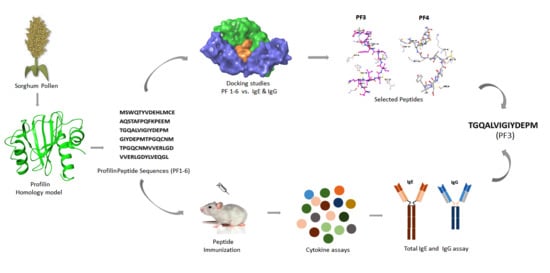
Peptide Mapping, In Silico and In Vivo Analysis of Allergenic Sorghum Profilin Peptides
Abstract
:1. Introduction
2. Materials and Methods
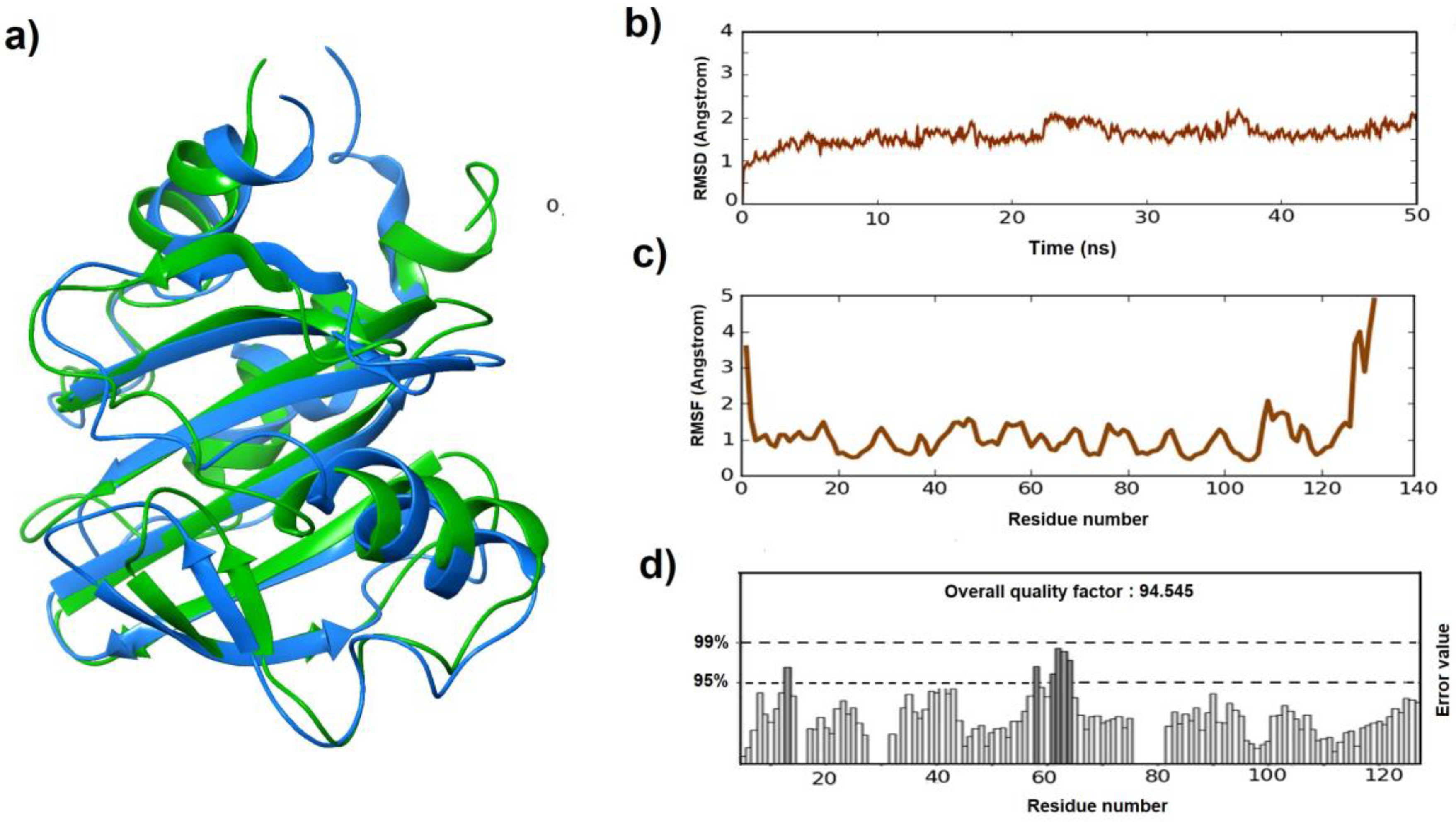
2.1. Homology Modeling and Evaluation
2.2. Model Refinement
2.3. Ag–Ab Docking Studies
2.4. Peptide Docking Studies
2.5. Animal Maintenance
2.6. Peptide Immunization
2.7. Overlapping Peptide Mapping and Synthesis
2.8. Cytokine Assays
2.9. Total IgE and IgG Assays
2.10. Statistical Analysis
3. Results
3.1. Homology Modeling, Evaluation, and Refinement
3.2. Antigen–antibody Binding Studies
3.3. Peptide Docking Studies
3.4. Cytokines
3.5. Total IgE and IgG
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singh, A. Pollen and fungal aeroallergens associated with allergy and asthma in India. Glob. J. Immunol. Allerg. Dis. 2014, 2, 19–28. [Google Scholar] [CrossRef]
- Skoner, D.P. Allergic rhinitis: Definition, epidemiology, pathophysiology, detection, and diagnosis. J. Allergy Clin. Immunol. 2001, 108, S2–S8. [Google Scholar] [CrossRef]
- Hauser, M.; Roulias, A. Panallergens and their impact on the allergic patient. Allergyasthma Clin. Immunol. 2010, 6, 1. [Google Scholar] [CrossRef]
- Asero, R.; Mistrello, G. Detection of clinical markers of sensitization to profilin in patients allergic to plant-derived foods. J. Allergy Clin. Immunol. 2003, 112, 427–432. [Google Scholar] [CrossRef]
- Kay, A.B.; Phipps, S. A role for eosinophils in airway remodelling in asthma. Trends Immunol. 2004, 25, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.E.; Hogan, S.P. The eosinophil. Annu. Rev. Immunol. 2006, 24, 147–174. [Google Scholar] [CrossRef]
- Asquith, K.L.; Ramshaw, H.S. The IL-3/IL-5/GM-CSF common β receptor plays a pivotal role in the regulation of Th2 immunity and allergic airway inflammation. J. Immunol. 2008, 180, 1199–1206. [Google Scholar] [CrossRef]
- Deo, S.S.; Mistry, K.J. Role played by Th2 type cytokines in IgE mediated allergy and asthma. Lung India Off. Organ Indian Chest Soc. 2010, 27, 66. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.; MacIntosh, S. Scientific advancement of novel protein allergenicity evaluation: An overview of work from the HESI Protein Allergenicity Technical Committee (2000–2008). Food Chem. Toxicol. 2009, 47, 1041–1050. [Google Scholar] [CrossRef]
- Anuradha, B.; Vijayalakshmi, V. Profile of pollen allergies in patients with asthma, allergic rhinitis and urticaria in Hyderabad. Indian J. Chest Dis. Allied Sci. 2006, 48, 221. [Google Scholar]
- Sekhar, B.C.; Sachin, C. Molecular characterization and in silico analysis of sorghum panallergens: profilin and polcalin. Indian J. Exp. Biol. 2015, 53, 726–731. [Google Scholar]
- Pomés, A. Relevant B cell epitopes in allergic disease. Int. Arch. Allergy Immunol. 2010, 152, 1–11. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, W. Homology modeling, molecular docking, and molecular dynamics simulations elucidated α-fetoprotein binding modes. BMC Bioinform. 2013, 14, S6. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laskowski, R.A.; MacArthur, M.W. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Veeramachaneni, G.K.; Raj, K.K. High-throughput virtual screening with e-pharmacophore and molecular simulations study in the designing of pancreatic lipase inhibitors. Drug Des. Dev. Ther. 2015, 9, 4397. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Schmidt, A.G. Key mutations stabilize antigen-binding conformation during affinity maturation of a broadly neutralizing influenza antibody lineage. Proteins Struct. Funct. Bioinform. 2015, 83, 771–780. [Google Scholar] [CrossRef]
- Schrödinger. Schrödinger Release 2018-1; BioLuminate: New York, NY, USA, 2018. [Google Scholar]
- Sastry, G.M.; Adzhigirey, M. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Conejero, L.; Higaki, Y. Pollen-induced airway inflammation, hyper-responsiveness and apoptosis in a murine model of allergy. Clin. Exp. Allergy 2007, 37, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Braag, S.A. Enhanced IgE allergic response to Aspergillus fumigatus in CFTR−/− mice. Lab. Investig. 2006, 86, 130. [Google Scholar] [CrossRef]
- Tisato, V.; Garrovo, C. Intranasal administration of recombinant TRAIL down-regulates CXCL-1/KC in an ovalbumin-induced airway inflammation murine model. PLoS ONE 2014, 9, e115387. [Google Scholar] [CrossRef]
- Asturias, J.A.; Gómez-Bayón, N. Molecular and structural analysis of the panallergen profilin B cell epitopes defined by monoclonal antibodies. Int. Immunol. 2002, 14, 993–1001. [Google Scholar] [CrossRef] [Green Version]
- López-Torrejón, G.; Díaz-Perales, A. An experimental and modeling-based approach to locate IgE epitopes of plant profilin allergens. J. Allergy Clin. Immunol. 2007, 119, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kong, H. Subsets of regulatory T cells and their roles in allergy. J. Transl. Med. 2014, 12, 125. [Google Scholar] [CrossRef]
- Raphael, I.; Nalawade, S. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 2015, 74, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, S. Immunologic influences on allergy and the TH1/TH2 balance. J. Allergy Clin. Immunol. 2004, 113, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D. Regulatory T cells and asthma. Clin. Exp. Allergy 2009, 39, 1314–1323. [Google Scholar] [CrossRef]

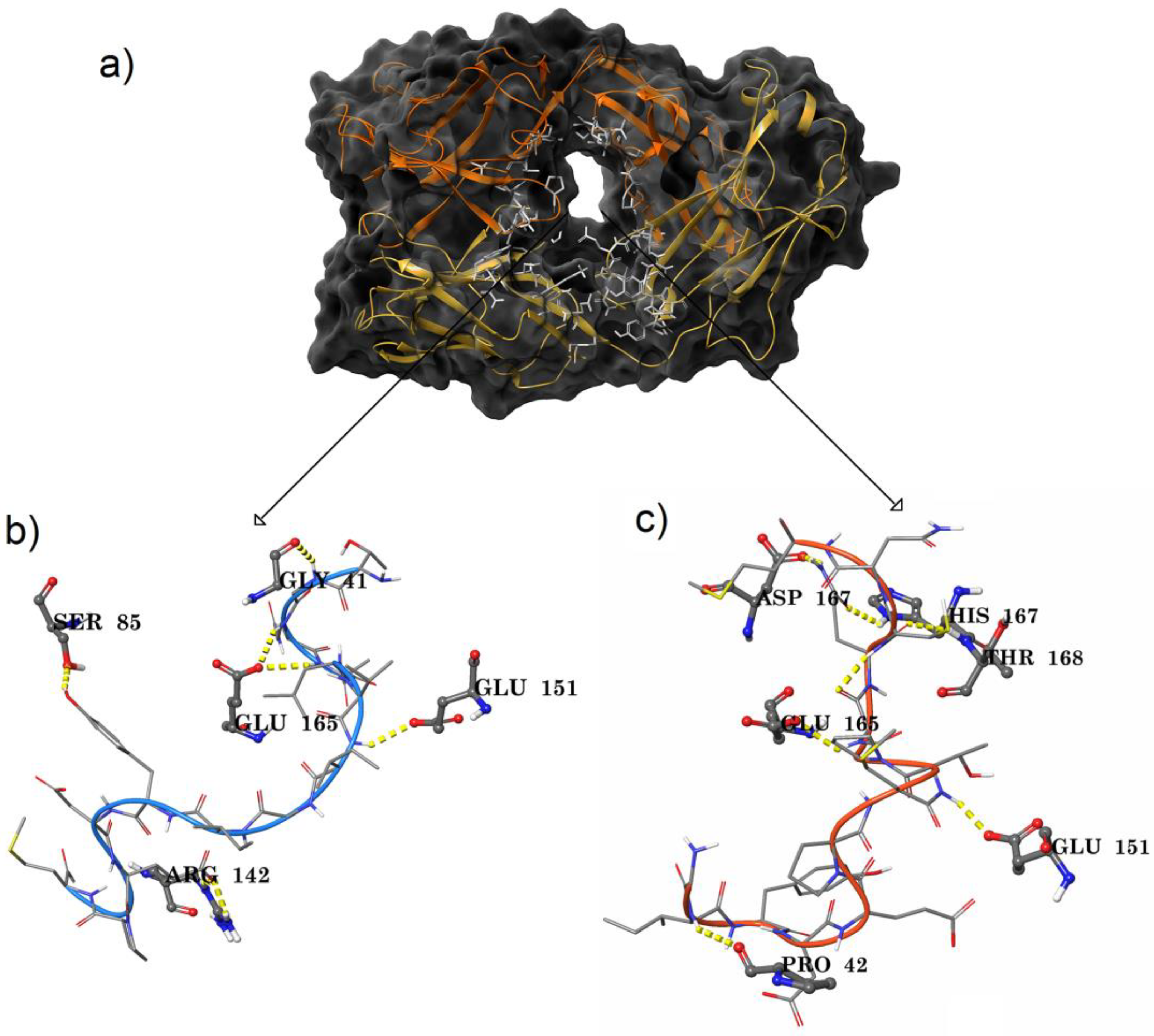

| Amino Acid Range | Heavy Chain | Light Chain |
|---|---|---|
| 0–50 | 40, 42 (PF4), 44, 45 | 12, 38, 40, 41 (PF3) |
| 51–100 | 91, 92, 93, 95 | 83, 84, 85 (PF3), 87, 100 |
| 101–150 | 112 | 101, 103, 104, 105, 140, 141, 142 (PF3), 143 |
| 151–200 | 151 (PF3 & PF4), 152, 153, 154, 155 163, 164, 165, 167 (PF4), 168 (PF4), 170, 171 | 163, 164, 165 (PF3 & PF4), 166, 167 (PF4), 168 (PF4), 173 |
| Peptide | IgG | IgE |
|---|---|---|
| MSWQTYVDEHLMCE (PF 1) | −11.62 | No binding |
| AQSTAFPQFKPEEM (PF 2) | −9.61 | −3.21 |
| TGQALVIGIYDEPM (PF 3) | −5.54 | −9.05 |
| GIYDEPMTPGQCNM (PF 4) | −5.37 | −9.05 |
| TPGQCNMVVERLGD (PF 5) | −12.60 | −12.05 |
| VVERLGDYLVEQGL (PF 6) | −9.25 | −10.63 |
| Serum | VC | PF3 | p-Value | ||
|---|---|---|---|---|---|
| D21 | D35 | D21 | D35 | (<0.05) | |
| IL2 | 35.20 ± 19.02 | 11.42 ± 3.62 | 608.46 ± 172.30 # | 26.76 ± 5.36 #,* | 0.02 |
| IL12 | 28.68 ± 4.91 | 26.07 ± 3.61 | 44.99 ± 10.20 | 47.39 ± 3.90 # | - |
| TNF-α | 143.33 ± 50.11 | 59.05 ± 15.54 | 12439.40 ± 445.69 # | 43.62 ± 6.87 * | 0.0004 |
| INF-γ | 22.24 ± 5.96 | 9.73 ± 4.20 | 0.97 ± 0.20 | 1.10 ± 0.34 # | - |
| IL5 | 114.87 ± 6.23 | 75.88 ± 6.23 | 1569.43 ± 375.16 # | 177.56 ± 19.73 #,* | 0.02 |
| IL10 | 18.99 ± 4.01 | 25.13 ± 6.59 | 31.96 ± 6.56 | 93.59 ± 7.52 #,* | 0.004 |
| GMCSF | 177.79 ± 70.58 | 82.72 ± 24.72 | 449.23 ± 286.95 | 173.52 ± 8.27 # | - |
| IgG | 6.77 ± 1.32 | 7.78 ± 1.52 | 4.01 ± 2.58 | 4.12 ± 1.35 | - |
| IgE | 13.57 ± 1.36 | 11.53 ± 1.15 | 65.63 ± 4.65 # | 3.74 ± 0.24 #,* | 0.002 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bokka, C.S.; Veeramachaneni, G.K.; Thunuguntla, V.B.S.C.; Bobbillapati, J.; Bondili, J.S. Peptide Mapping, In Silico and In Vivo Analysis of Allergenic Sorghum Profilin Peptides. Medicina 2019, 55, 178. https://doi.org/10.3390/medicina55050178
Bokka CS, Veeramachaneni GK, Thunuguntla VBSC, Bobbillapati J, Bondili JS. Peptide Mapping, In Silico and In Vivo Analysis of Allergenic Sorghum Profilin Peptides. Medicina. 2019; 55(5):178. https://doi.org/10.3390/medicina55050178
Chicago/Turabian StyleBokka, Chandra Sekhar, Ganesh Kumar Veeramachaneni, V. B. S. C. Thunuguntla, Janakiram Bobbillapati, and Jayakumar Singh Bondili. 2019. "Peptide Mapping, In Silico and In Vivo Analysis of Allergenic Sorghum Profilin Peptides" Medicina 55, no. 5: 178. https://doi.org/10.3390/medicina55050178