 27 | 925 | 34 | Yes | No |
| 3 | M | W2 | W4 | W6 | No | CS | 26 | 565 | 31 | Yes | No |
| 4 | 0 | 0 | W4 | 0 | No | VD | 25 | 820 | 34 | Yes | No |
| 5 | M | W2 | W4 | 0 | No | CS | 32 | 1260 | 39 | Yes | No |
| 6 | M | W2 | W4 | W6 | Yes | CS | 27 | 680 | 31 | Yes | No |
| 7 | M | 0 | W4 | W6 | No | CS | 29 | 1565 | 42 | No | No |
| 8 | M | W2 | W4 | W6 | No | CS | 28 | 890 | 33 | No | No |
| 9 | 0 | 0 | 0 | W6 | Yes | CS | 30 | 1150 | 38 | No | No |
| 10 | M | W2 | 0 | 0 | No | CS | 31 | 1570 | 43 | No | No |
| 11 | M | W2 | W4 | 0 | No | CS | 32 | 1575 | 44 | No | No |
| 12 | M | W2 | W4 | 0 | No | CS | 30 | 1360 | 39 | No | Yes |
| 13 | M | W2 | 0 | W6 | No | CS | 25 | 870 | 34 | Yes | Yes |
| 14 | M | W2 | W4 | 0 | No | CS | 32 | 1155 | 39 | Yes | No |
| 15 | 0 | W2 | W4 | W6 | No | CS | 25 | 440 | 28 | No | No |
| 16 | 0 | 0 | 0 | W6 | No | VD | 30 | 1590 | 41 | No | No |
| 17 | M | W2 | W4 | W6 | No | CS | 24 | 530 | 31 | No | No |
| 18 | M | W2 | 0 | 0 | No | CS | 26 | 925 | 35 | No | No |
| 19 | M | W2 | W4 | 0 | No | VD | 30 | 1480 | 41 | No | No |
| 20 | M | W2 | W4 | 0 | No | VD | 30 | 1460 | 38 | No | No |
| 21 | M | W2 | W4 | W6 | No | CS | 29 | 880 | 35 | No | No |
| 22 | 0 | W2 | W4 | W6 | No | CS | 28 | 840 | 35 | No | No |
| 23 | M | W2 | W4 | 0 | Yes | VD | 30 | 1670 | 43 | No | No |
| 24 | M | W2 | W4 | W6 | No | CS | 31 | 1120 | 38 | No | No |
| 25 | 0 | W2 | 0 | 0 | No | CS | 28 | 915 | 36 | No | No |
| 26 | M | 0 | W4 | W6 | No | CS | 26 | 925 | 35 | No | No |
| 27 | M | W2 | W4 | W6 | Yes | CS | 30 | 1335 | 39 | No | No |
| 28 | M | W2 | W4 | W6 | Yes | CS | 30 | 1355 | 47 | No | No |
| 29 | M | W2 | 0 | W6 | No | CS | 30 | 1480 | 39 | No | No |
| 30 | 0 | W2 | W4 | W6 | Yes | CS | 28 | 1010 | 35 | No | No |
| 31 | M | W2 | W4 | W6 | No | CS | 29 | 1050 | 39 | No | No |
| 32 | M | W2 | W4 | 0 | No | CS | 29 | 1190 | 38 | No | No |
| 33 | 0 | W2 | W4 | W6 | No | CS | 30 | 1175 | 39 | No | No |
| 34 | M | 0 | 0 | 0 | No | CS | 32 | 1930 | 44 | No | No |
| 35 | 0 | W2 | W4 | 0 | Yes | CS | 29 | 1430 | 30 | No | No |
| 36 | M | W2 | W4 | 0 | No | CS | 30 | 1750 | 43 | No | No |
| 37 | M | 0 | 0 | 0 | No | CS | 27 | 600 | 29 | No | No |
| 38 | M | W2 | W4 | W6 | No | VD | 25 | 750 | 32 | No | No |
| 39 | M | W2 | 0 | W6 | No | CS | 31 | 980 | 36 | No | No |
| 40 | M | W2 | 0 | W6 | No | CS | 31 | 1410 | 39 | No | No |
| 41 | M | 0 | 0 | 0 | No | CS | 30 | 770 | 33 | No | No |
| 42 | M | 0 | 0 | 0 | Yes | VD | 32 | 1568 | 41 | No | No |
| 43 | M | 0 | 0 | 0 | No | CS | 30 | 1680 | 39 | No | No |
| Meconium | Two Weeks | Four Weeks | Six Weeks | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n = 33 | n = 33 | n = 29 | n = 24 | |||||||
| n (%) Detectable | Median IQR | n (%) Detectable | Median IQR | n (%) Detectable | Median IQR | n (%) Detectable | Median IQR | p-Value (Frequency) | p-Value (Levels) | |
| Total Proteins (g/L) | 33 (100) | 9.18 (4.51–13.54) | 33 (10,055) | 5. 4.23–6.05) | 29 (100) | 4.53 (3.00–5.52) | 24 (100) | 6.46 (5.39–7.76) | NS | 0.10 |
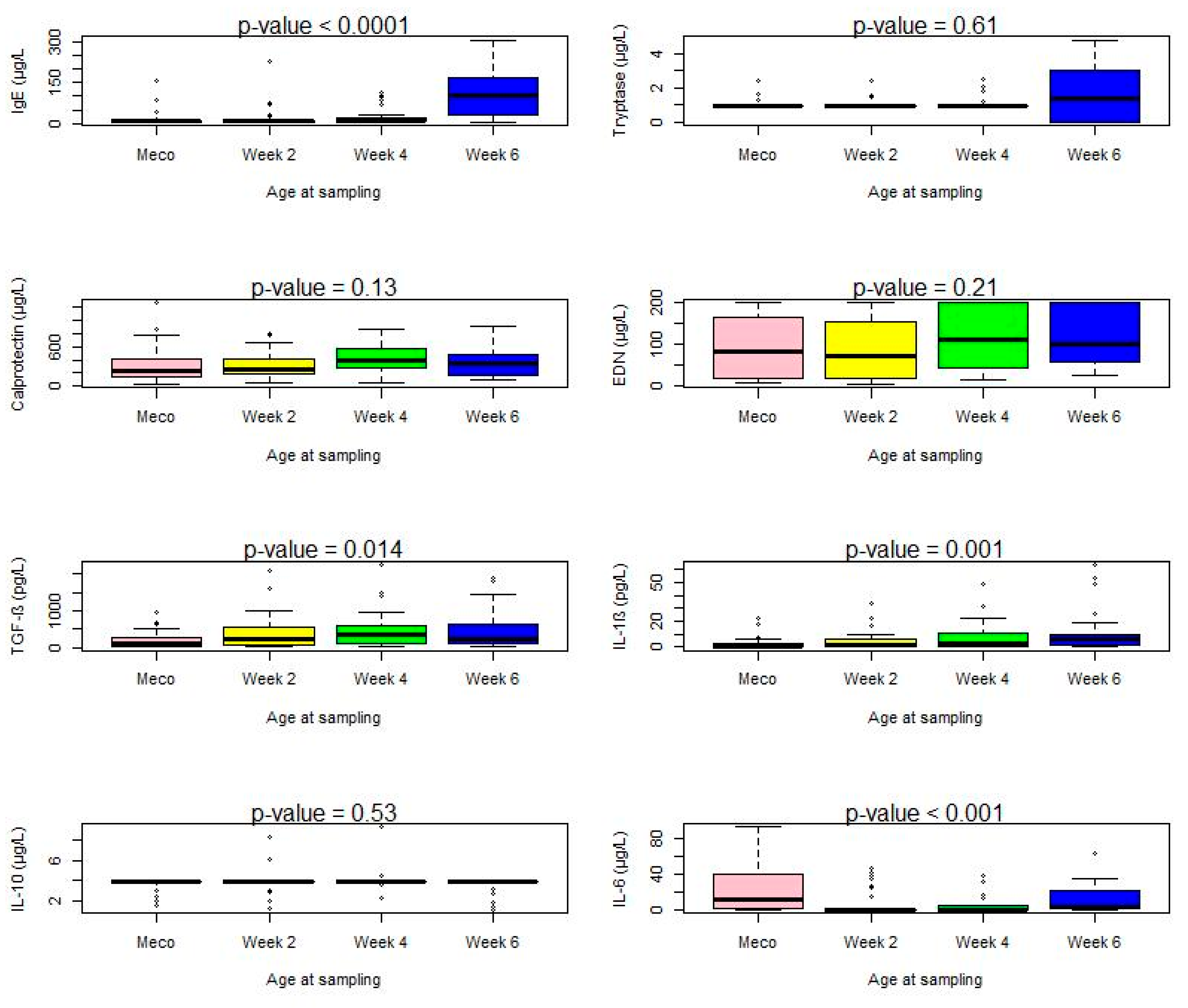
| Total IgE (µg/L) | 30 (90.90) | 7.3 (6.4–9.9) | 32 (97) | 8.47 (6.8–9.8) | 27 (93.10) | 9.74 (3.39–0.26) | 24 (100) | 115.08 (41.00–193.70) | 0.41 | <0.0001 |
| Tryptase (µg/L) | 3 (9.1) | <1 | 3 (9.1) | <1 | 4 (13.79) | <1 | 14 (58.33) | 1.8 (0.0–3.4) | <0.0001 | 0.61 |
| Calprotectin (µg/L) | 33 (100) | 310.4 (151.1–771.3) | 33 (100) | 291.23 (189.41–487.87) | 29 (100) | 402.44 (300.06–607.3) | 24 (100) | 422.37 (335.53–823.30) | NC | 0.13 |
|
EDN (µg/L) | 33 (100) | 83.2 (19.3–165.0) | 33 (100) | 70.1 (17.8–152.5) | 29 (100) | 109.0 (44.2–200.0) | 24 (100) | 98.1 (57.5–200.0) | NC | 0.21 |
| TGF-β (pg/L) | 24 (72.7) | 121.3 (4.6–258.9) | 30 (91) | 267.43 (61.71–1000) | 26 (89.65) | 384.57 (129.60–936) | 22 (91.66) | 466 (104.36–1430.29) | 0.09 | 0.014 |
|
IL-1β (pg/L) | 13 (39.4) | 0.12 (0.1–2.7) | 28 (84.8) | 1.53 (0.37–6.53) | 25 (86.20) | 3.27 (0.31–10.76) | 22 (91.66) | 6.23 (1.66–20.84) | <0.0001 | 0.001 |
|
IL-10 (pg/L) | 4 (12.12) | 3.9 (3.9–3.9) | 6 (18.18) | 3.9 (3.9–3.9) | 5 (17.24) | 3.9 (3.9–3.9) | 5 (20.83) | 3.9 (3.9–3.9) | 0.85 | 0.53 |
|
IL-6 (pg/L) | 25 (75.75) | 11.6 (0.5–43.7) | 7 (21.21) | 0.2 (0.2–0.2) | 20 (68.96) | 0.2 (0.2–0.2) | 20 (83.33) | 3.77 (1.66–19.25) | <0.0001 | <0.001 |
| Variables | Allergic Condition (APLV and Asthma) | p-Value | |||
|---|---|---|---|---|---|
| Meconium | 2 Weeks | 4 Weeks | 6 Weeks | ||
| IgE | Yes | 0.27 | 0.06 | 0.12 | 0.03 |
| No | |||||
| Calprotectin | Yes | 0.27 | 0.18 | 0.91 | 0.61 |
| No | |||||
| EDN | Yes | 0.59 | 0.19 | 0.41 | 0.87 |
| No | |||||
| TGF-β | Yes | 0.09 | 0.18 | 0.76 | 0.76 |
| No | |||||
| IL-1β | Yes | 0.37 | 1.00 | 0.28 | 0.91 |
| No | |||||
| IL-10 | Yes | 0.62 | 1.00 | 0.89 | 0.13 |
| No | |||||
| IL-6 | Yes | 0.61 | 0.28 | 0.37 | 0.75 |
| Meconium (n = 33) | Two Weeks (n = 33) | Four Weeks (n = 29) | Six Weeks (n = 26) | p-Value (Frequency) | p-Value (CT) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | Median IQR | n (%) | Median IQR | n (%) | Median IQR | n (%) | Median IQR | |||
| CT qPCR | 30 (90.9) | 36.74 (33.85–38.24) | 27 (81.81) | 37.20 (36.07–38.33) | 23 (79.31) | 37.75 (36.13–38.50) | 19 (73.03) | 38.28 (37.27–39.96) | 0.34 | 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sereme, Y.; Michel, M.; Mezouar, S.; Guindo, C.O.; Kaba, L.; Grine, G.; Mura, T.; Mège, J.-L.; Tran, T.A.; Corbeau, P.; et al. A Non-Invasive Neonatal Signature Predicts Later Development of Atopic Diseases. J. Clin. Med. 2022, 11, 2749. https://doi.org/10.3390/jcm11102749
Sereme Y, Michel M, Mezouar S, Guindo CO, Kaba L, Grine G, Mura T, Mège J-L, Tran TA, Corbeau P, et al. A Non-Invasive Neonatal Signature Predicts Later Development of Atopic Diseases. Journal of Clinical Medicine. 2022; 11(10):2749. https://doi.org/10.3390/jcm11102749
Chicago/Turabian StyleSereme, Youssouf, Moïse Michel, Soraya Mezouar, Cheick Oumar Guindo, Lanceï Kaba, Ghiles Grine, Thibault Mura, Jean-Louis Mège, Tu Anh Tran, Pierre Corbeau, and et al. 2022. "A Non-Invasive Neonatal Signature Predicts Later Development of Atopic Diseases" Journal of Clinical Medicine 11, no. 10: 2749. https://doi.org/10.3390/jcm11102749
APA StyleSereme, Y., Michel, M., Mezouar, S., Guindo, C. O., Kaba, L., Grine, G., Mura, T., Mège, J.-L., Tran, T. A., Corbeau, P., Filleron, A., & Vitte, J. (2022). A Non-Invasive Neonatal Signature Predicts Later Development of Atopic Diseases. Journal of Clinical Medicine, 11(10), 2749. https://doi.org/10.3390/jcm11102749









