How Genetics and Genomics Advances Are Rewriting Pediatric Cancer Research and Clinical Care
Abstract
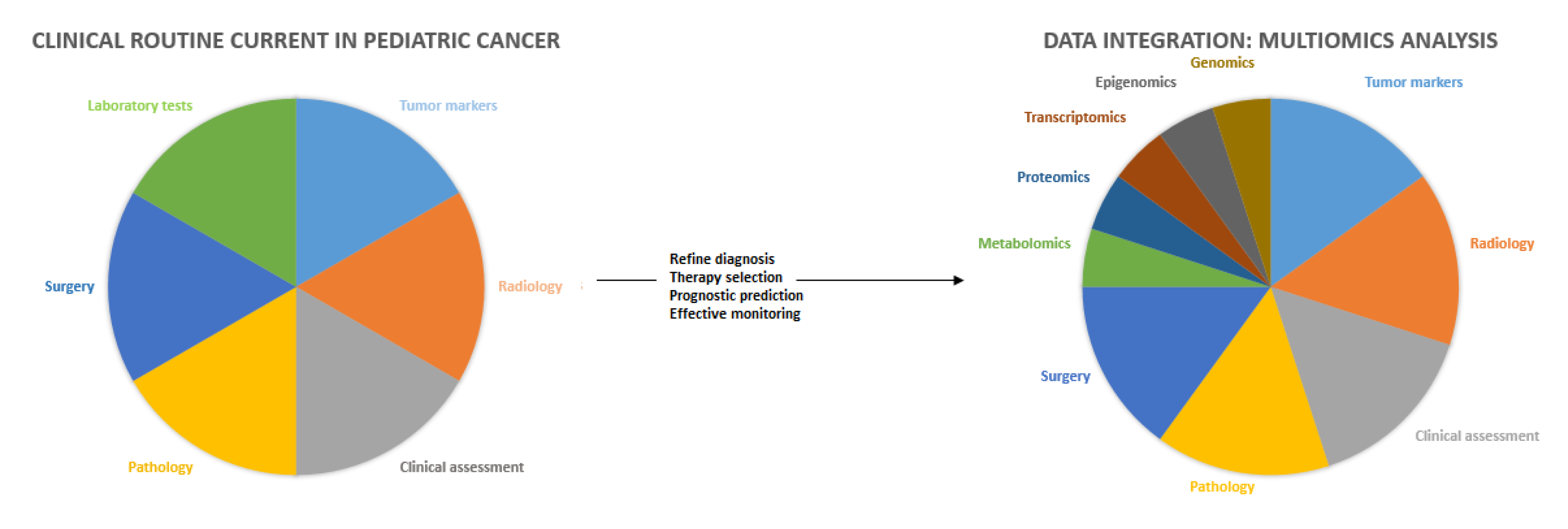
1. Introduction
2. Next-Generation Sequencing Promises a New Era in Pediatric Oncology: Impact on the Patient from Diagnosis to Prognosis
2.1. Large-Scale Next-Generation Sequencing in Pediatric Cancer Patients
2.2. Combined NGS Approach, a Strong Strategy Underlying the Synergy between Research and Diagnostics
3. Germline Variants and Cancer Predisposition Genes
4. Alterations in the Genes That Encode Proteins Involved in Epigenetic Regulation
5. From the Origin of Carcinogenesis to Relapse: The Importance of the Mutational Signature
6. Genomic Studies on Pediatric and Adult Cancers
7. The Road to Personalized Medicine
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steliarova-Foucher, E.; Colombet, M.; Ries, L.A.G.; Moreno, F.; Dolya, A.; Bray, F.; Hesseling, P.; Shin, H.Y.; Stiller, C.A. International incidence of childhood cancer, 2001–2010: A population-based registry study. Lancet Oncol. 2017, 18, 719–731. [Google Scholar] [CrossRef]
- World Health Organization. Available online: https://apps.who.int/iris/handle/10665/347370 (accessed on 29 July 2022).
- Hunger, S.P.; Mullighan, C.G. Acute Lymphoblastic Leukemia in Children. N. Engl. J. Med. 2015, 373, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Pourfeizi, A.; Dastgiri, S. Childhood Cancer in Iran. J. Pediatr. Hematol. Oncol. 2010, 32, 376–382. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Available online: https://seer.cancer.gov (accessed on 29 July 2022).
- Saletta, F.; Seng, M.S.; Lau, L.M. Advances in paediatric cancer treatment. Transl. Pediatr. 2014, 3, 156–182. [Google Scholar] [CrossRef]
- Blaney, S. Pizzo and Poplack’s Pediatric Oncology; Blaney, S., APaHL, Eds.; Wolters Kluwer: Alphen aan den Rijn, the Netherlands, 2021. [Google Scholar]
- Gatta, G.; Capocaccia, R.; Stiller, C.; Kaatsch, P.; Berrino, F.; Terenziani, M.; EUROCARE Working Group. Childhood cancer survival trends in Europe: A EUROCARE Working Group study. J. Clin. Oncol. 2005, 23, 3742–3751. [Google Scholar] [CrossRef]
- Gatta, G.; Zigon, G.; Capocaccia, R.; Coebergh, J.W.; Desandes, E.; Kaatsch, P.; Patore, G.; Peris-Bonet, R.; Stiller, C.A. Survival of European children and young adults with cancer diagnosed 1995–2002. Eur. J. Cancer 2009, 45, 992–1005. [Google Scholar] [CrossRef]
- Smith, M.A.; Seibel, N.L.; Altekruse, S.F.; Ries, L.A.; Melbert, D.L.; O’Leary, M.; Smith, F.O.; Reaman, G.H. Outcomes for children and adolescents with cancer: Challenges for the twenty-first century. J. Clin. Oncol. 2010, 28, 2625–2634. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pancancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018, 555, 371–376. [Google Scholar] [CrossRef]
- Gröbner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef]
- Chatsirisupachai, K.; Lagger, C.; de Magalhães, J.P. Age-associated differences in the cancer molecular landscape. Trends. Cancer 2022, 8033, 00135-2. [Google Scholar] [CrossRef]
- Mody, R.J.; Wu, Y.M.; Lonigro, R.J.; Cao, X.; Roychowdhury, S.; Vats, P.; Frank, K.M.; Prensner, J.R.; Asangani, I.; Palanisamy, N.; et al. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA 2015, 314, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Roy, A.; Yang, Y.; Wang, T.; Scollon, S.; Bergstrom, K.; Kerstein, R.A.; Gutierrez, S.; Petersen, A.K.; Bavle, A.; et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. 2016, 2, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Walsh, M.F.; Wu, G.; Edmonson, M.N.; Gruber, T.A.; Easton, J.; Hedges, D.; Ma, X.; Zhou, X.; Yergeau, D.A.; et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med. 2015, 373, 2336–2346. [Google Scholar] [CrossRef] [PubMed]
- Green, R.C.; Berg, J.S.; Grody, W.W.; Kalia, S.S.; Korf, B.R.; Martin, C.L.; McGuire, A.L.; Nussbaum, R.L.; O’Daniel, J.M.; Ormond, K.E.; et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013, 15, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Bombard, Y.; Offit, K.; Robson, M.E. Risks to relatives in genomic research: A duty to warn? Am. J. Bioeth. 2012, 12, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, D.E.; Chen, Y.; Jamieson, R.V.; Dalla-Pozza, L.; Byrne, J.A. Investigation of clinically relevant germline variants detected by next-generation sequencing in patients with childhood cancer: A review of the literature. J. Med. Genet. 2018, 55, 785–793. [Google Scholar] [CrossRef]
- Knudson, A.G., Jr. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc. Natl Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef]
- Machiela, M.J.; Ho, B.M.; Fisher, V.A.; Hua, X.; Chanock, S.J. Limited evidence that cancer susceptibility regions are preferential targets for somatic mutation. Genome Biol. 2015, 16, 193. [Google Scholar] [CrossRef]
- Vladoiu, M.C.; El-Hamamy, I.; Donovan, L.K.; Farooq, H.; Holgado, B.L.; Sundaravadanam, Y.; Ramaswamy, V.; Hendrikse, L.D.; Kumar, S.; Mack, S.C.; et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 2019, 572, 67–73. [Google Scholar] [CrossRef]
- Hudson, M.M.; Neglia, J.P.; Woods, W.G.; Sandlund, J.T.; Pui, C.H.; Kun, L.E.; Robison, L.L.; Green, D.M. Lessons from the past: Opportunities to improve childhood cancer survivor care through outcomes investigations of historical therapeutic approaches for pediatric hematological malignancies. Pediatr. Blood Cancer 2012, 58, 334–343. [Google Scholar] [CrossRef]
- Garraway, L.A. Genomics-driven oncology: Framework for an emerging paradigm. J. Clin Oncol. 2013, 31, 1806–1814. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.E.; Wheatley, K.; Hann, I.M.; Stevens, R.F.; Webb, D.; Hills, R.K.; De Graaf, S.S.N.; Harrison, C.J. Treatment strategy and long-term results in paediatric patients treated in consecutive UK AML trials. Leukemia 2005, 19, 2130–2138. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, D.; Walker, H.; Oliver, F.; Wheatley, K.; Harrison, C.; Harrison, G.; Rees, J.; Hann, I.; Stevens, R.; Burnett, A.; et al. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood 1998, 92, 2322–2333. [Google Scholar] [CrossRef] [PubMed]
- Rubnitz, J.E. Childhood acute myeloid leukemia. Curr. Treat. Options Oncol. 2008, 9, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Sabittini, E.; Bacci, F.; Sagramoso, C.; Pileri, S. WHO classification of tumours of the haematopoietic and lymphoid tissues in 2008: An overview. Pathologica 2010, 102, 83–88. [Google Scholar]
- Sahm, F.; Schrimpf, D.; Jones, D.T.; Meyer, J.; Kratz, A.; Reuss, D.; Capper, D.; Koelsche, C.; Korshunov, A.; Wiestler, B.; et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016, 131, 903–910. [Google Scholar] [CrossRef]
- Worst, B.C.; van Tilburg, C.M.; Balasubramanian, G.P.; Fiesel, P.; Witt, R.; Freitag, A.; Boudalil, M.; Previti, C.; Wolf, S.; Schmidt, S.; et al. Next-generation personalised medicine for high-risk paediatric cancer patients—The INFORM pilot study. Eur. J. Cancer 2016, 65, 91–101. [Google Scholar] [CrossRef]
- Wong, M.; Mayoh, C.; Lau, L.M.S.; Khuong-Quang, D.A.; Pinese, M.; Kumar, A.; Barahona, P.; Wilkie, E.E.; Sullivan, P.; Bowen-Jameset, R.; et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat. Med. 2020, 26, 1742–1753. [Google Scholar] [CrossRef]
- Newman, S.; Nakitandwe, J.; Kesserwan, C.A.; Azzato, E.M.; Wheeler, D.A.; Rusch, M.; Shurtleff, S.; Hedges, D.J.; Hamilton, K.V.; Foy, S.G.; et al. Genomes for Kids: The scope of pathogenic mutations in pediatric cancer revealed by comprehensive DNA and RNA sequencing. Cancer Discov. 2021, 11, 3008–3027. [Google Scholar] [CrossRef]
- Pikman, Y.; Tasian, S.K.; Sulis, M.L.; Stevenson, K.; Blonquist, T.M.; Apsel Winger, B.; Cooper, T.M.; Pauly, M.; Maloney, K.W.; Burke, M.J.; et al. Matched targeted therapy for pediatric patients with relapsed, refractory, or high-risk leukemias: A report from the LEAP Consortium. Cancer Discov. 2021, 11, 1424–1439. [Google Scholar] [CrossRef] [PubMed]
- Van Tilburg, C.M.; Pfaff, E.; Pajtler, K.W.; Langenberg, K.P.S.; Fiesel, P.; Jones, B.C.; Balasubramanian, G.P.; Stark, S.; Johann, P.D.; Blattner-Johnson, M.; et al. The pediatric precision oncology INFORM registry: Clinical outcome and benefit for patients with very high-evidence targets. Cancer Discov. 2021, 11, 2764–2779. [Google Scholar] [CrossRef] [PubMed]
- Forrest, S.J.; Geoerger, B.; Janeway, K.A. Precision medicine in pediatric oncology. Curr. Opin. Pediatr. 2018, 30, 17–24. [Google Scholar] [CrossRef]
- Mody, R.J.; Prensner, J.R.; Everett, J.; Parsons, D.W.; Chinnaiyan, A.M. Precision medicine in pediatric oncology: Lessons learned and next steps. Pediatr. Blood Cancer. 2017, 64. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J.; Morozova, O.; Attiyeh, E.F.; Asgharzadeh, S.; Wei, J.S.; Auclair, D.; Carter, S.L.; Cibulskis, K.; Hanna, M.; Kiezun, A.; et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013, 45, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Downing, J.R.; Wilson, R.K.; Zhang, J.; Mardis, E.R.; Pui, C.H.; Ding, L.; Ley, T.J.; Evans, W.E. The Pediatric Cancer Genome Project. Nat. Genet. 2012, 44, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012, 44, 251–253. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef]
- Chang, W.; Brohl, A.S.; Patidar, R.; Sindiri, S.; Shern, J.F.; Wei, J.S.; Song, Y.K.; Yohe, M.E.; Gryder, B.; Zhanget, S.; et al. MultiDimensional ClinOmics for precision therapy of children and adolescent young adults with relapsed and refractory cancer: A report from the center for cancer research. Clin. Cancer Res. 2016, 22, 3810–3820. [Google Scholar] [CrossRef]
- Surrey, L.F.; MacFarland, S.P.; Chang, F.; Cao, K.; Rathi, K.S.; Akgumus, G.T.; Gallo, D.; Lin, F.; Gleason, A.; Raman, P.; et al. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Gen. Med. 2019, 11, 32. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymondet, V.; et al. Integrative clinical genomics of metastatic cancer. Nature 2017, 548, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Rusch, M.; Nakitandwe, J.; Shurtleff, S.; Newman, S.; Zhang, Z.; Edmonson, M.N.; Parker, M.; Jiao, Y.; Ma, X.; Liu, Y.; et al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat. Commun. 2018, 9, 3962. [Google Scholar] [CrossRef] [PubMed]
- Gadd, S.; Huff, V.; Walz, A.L.; Ooms, A.H.A.G.; Armstrong, A.E.; Gerhard, D.S.; Smith, M.A.; Auvil, J.M.G.; Meerzaman, D.; Chen, Q.R.; et al. A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat. Genet. 2017, 49, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Easton, J.; Shao, Y.; Maciaszek, J.; Wang, Z.; Wilkinson, M.R.; McCastlain, K.; Edmonson, M.; Pounds, S.B.; Shi, L.; et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017, 49, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, H.; Farrar, J.E.; Triche Jr, T.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef]
- Jia, D.; Dong, R.; Jing, Y.; Xu, D.; Wang, Q.; Chen, L.; Li, Q.; Huang, Y.; Zhang, Y.; Zhang, Z.; et al. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology 2014, 60, 1686–1696. [Google Scholar] [CrossRef]
- Sumazin, P.; Chen, Y.; Treviño, L.R.; Sarabia, S.F.; Hampton, O.A.; Patel, K.; Mistretta, T.A.; Zorman, B.; Thompson, P.; Hec-zey, A.; et al. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology 2017, 65, 104–121. [Google Scholar] [CrossRef]
- Carrillo-Reixach, J.; Torrens, L.; Simon-Coma, M.; Royo, L.; Domingo-Sàbat, M.; Abril-Fornaguera, J.; Akers, N.; Sala, M.; Ragull, S.; Arnal, M.; et al. Epigenetic footprint enables molecular risk stratification of hepato-blastoma with clinical implications. J. Hepatol. 2020, 73, 328–341. [Google Scholar] [CrossRef]
- Vilarinho, S.; Erson-Omay, E.Z.; Harmanci, A.S.; Morotti, R.; Carrion-Grant, G.; Baranoski, J.; Knisely, A.S.; Ekong, U.; Emre, S.; Yasuno, K.; et al. Paediatric hepatocellular carcinoma due to somatic CTNNB1 and NFE2L2 mutations in the setting of inherited bi-allelic ABCB11 mutations. J. Hepatol. 2014, 61, 1178–1183. [Google Scholar] [CrossRef]
- Lee, R.S.; Stewart, C.; Carter, S.L.; Ambrogio, L.; Cibulskis, K.; Sougnez, C.; Lawrence, M.S.; Auclair, D.; Mora, J.; Golub, T.R.; et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J. Clin. Investig. 2012, 122, 2983–2988. [Google Scholar] [CrossRef]
- Stichel, D.; Schrimpf, D.; Casalini, B.; Meyer, J.; Wefers, A.K.; Sievers, P.; Korshunov, A.; Koelsche, C.; Reuss, D.E.; Reinhardt, A.; et al. Routine RNA sequencing of formalin-fixed paraffin-embedded specimens in neuropathology diagnostics identifies diagnostically and therapeutically relevant gene fusions. Acta Neuropathol. 2019, 138, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Maraka, S.; Janku, F. BRAF alterations in primary brain tumors. Discov. Med. 2018, 26, 51–60. [Google Scholar] [PubMed]
- Boer, J.M.; Valsecchi, M.G.; Hormann, F.M.; Antić, Z.; Zaliova, M.; Schwab, C.; Cazzaniga, G.; Arfeuille, C.; Cavé, H.; At-tarbaschi, A.; et al. Favorable outcome of NUTM1-rearranged infant and pediatric B cell precursor acute lymphoblastic leu-kemia in a collaborative international study. Leukemia 2021, 35, 2978–2982. [Google Scholar] [CrossRef] [PubMed]
- Ricarte-Filho, J.C.; Li, S.; Garcia-Rendueles, M.E.R.; Montero-Conde, C.; Voza, F.; Knauf, J.A.; Heguy, A.; Viale, A.; Bogda-nova, T.; Thomas, G.A.; et al. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid can-cers. J. Clin. Investig. 2013, 123, 4935–4944. [Google Scholar] [CrossRef]
- Northcott, P.A.; Lee, C.; Zichner, T.; Stütz, A.M.; Erkek, S.; Kawauchi, D.; Shih, D.J.H.; Hovestadt, V.; Zapatka, M.; Sturm, D.; et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 2014, 511, 428–434. [Google Scholar] [CrossRef]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L.; et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef]
- LaHaye, S.; Fitch, J.R.; Voytovich, K.J.; Herman, A.C.; Kelly, B.J.; Lammi, G.E.; Arbesfeld, J.A.; Wijeratne, S.; Franklin, S.J.; Schieffer, K.M.; et al. Discovery of clinically relevant fusions in pediatric cancer. BMC Genom. 2021, 22, 872. [Google Scholar] [CrossRef]
- Kline, C.N.; Joseph, N.M.; Grenert, J.P.; van Ziffle, J.; Talevich, E.; Onodera, C.; Aboian, M.; Cha, S.; Raleigh, S.R.; Braunstein, S.; et al. Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies patho-genic germline mutations, and directs targeted therapy. Neuro. Oncol. 2017, 19, 699–709. [Google Scholar] [CrossRef]
- Oberg, J.A.; Bender, J.L.; Sulis, M.L.; Pendrick, D.; Sireci, A.N.; Hsiao, S.J.; Turk, A.T.; Cruz, F.S.D.; Hibshoosh, H.; Remotti, H.; et al. Implementation of next generation sequencing into pediatric hematology-oncology practice: Moving beyond ac-tionable alterations. Gen. Med. 2016, 8, 133. [Google Scholar] [CrossRef]
- Marks, L.J.; Oberg, J.A.; Pendrick, D.; Sireci, A.N.; Glasser, C.; Coval, C.; Zylber, R.J.; Chung, W.K.; Pang, J.; Turk, A.J.; et al. Precision medicine in children and young adults with hematologic malignancies and blood disorders: The Columbia Uni-versity Experience. Front. Pediatr. 2017, 5, 265. [Google Scholar] [CrossRef]
- Lin, J.J.; Riely, G.J.; Shaw, A.T. Targeting ALK: Precision medicine takes on drug resistance. Cancer Discov. 2017, 7, 137–155. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Hübner, J.M.; Sharma, T.; Luu, B.; Sill, M.; Zapotocky, M.; Mack, S.C.; Witt, H.; Lin, T.; Shih, D.J.H.; et al. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol. 2018, 136, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.; Mohankumar, K.M.; Punchihewa, C.; Weinlich, R.; Dalton, J.D.; Li, Y.; Lee, R.; Tatevossian, R.G.; Phoenix, T.N.; Thiruvenkatam, R.; et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature 2014, 506, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Mack, S.C.; Pajtler, K.W.; Chavez, L.; Okonechnikov, K.; Bertrand, K.C.; Wang, X.; Erkek, S.; Federation, A.; Song, A.; Lee, C.; et al. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 2018, 553, 101–105. [Google Scholar] [CrossRef]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef]
- Robinson, G.W.; Rudneva, V.A.; Buchhalter, I.; Billups, C.A.; Waszak, S.M.; Smith, K.S.; Bowers, D.C.; Bendel, A.; Fisher, P.G.; Partap, S.; et al. Risk-adapted therapy for young children with medulloblastoma (SJYC07): Therapeutic and molecular out-comes from a multicentre, phase 2 trial. Lancet Oncol. 2018, 19, 768–784. [Google Scholar] [CrossRef]
- Clarke, M.; Mackay, A.; Ismer, B.; Pickles, J.C.; Tatevossian, R.G.; Newman, S.; Bale, T.A.; Stoler, I.; Izquierdo, E.; Temelso, S.; et al. Infant High-Grade Gliomas Comprise Multiple Subgroups Characterized by Novel Targetable Gene Fusions and Fa-vorable Outcomes. Cancer Discov. 2020, 10, 942–963. [Google Scholar] [CrossRef]
- Eaton, K.W.; Tooke, L.S.; Wainwright, L.M.; Judkins, A.R.; Biegel, A.J. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr. Blood Cancer 2011, 56, 7–15. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Kamihara, J.; Evans, D.G.R.; Brugières, L.; Bourdeaut, F.; Molenaar, J.J.; Walsh, M.F.; Brodeur, G.M.; Diller, L. Cancer Surveillance in Gorlin Syndrome and Rhabdoid Tumor Predisposition Syndrome. Clin. Cancer Res. 2017, 23, e62–e67. [Google Scholar] [CrossRef]
- Bruggers, C.S.; Bleyl, S.B.; Pysher, T.; Barnette, P.; Afify, Z.; Walker, M.; Biegel, A.J. Clinicopathologic Comparison of Fa-milial Versus Sporadic Atypical teratoid/rhabdoid Tumors (AT/RT) of the Central Nervous System. Pediatr. Blood Cancer 2011, 56, 1026–1031. [Google Scholar] [CrossRef]
- Sévenet, N.; Sheridan, E.; Amram, D.; Schneider, P.; Handgretinger, R.; Delattre, O. Constitutional Mutations of the hSNF5/INI1 Gene Predispose to a Variety of Cancers. Am. J. Hum. Genet. 1999, 65, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Biegel, J.A.; Busse, T.M.; Weissman, B.E. SWI/SNF Chromatin Remodeling Complexes and Cancer. Am. J. Med. Genet. 2014, 166C, 350–366. [Google Scholar] [CrossRef] [PubMed]
- Gigante, L.; Paganini, I.; Frontali, M.; Ciabattoni, S.; Sangiuolo, F.C.; Papi, L. Rhabdoid tumor predisposition syndrome caused by SMARCB1 constitutional deletion: Prenatal detection of new case of recurrence in siblings due to gonadal mosai-cism. Fam. Cancer 2016, 15, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Biegel, J.A.; Zhou, J.Y.; Rorke, L.B.; Stenstrom, C.; Wainwright, L.M.; Fogelgren, B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999, 59, 74–79. [Google Scholar] [PubMed]
- Paparella, R.; Caroleo, A.M.; Agolini, E.; Chillemi, G.; Miele, E.; Pedace, L.; Rinelli, M.; Pizzi, S.; Boccuto, L.; Colafati, G.S.; et al. Posterior fossa ependymoma in neurodevelopmental syndrome caused by a de novo germline pathogenic Polr2a vari-ant. Am. J. Med. Gen. 2022, 188, 2796–2802. [Google Scholar] [CrossRef]
- Palculict, T.B.; Ruteshouser, E.C.; Fan, Y.; Wang, W.; Strong, L.; Huff, V. Identification of germline DICER1 mutations and loss of heterozygosity in familial Wilms tumour. J. Med. Genet. 2016, 53, 385–388. [Google Scholar] [CrossRef]
- Huff, V. Wilms tumor genetics. Am. J. Med. Genet. 1998, 79, 260–267. [Google Scholar] [CrossRef]
- Mahamdallie, S.; Yost, S.; Poyastro-Pearson, E.; Holt, E.; Zachariou, A.; Seal, S.; Elliott, A.; Clarke, M.; Warren-Perry, M.; Hanks, S.; et al. Identification of new Wilms tumour predisposition genes: An exome sequencing study. Lancet Child. Ado-lesc. Health 2019, 3, 322–331. [Google Scholar] [CrossRef]
- Mahamdallie, S.S.; Hanks, S.; Karlin, K.L.; Zachariou, A.; Perdeaux, E.R.; Ruark, E.; Shaw, C.A.; Renwick, A.; Ramsay, E.; Yost, S.; et al. Mutations in the transcriptional repressor REST predispose to Wilms tumor. Nat. Genet. 2015, 47, 1471–1474. [Google Scholar] [CrossRef]
- Boni, A.; Ranalli, M.; Del Baldo, G.; Carta, R.; Lodi, M.; Agolini, E.; Rinelli, M.; Valentini, D.; Rossi, S.; Alesi, V.; et al. Me-dulloblastoma Associated with Down Syndrome: From a Rare Event Leading to a Pathogenic Hypothesis. Diagnostics 2021, 11, 254. [Google Scholar] [CrossRef]
- Miele, E.; Di Giannatale, A.; Crocoli, A.; Cozza, R.; Serra, A.; Castellano, A.; Cacchione, A.; Cefalo, M.G.; Alaggio, R.; De Pasquale, M.D. Clinical, Genetic, and Prognostic Features of Adrenocortical Tumors in Children: A 10-Year Single-Center Experience. Front. Oncol. 2020, 13, 5266. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Zhu, B.; Koster, R.; Karlins, E.; Dean, M.; Yeager, M.; Gianferante, M.; Spector, L.G.; Morton, L.M.; Karyadi, D.; et al. Frequency of Pathogenic Germline Variants in Cancer-Susceptibility Genes in Patients with Osteosarcoma. JAMA Oncol. 2020, 6, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Fiala, E.M.; Jayakumaran, G.; Mauguen, A.; Kennedy, J.A.; Bouvier, N.; Kemel, Y.; Fleischut, M.H.; Maio, A.; Salo-Mullen, E.E.; Sheehan, M.; et al. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 pa-tients with pediatric solid tumors. Nat. Cancer 2021, 2, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wilson, C.L.; Easton, J.; Thrasher, A.; Mulder, H.; Liu, Q.; Hedges, D.J.; Wang, S.; Rusch, M.C.; Edmonson, M.N.; et al. Genetic risk for subsequent neoplasms among long-term survivors of childhood cancer. J. Clin. Oncol. 2018, 36, 2078–2087. [Google Scholar] [CrossRef]
- Wang, Z.; Wilson, C.L.; Armstrong, G.T.; Hudson, M.M.; Zhang, J.; Nichols, K.E.; Robison, L.L. Association of germline BRCA2 mutations with the risk of pediatric or adolescent non-Hodgkin lymphoma. JAMA Oncol. 2019, 5, 1362–1364. [Google Scholar] [CrossRef]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Versteege, I.; Sévenet, N.; Lange, J.; Rousseau-Merck, M.F.; Ambros, P.; Handgretinger, R.; Aurias, A.; Delattre, O. Truncat-ing mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998, 394, 203–206. [Google Scholar] [CrossRef]
- Scott, R.H.; Murray, A.; Baskcomb, L.; Turnbull, C.; Loveday, C.; Al-Saadi, R.; Williams, R.; Breatnach, F.; Gerrard, M.; Hale, J.; et al. Stratification of Wilms tumor by genetic and epigenetic analysis. Oncotarget 2012, 3, 327–335. [Google Scholar] [CrossRef]
- Wegert, J.; Wittmann, S.; Leuschner, I.; Geissinger, E.; Graf, N.; Gessler, M. WTX inactivation is a frequent, but late event in Wilms tumors without apparent clinical impact. Genes Chrom. Cancer 2009, 48, 1102–1111. [Google Scholar] [CrossRef]
- Scott, R.H.; Douglas, J.; Baskcomb, L.; Huxter, N.; Barker, K.; Hanks, S.; Craft, A.; Gerrard, M.; Kohler, J.A.; Levitt, G.A.; et al. Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat. Genet. 2008, 40, 1329–1334. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.W.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.K.; Tönjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Castel, D.; Philippe, C.; Calmon, R.; Le Dret, L.; Truffaux, N.; Boddaert, N.; Pagès, M.; Taylor, K.R.; Saulnier, P.; Lacroix, L.; et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropath. 2015, 130, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Malinowska-Ozdowy, K.; Frech, C.; Schönegger, A.; Eckert, C.; Cazzaniga, G.; Stanulla, M.; zur Stadt, U.; Mecklenbräuker, A.; Schuster, M.; Kneidinger, D.; et al. KRAS and CREBBP mutations: A relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia 2015, 29, 1656–1667. [Google Scholar] [CrossRef] [PubMed]
- Mar, B.G.; Bullinger, L.B.; McLean, K.M.; Grauman, P.V.; Harris, M.H.; Stevenson, K.; Neuberg, D.S.; Sinha, A.U.; Sallan, S.E.; Silverman, L.B.; et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat. Commun. 2014, 5, 3469. [Google Scholar] [CrossRef]
- Jacob, J.D.; Wang, Y.; Chan, H.-M.; Zhang, J.; Huether, R.; Kryukov, G.V.; Bhang, H.C.; Taylor, J.E.; Hu, M.; Englund, N.P.; et al. Global Chromatin Profiling Reveals NSD2 Mutations in Pediatric Acute Lymphoblastic Leukemia. Nat. Genet. 2013, 45, 1386–1391. [Google Scholar] [CrossRef]
- Jianping, L.; Hlavka-Zhang, J.; Shrimp, J.; Piper, C.; Dupéré-Richér, D.; Roth, J.S.; Jing, D.; Román, H.L.C.; Troche, C.; Swaroop, A.; et al. PRC2 Inhibitors Overcome Glucocorticoid Resistance Driven by NSD2 Mutation in Pediatric Acute Lymphoblastic Leukemia. Cancer Discov. 2022, 12, 186–203. [Google Scholar] [CrossRef]
- Pui, C.-H.; Chessells, J.M.; Camitta, B.; Baruchel, A.; Biondi, A.; Boyett, J.M.; Carroll, A.; Eden, O.B.; Evans, W.E.; Gadner, H.; et al. Clinical Heterogeneity in Childhood Acute Lymphoblastic Leukemia with 11q23 Rearrangements. Leukemia 2003, 17, 700–706. [Google Scholar] [CrossRef]
- Johansson, B.; Moorman, A.V.; Haas, O.A.; Watmore, A.E.; Cheung, K.l.; Swanton, S.; Secker-Walker, L.M. Hematologic Malig-nancies with t(4;11)(Q21;Q23)—A Cytogenetic, Morphologic, Immunophenotypic and Clinical Study of 183 Cases. Leukemia 1998, 12, 779–787. [Google Scholar] [CrossRef]
- Raimondi, S.C.; Peiper, S.C.; Kitchingman, G.R.; Behm, F.G.; Williams, D.L.; Hancock, M.L.; Mirro, J. Childhood Acute Lymphoblastic Leukemia with Chromosomal Breakpoints at 11q23. Blood 1989, 73, 1627–1634. [Google Scholar] [CrossRef]
- Christine, J.H.; Moorman, A.V.; Barber, K.E.; Broadfield, Z.J.; Cheung, K.L.; Harris, R.L.; Reza Jalali, G.; Robinson, H.M.; Strefford, J.C.; Stewart,, A.; et al. Interphase Molecular Cytogenetic Screening for Chromosomal Abnormalities of Prognostic Significance in Childhood Acute Lymphoblastic Leukaemia: A UK Cancer Cytogenetics Group Study. Br. J. Haematol. 2005, 129, 520–530. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.W.; Liu, Y.; Ma, X.; Gout, A.M.; Hagiwara, K.; Zhou, X.; Wang, J.; Macias, M.; Chen, X.; Easton, J.; et al. Panneuro-blastoma analysis reveals age- and signature-associated driver alterations. Nat. Commun. 2020, 11, 5183. [Google Scholar] [CrossRef] [PubMed]
- McLeod, C.; Gout, A.M.; Zhou, X.; Thrasher, A.; Rahbarinia, D.; Brady, S.W.; Macias, M.; Birch, K.; Finkelstein, D.; Sunny, J.; et al. St. Jude cloud-a pediatric cancer genomic data sharing ecosystem. Cancer Discov. 2021, 11, 1082–1099. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Brady, S.W.; Ma, X.; Shen, S.; Zhang, Y.; Li, Y.; Szlachta, K.; Dong, L.; Liu, Y.; Yang, F.; et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 2020, 135, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Klapper, W.; Szczepanowski, M.; Burkhardt, B.; Berger, H.; Rosolowski, M.; Bentink, S.; Schwaenen, C.; Wessendorf, S.; Spang, R.; Möller, P.; et al. Molecular profiling of pediatric mature B-cell lymphoma treated in population-based prospec-tive clinical trials. Blood 2008, 112, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Deffenbacher, K.E.; Iqbal, J.; Sanger, W.; Shen, Y.; Lachel, C.; Liu, Z.; Liu, Y.; Lim, M.S.; Perkins, S.L.; Fu, K.; et al. Molecular distinctions between pediatric and adult mature B-cell non-Hodgkin lymphomas identified through genomic profiling. Blood 2012, 119, 3757–3766. [Google Scholar] [CrossRef]
- Brady, S.W.; Ma, X.; Bahrami, A.; Satas, G.; Wu, G.; Newman, S.; Rusch, M.; Putnam, D.K.; Mulder, H.L.; Yergeau, D.A.; et al. The clonal evolution of metastatic osteosarcoma as shaped by cisplatin treatment. Mol. Cancer Res. 2019, 17, 895–906. [Google Scholar] [CrossRef]
- Tarlock, K.; Lamble, A.J.; Wang, Y.C.; Gerbing, R.B.; Ries, R.E.; Loken, M.R.; Brodersen, L.E.; Pardo, L.; Leonti, A.; Smith, J.L.; et al. CEBPA-bZip mutations are associated with favorable prognosis in de novo AML: A report from the ChildreN’s Oncology Group. Blood 2021, 138, 1137–1147. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Tzoneva, G.; Dieck, C.L.; Oshima, K.; Ambesi-Impiombato, A.; Sánchez-Martín, M.; Madubata, C.J.; Khiabanian, H.; Yu, J.; Waanders, E.; Iacobucci, I.; et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature 2018, 553, 511–514. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Zhang, J.; Kasper, L.H.; Lerach, S.; Payne-Turner, D.; Phillips, L.A.; Heatley, S.L.; Holmfeldt, L.; Col-lins-Underwood, J.R.; Ma, J.; et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011, 471, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Zamora, A.E.; Crawford, J.C.; Allen, E.K.; Guo, X.J.; Bakke, J.; Carter, R.A.; Abdelsamed, H.A.; Moustaki, A.; Li, Y.; Chang, T.C.; et al. Pediatric patients with acute lymphoblastic leukemia generate abundant and functional neoantigen-specific CD8(+) T cell responses. Sci. Transl. Med. 2019, 11, 8549. [Google Scholar] [CrossRef] [PubMed]
- Schmiegelow, K.; Björk, O.; Glomstein, A.; Gustafsson, G.; Keiding, N.; Kristinsson, J.; Mäkipernaa, A.; Rosthøj, S.; Szumlanski, C.; Sørensen, T.M.; et al. Intensification of mercaptopurine/methotrexate maintenance chemotherapy may in-crease the risk of relapse for some children with acute lymphoblastic leukemi. J. Clin. Oncol. 2003, 21, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Waanders, E.; Zhaohui, G.; Dobson, S.M.; Antić, Ž.; Crawford, J.C.; Ma, X.; Edmonson, M.N.; Payne-Turner, D.; van de Vorst, M.; Jongmans, M.C.J.; et al. Mutational Landscape and Patterns of Clonal Evolution in Relapsed Pediatric Acute Lymphoblastic Leukemia. Blood Cancer Discov. 2020, 1, 96–111. [Google Scholar] [CrossRef]
- Evensen, N.A.; Madhusoodhan, P.P.; Meyer, J.; Saliba, J.; Chowdhury, A.; Araten, D.J.; Nersting, J.; Bhatla, T.; Vincent, T.L.; Teachey, D.; et al. MSH6 Haploinsufficiency at Relapse Contributes to the Development of Thiopurine Resistance in Pediatric B-Lymphoblastic Leukemia. Haematologica 2018, 103, 830–839. [Google Scholar] [CrossRef]
- Fan, Y.; Brady, S.W.; Tang, C.; Sun, H.; Du, L.; Barz, M.J.; Ma, X.; Chen, Y.; Fang, H.; Li, X.; et al. Chemotherapy and Mismatch Repair Deficiency Cooperate to Fuel TP53 Mutagenesis and ALL Relapse. Nat. Cancer 2021, 2, 819–834. [Google Scholar] [CrossRef]
- Schwartz, J.R.; Ma, J.; Kamens, J.; Westover, T.; Walsh, M.P.; Brady, S.W.; Robert Michael, J.; Chen, X.; Montefiori, L.; Song, G.; et al. The acquisition of molecular drivers in pediatric therapy-related myeloid neoplasms. Nat. Commun. 2021, 12, 985. [Google Scholar] [CrossRef]
- Ma, X.; Edmonson, M.; Yergeau, D.; Muzny, D.M.; Hampton, O.A.; Rusch, M.; Song, G.; Easton, J.; Harvey, R.C.; Wheeler, D.A.; et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat. Commun. 2015, 6, 6604. [Google Scholar] [CrossRef]
- Antić, Ž.; Yu, J.; van Reijmersdal, S.V.; van Dijk, A.; Dekker, L.; Segerink, W.H.; Sonneveld, E.; Fiocco, M.; Pieters, R.; Hoogerbrugge, P.M.; et al. Multiclonal Complexity of Pediatric Acute Lymphoblastic Leukemia and the Prognostic Relevance of Subclonal Mutations. Haematologica 2021, 106, 3046–3055. [Google Scholar] [CrossRef]
- Jerchel, I.S.; Hoogkamer, A.Q.; Ariës, I.M.; Steeghs, E.M.P.; Boer, N.J.M.; Besselink, A.B.; Boeree, A.; van de Ven, C.; de Groot-Kruseman, H.A.; de Haas, V.; et al. RAS Pathway Mutations as a Predictive Biomarker for Treatment Adaptation in Pediatric B-Cell Precursor Acute Lymphoblastic Leukemia. Leukemia 2018, 32, 931–940. [Google Scholar] [CrossRef]
- D’Angelo, F.; Ceccarelli, M.; Tala; Garofano, L.; Zhang, J.; Frattini, V.; Caruso, F.P.; Lewis, G.; Alfaro, K.D..; Bauchet, L.; et al. The molecular landscape of glioma in patients with Neurofibromatosis 1. Nat. Med. 2019, 25, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Wang, Z.; Saffern, M.; Jun, T.; Huang, K.I. Genomic and molecular features distinguish young adult cancer from later-onset cancer. Cell Rep. 2021, 37, 110005. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Chatsirisupachai, K.; Lesluyes, T.; Paraoan, L.; Van Loo, P.; de Magalhães, J.P. An integrative analysis of the age-associated multi-omic landscape across cancers. Nat. Commun. 2021, 12, 2345. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Haider, S.; Boutros, P.C. Age influences on the molecular presentation of tumours. Nat. Commun. 2022, 13, 208. [Google Scholar] [CrossRef] [PubMed]
- Von Bueren, A.O.; Kortmann, R.D.; von Hoff, K.; Friedrich, C.; Mynarek, M.; Müller, K.; Goschzik, T.; Mühlen, A.z.; Gerber, N.; Warmuth-Metz, M.; et al. Treatment of Children and Adolescents With Metastatic Medulloblastoma and Prognostic Relevance of Clinical and Biologic Parameters. J. Clin. Oncol. 2016, 34, 4151–4160. [Google Scholar] [CrossRef]
- Vijay, R.; Remke, M.; Bouffet, E.; Bailey, S.; Clifford, S.C.; Doz, F.; Kool, M.; Dufour, C.; Vassal, G.; Milde, T.; et al. Risk Stratification of Childhood Medulloblastoma in the Molecular Era: The Current Consensus. Acta Neuropathol. 2016, 131, 821–831. [Google Scholar] [CrossRef]
- The SIOP-Europe PNET Group; Clifford, S.C.; Lannering, B.; Schwalbe, E.C.; Hicks, D.; Toole, K.O.; Nicholson, S.L.; Goschzik, T.; Mühlen, A.z.; Figarella-Branger, D.; et al. Biomarker-Driven Stratification of Disease-Risk in Non-Metastatic Medulloblastoma: Results from the Multi-Center HIT-SIOP-PNET4 Clinical Trial. Oncotarget 2015, 6, 38827–38839. [Google Scholar] [CrossRef]
- Phipps, A.I.; Limburg, P.J.; Baron, J.A.; Burnett-Hartman, A.N.; Weisenberger, D.J.; Laird, P.W.; Sinicrope, F.A.; Rosty, C.; Buchanan, D.D.; Potter, J.D.; et al. Association Between Molecular Subtypes of Colorectal Cancer and Patient Survival. Gastroenterology 2015, 148, 77–87.e2. [Google Scholar] [CrossRef]
- Lourenço, S.V.; Boggio, P.; Nico, M.M.S. Inflammatory Myofibroblastic Tumor of the Tongue: Report of an Unusual Case in a Teenage Patient. Dermatol. Online J. 2012, 18, 6. [Google Scholar] [CrossRef]
- Balachandran, V.P.; DeMatteo, R.P. Gastrointestinal Stromal Tumors. Adv. Surg. 2014, 48, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational Landscape and Significance across 12 Major Cancer Types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and Saturation Analysis of Cancer Genes across 21 Tumour Types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Leiserson, M.D.M.; Vandin, F.; Wu, H.; Dobson, J.R.; Eldridge, J.V.; Thomas, J.L.; Papoutsaki, A.; Kim, Y.; Niu, B.; McLellan, M.; et al. Pan-Cancer Network Analysis Identifies Combinations of Rare Somatic Mutations across Pathways and Protein Complexes. Nat. Genet. 2015, 47, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.; Wala, J.; Mermel, C.H.; et al. Pan-Cancer Patterns of Somatic Copy Number Alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef]
- de Rooij, J.D.; Branstetter, C.; Ma, J.; Li, Y.; Walsh, M.P.; Cheng, J.; Obulkasim, A.; Dang, J.; Easton, J.; Verboon, L.J.; et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat. Genet. 2017, 49, 451–456. [Google Scholar] [CrossRef]
- Iacobucci, I.; Wen, J.; Meggendorfer, M.; Choi, J.K.; Shi, L.; Pounds, S.B.; Carmichael, C.L.; Masih, K.E.; Morris, S.M.; Lindsley, R.C.; et al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat. Genet. 2019, 51, 694–704. [Google Scholar] [CrossRef]
- Andersson, A.K.; Ma, J.; Wang, J.; Chen, X.; Gedman, A.L.; Dang, J.; Nakitandwe, J.; Holmfeldt, L.; Parker, M.; Easton, J.; et al. The Landscape of Somatic Mutations in Infant MLL-Rearranged Acute Lymphoblastic Leukemias. Nat. Genet. 2015, 47, 330–337. [Google Scholar] [CrossRef]
- Roberts, K.G.; Gu, Z.; Payne-Turner, D.; McCastlain, K.; Harvey, R.C.; Chen, I.-M.; Pei, D.; Iacobucci, I.; Valentine, M.; Pounds, S.B.; et al. High Frequency and Poor Outcome of Philadelphia Chromosome–Like Acute Lymphoblastic Leukemia in Adults. J. Clin. Oncol. 2017, 35, 394–401. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Miller, C.B.; Radtke, I.; Phillips, L.A.; Dalton, J.; Ma, J.; White, D.; Hughes, T.P.; le Beau, M.M.; Pui, C.; et al. BCR–ABL1 Lymphoblastic Leukaemia Is Characterized by the Deletion of Ikaros. Nature 2008, 453, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Berlanga, P.; Pierron, G.; Lacroix, L.; Chicard, M.; de Beaumais, T.A.; Marchais, A.; Harttrampf, A.C.; Iddir, Y.; Larive, A.; Fernandez, A.S.; et al. The European MAPPYACTS Trial: Precision Medicine Program in Pediatric and Adolescent Patients with Recurrent Malignancies. Cancer Discov. 2022, 12, 1266–1281. [Google Scholar] [CrossRef] [PubMed]
- Janoueix-Lerosey, I.; Lequin, D.; Brugières, L.; Ribeiro, A.; de Pontual, L.; Combaret, V.; Raynal, V.; Puisieux, A.; Schleier-macher, G.; Pierron, G.; et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 2008, 455, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Takita, J.; Choi, Y.L.; Kato, M.; Ohira, M.; Sanada, M.; Wang, L.; Soda, M.; Kikuchi, A.; IgarashI, T.; et al. Oncogenic mutations of ALK kinase in Neuroblastoma. Nature 2008, 455, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Mosse, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.L.; Perri, P.; et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef] [PubMed]
- George, R.E.; Sanda, T.; Hanna, M.; Fröhling, S.; Luther, W., 2nd; Zhang, J.; Ahn, Y.; Zhou, W.; London, W.B.; McGrady, P.; et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 2008, 455, 975–978. [Google Scholar] [CrossRef]
- Next Generation Personalized Neuroblastoma Therapy (NEPENTHE). Available online: https://clinicaltrials.gov/ct2/show/NCT02780128 (accessed on 16 September 2022).
- Wood, A.C.; Krytska, K.; Ryles, H.T.; Infarinato, N.R.; Sano, R.; Hansel, T.D.; Hart, L.S.; King, F.J.; Smith, T.R.; Ainscow, E.; et al. Dual ALK and CDK4/6 inhibition demonstrates synergy against neuroblastoma. Clin. Cancer Res. 2017, 23, 2856–2868. [Google Scholar] [CrossRef]
- Sussman, R.T.; Rokita, J.L.; Huang, K.; Raman, P.; Rathi, K.S.; Martinez, D.; Bosse, K.R.; Lane, M.; Hart, L.S.; Bhatti, T.; et al. CAMKV is a candidate immunotherapeutic target in MYCN amplified neuroblastoma. Front. Oncol. 2020, 10, 302. [Google Scholar] [CrossRef]
- Bosse, K.R.; Raman, P.; Zhu, Z.; Lane, M.; Martinez, D.; Heitzeneder, S.; Rathi, K.S.; Kendsersky, N.M.; Randall, M.; Donovan, L.; et al. Identification of GPC2 as an oncoprotein and candidate immunotherapeutic target in high-risk neuroblastoma. Cancer Cell 2017, 32, 295–309.e12. [Google Scholar] [CrossRef]
- Cacchione, A.; Lodi, M.; Carai, A.; Miele, E.; Tartaglia, M.; Megaro, G.; Del Baldo, G.; Alessi, I.; Colafati, G.S.; Carboni, A.; et al. Upfront treatment with mTOR inhibitor everolimus in pediatric low-grade gliomas: A single-center experience. Int. J. Cancer. 2020, 148, 2522–2534. [Google Scholar] [CrossRef]
- Lodi, M.; Boccuto, L.; Carai, A.; Cacchione, A.; Miele, E.; Colafati, G.S.; Diomedi Camassei, F.; De Palma, L.; De Benedictis, A.; Ferretti, E.; et al. Low-Grade Gliomas in Patients with Noonan Syndrome: Case-Based Review of the Literature. Diagnostics 2020, 10, 582. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Carter, R.A.; Li, Y.; Li, Y.; Wang, H.; Edmonson, M.N.; Chen, X.; Arnold, P.; Geiger, T.L.; Wu, G.; et al. The ne-oepitope landscape in pediatric cancers. Genome Med. 2017, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Biernacki, M.A.; Foster, K.A.; Woodward, K.B.; Coon, M.E.; Cummings, C.; Cunningham, T.M.; Dossa, R.G.; Brault, M.; Stokke, J.; Olsen, T.M.; et al. CBFB- MYH11 fusion neoantigen enables T cell recognition and killing of acute myeloid leukemia. J. Clin. Investig. 2020, 130, 5127–5141. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Li, Y.; Edmonson, M.N.; Zhou, X.; Newman, S.; McLeod, C.; Thrasher, A.; Liu, Y.; Tang, B.; Rusch, M.C.; et al. CICE-RO: A versatile method for detecting complex and diverse driver fusions using cancer RNA sequencing data. Genome Biol. 2020, 21, 126. [Google Scholar] [CrossRef]
- Nicorici, D.; Satalan, M.; Edgren, H.; Kangaspeska, S.; Murumagi, A.; Kallioniemi, O.; Virtanen, S.; Kilkku, O. FusionCatcher—A tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014. [Google Scholar] [CrossRef]
- Wang, J.; Mullighan, C.G.; Easton, J.; Roberts, S.; Heatley, S.L.; Ma, J.; Rusch, M.C.; Chen, K.; Harris, C.C.; Ding, L.; et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods 2011, 8, 652–654. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated-paired-end and split-read analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Biondi, A.; Schrappe, M.; De Lorenzo, P.; Castor, A.; Lucchini, G.; Gandemer, V.; Pieters, R.; Stary, J.; Escherich, G.; Campbell, M.; et al. Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): A randomised, open-label, intergroup study. Lancet Oncol. 2012, 13, 936–945. [Google Scholar] [CrossRef]
- Schultz, K.R.; Bowman, W.P.; Aledo, A.; Slayton, W.B.; Sather, H.; Devidas, M.; Wang, C.; Davies, S.M.; Gaynon, P.S.; Trigg, M.; et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leu-kemia: A children’s oncology group study. J. Clin. Oncol. 2009, 27, 5175–5181. [Google Scholar] [CrossRef]
- Schultz, K.R.; Carroll, A.; Heerema, N.A.; Bowman, W.P.; Aledo, A.; Slayton, W.B.; Sather, H.; Devidas, M.; Zheng, H.W.; Davies, S.M.; et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia 2014, 28, 1467–1471. [Google Scholar] [CrossRef]
- Ansuinelli, M.; Cesini, L.; Chiaretti, S.; Foà, R. Emerging tyrosine kinase inhibitors for the treatment of adult acute lymphoblastic leukemia. Expert Opin Emerg Drugs. 2021, 26, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Reshmi, S.C.; Harvey, R.C.; Roberts, K.G.; Stonerock, E.; Smith, A.; Jenkins, H.; Chen, I.M.; Valentine, M.; Liu, Y.; Li, Y.; et al. Targetable kinase gene fusions in high-risk B-ALL: A study from the Children’s Oncology Group. Blood 2017, 129, 3352–3361. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, I.; Lasorsa, V.A.; Manna, F.; Rosato, B.E.; Formicola, D.; Iolascon, A.; Capasso, M. Kinome multigenic panel identi-fied novel druggable EPHB4-V871I somatic variant in high-risk neuroblastoma. J. Cell. Mol. Med. 2020, 24, 6459–6471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, G.; Miller, C.P.; Tatevossian, R.G.; Dalton, J.D.; Tang, B.; Orisme, W.; Punchihewa, C.; Parker, M.; Qaddoumi, I.; et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013, 45, 602–612. [Google Scholar] [CrossRef]
- Jones, D.T.; Hutter, B.; Jäger, N.; Korshunov, A.; Kool, M.; Warnatz, H.J.; Zichner, T.; Lambert, S.R.; Ryzhova, M.; Quang, D.A.; et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet. 2013, 45, 927–932. [Google Scholar] [CrossRef]
- Wu, G.; Diaz, A.K.; Paugh, B.S.; Rankin, S.L.; Ju, B.; Li, Y.; Zhu, X.; Qu, C.; Chen, X.; Zhang, J.; et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem highgrade glioma. Nat. Genet. 2014, 46, 444–450. [Google Scholar] [CrossRef]
- Ziegler, D.S.; Wong, M.; Mayoh, C.; Kumar, A.; Tsoli, M.; Mould, E.; Tyrrell, V.; Khuong-Quang, D.A.; Pinese, M.; Gayevskiy, V.; et al. Brief report: Potent clinical andradiological response to larotrectinib in TRK fusion driven high-grade glioma. Br. J. Cancer 2018, 119, 693–696. [Google Scholar] [CrossRef]
- FDA Approves Larotrectinib for Solid Tumors with NTRK Gene Fusions. Available online:https://www.fda.gov/drugs/fda-approves-larotrectinib-solid-tumors-ntrk-gene-fusions (accessed on 18 September 2022).
- Di Ruscio, V.; Carai, A.; Del Baldo, G.; Vinci, M.; Cacchione, A.; Miele, E.; Rossi, S.; Antonelli, M.; Barresi, S.; Caulo, M.; et al. Molecular Landscape in Infant High-Grade Gliomas: A Single Center Experience. Diagnostics 2022, 12, 372. [Google Scholar] [CrossRef]
- FDA Approves Entrectinib Based on Tumor Genetics Rather Than Cancer Type. Available online: https://www.cancer.gov/news-events/cancer-currents-blog/2019/fda-entrectinib-ntrk-fusion (accessed on 20 September 2022).
- Taylor, J.; Pavlick, D.; Yoshimi, A.; Marcelus, C.; Chung, S.S.; Hechtman, J.F.; Benayed, R.; Cocco, E.; Durham, B.H.; Bitner, L.; et al. Oncogenic TRK fusions are amenable to inhibition in hematologic malignancies. J. Clin. Investig. 2018, 128, 3819–3825. [Google Scholar] [CrossRef]
- Roberts, K.G.; Janke, L.J.; Zhao, Y.; Seth, A.; Ma, J.; Finkelstein, D.; Smith, S.; Ebata, K.; Tuch, B.B.; Hunger, S.P.; et al. ETV6-NTRK3 induces aggressive acute lymphoblastic leukemia highly sensitive to selective TRK inhibition. Blood 2018, 132, 861–865. [Google Scholar] [CrossRef]
- Murakami, N.; Okuno, Y.; Yoshida, K.; Shiraishi, Y.; Nagae, G.; Suzuki, K.; Narita, A.; Sakaguchi, H.; Kawashima, N.; Wang, X.; et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood 2018, 131, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Hancock, M.L.; Rivera, G.K.; Sandlund, J.T.; Ribeiro, R.C.; Krynestski, E.Y.; Pui, C.H.; Evans, W.E. Mercapto-purine therapy intolerance and heterozygosity at the thiopurine S- methyltransferase gene locus. J. Natl. Cancer Inst. 1999, 91, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Bhatia, P.; Khera, S.; Trehan, A. Emerging role of NUDT15 polymorphisms in 6-mercaptopurine metabolism and dose related toxicity in acute lymphoblastic leukaemia. Leuk. Res. 2017, 62, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Sterner, R.C.; Sterner, R.M. CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Mount, C.W.; Majzner, R.G.; Sundaresh, S.; Arnold, E.P.; Kadapakkam, M.; Haile, S.; Labanieh, L.; Hulleman, E.; Woo, P.J.; Rietberg, S.P.; et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M + diffuse midline gliomas. Nat. Med. 2018, 24, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Majzner, R.G.; Ramakrishna, S.; Yeom, K.W.; Patel, S.; Chinnasamy, H.; Schultz, L.M.; Richards, R.M.; Jiang, L.; Barsan, V.; Mancusi, R.; et al. GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature 2022, 603, 934–941. [Google Scholar] [CrossRef]
- Theruvath, J.; Sotillo, E.; Mount, C.W.; Graef, C.M.; Delaidelli, A.; Heitzeneder, S.; Labanieh, L.; Dhingra, S.; Leruste, A.; Majzner, R.G.; et al. Locoregionally administered B7-H3-targeted CAR T cells for treatment of atypical teratoid/rhabdoid tumors. Nat. Med. 2020, 26, 712–719. [Google Scholar] [CrossRef]
- Garcia-Prieto, C.A.; Villanueva, L.; Bueno-Costa, A.; Davalos, V.; González-Navarro, E.A.; Juan, M.; Urbano-Ispizua, Á.; Delgado, J.; Ortiz-Maldonado, V.; Del Bufalo, F.; et al. Epigenetic Profiling and Response to CD19 Chimeric Antigen Recep-tor T-Cell Therapy in B-Cell Malignancies. Natl. Cancer Inst. 2022, 114, 436–445. [Google Scholar] [CrossRef]
- Dulbecco, R. A Turning Point in Cancer Research: Sequencing the Human Genome. Science 1986, 231, 1055–1056. [Google Scholar] [CrossRef]
- Abenavoli, L.; Boccuto, L. The Legacy of Renato Dulbecco in the Post-Genomic Era. Medicina 2022, 58, 974. [Google Scholar] [CrossRef]
- Boccuto, L.; Mitz, A.; Abenavoli, L.; Sarasua, S.M.; Bennett, W.; Rogers, C.; DuPont, B.; Phelan, K. Phenotypic Variability in Phelan-McDermid Syndrome and Its Putative Link to Environmental Factors. Genes 2022, 13, 528. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cipri, S.; Abenavoli, L.; Boccuto, L.; Del Baldo, G.; Mastronuzzi, A. How Genetics and Genomics Advances Are Rewriting Pediatric Cancer Research and Clinical Care. Medicina 2022, 58, 1386. https://doi.org/10.3390/medicina58101386
Cipri S, Abenavoli L, Boccuto L, Del Baldo G, Mastronuzzi A. How Genetics and Genomics Advances Are Rewriting Pediatric Cancer Research and Clinical Care. Medicina. 2022; 58(10):1386. https://doi.org/10.3390/medicina58101386
Chicago/Turabian StyleCipri, Selene, Ludovico Abenavoli, Luigi Boccuto, Giada Del Baldo, and Angela Mastronuzzi. 2022. "How Genetics and Genomics Advances Are Rewriting Pediatric Cancer Research and Clinical Care" Medicina 58, no. 10: 1386. https://doi.org/10.3390/medicina58101386
APA StyleCipri, S., Abenavoli, L., Boccuto, L., Del Baldo, G., & Mastronuzzi, A. (2022). How Genetics and Genomics Advances Are Rewriting Pediatric Cancer Research and Clinical Care. Medicina, 58(10), 1386. https://doi.org/10.3390/medicina58101386








