Gene Expression Profiling of Peripheral Blood Mononuclear Cells in Type 2 Diabetes: An Exploratory Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Measurement of Plasma Insulin, LDL, HDL, Triglycerides, HbA1c and HOMA-IR
2.3. Isolation of PBMCs
2.4. Quantitative Real-Time Polymerase Chain Reaction Analysis
2.5. Statistical Analysis
3. Results
3.1. Demographic Data of Study Participants
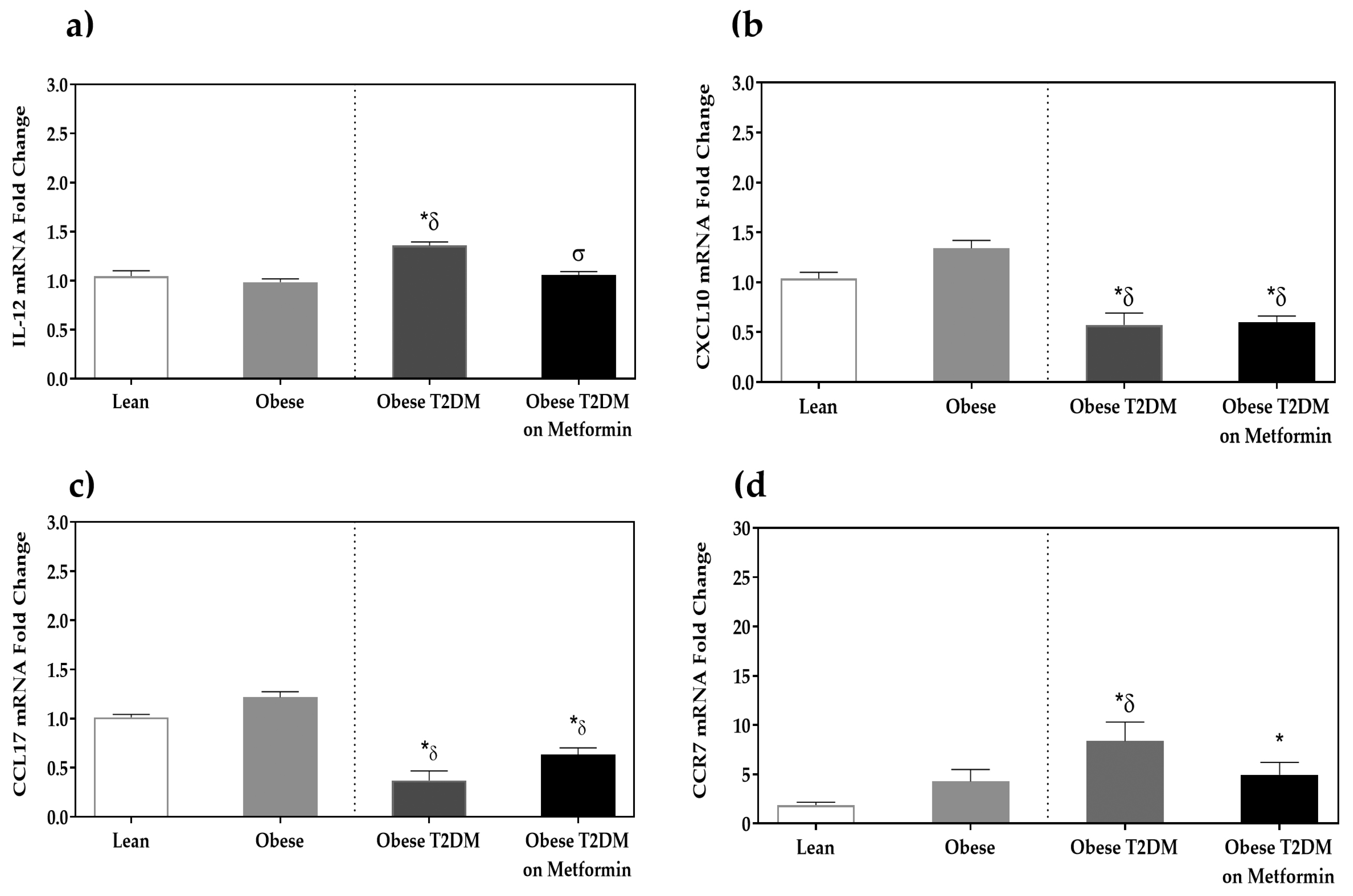
3.2. Macrophage Phenotypic Markers’ Expression
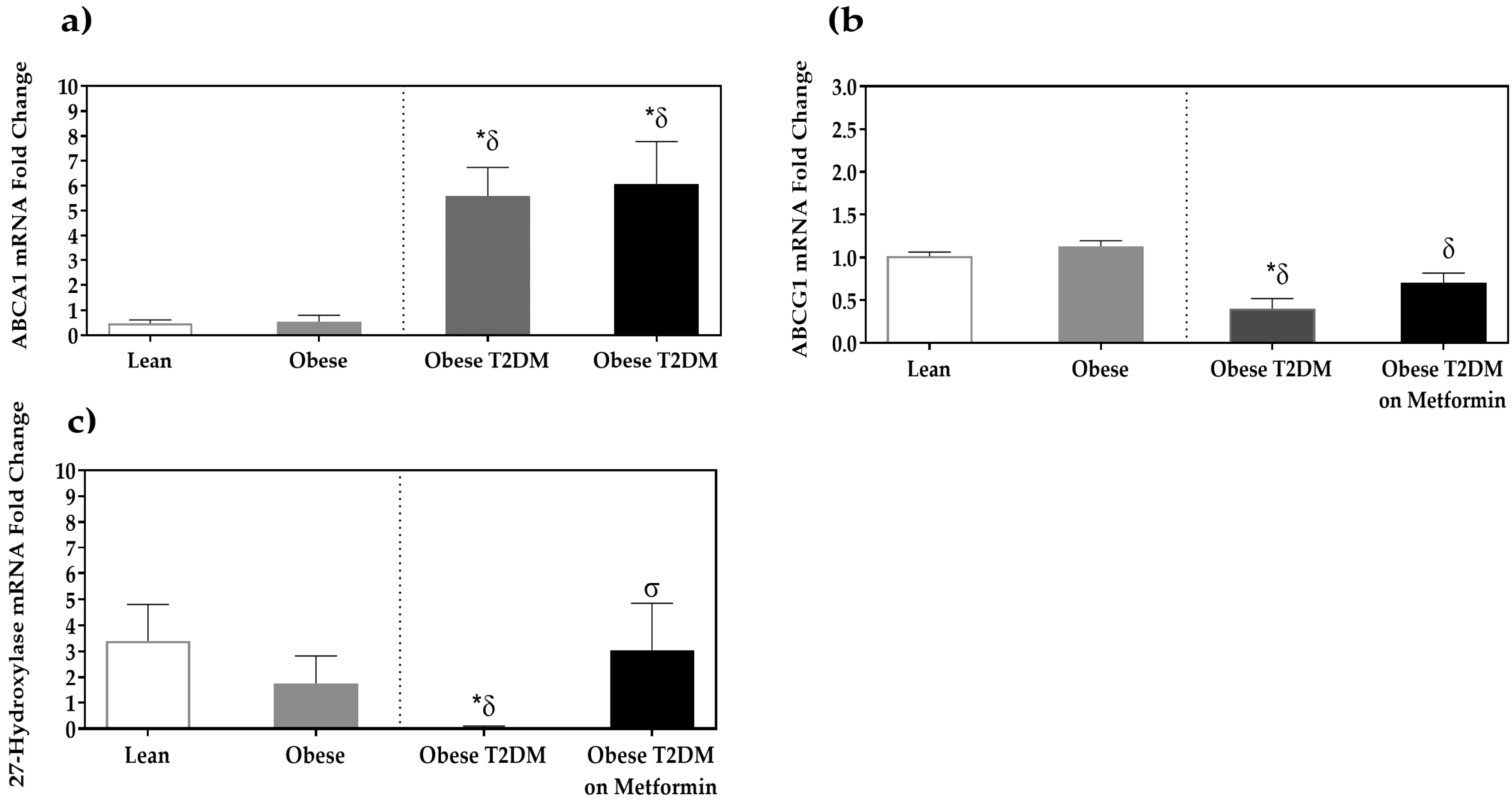
3.3. Expression of Cholesterol Efflux Proteins
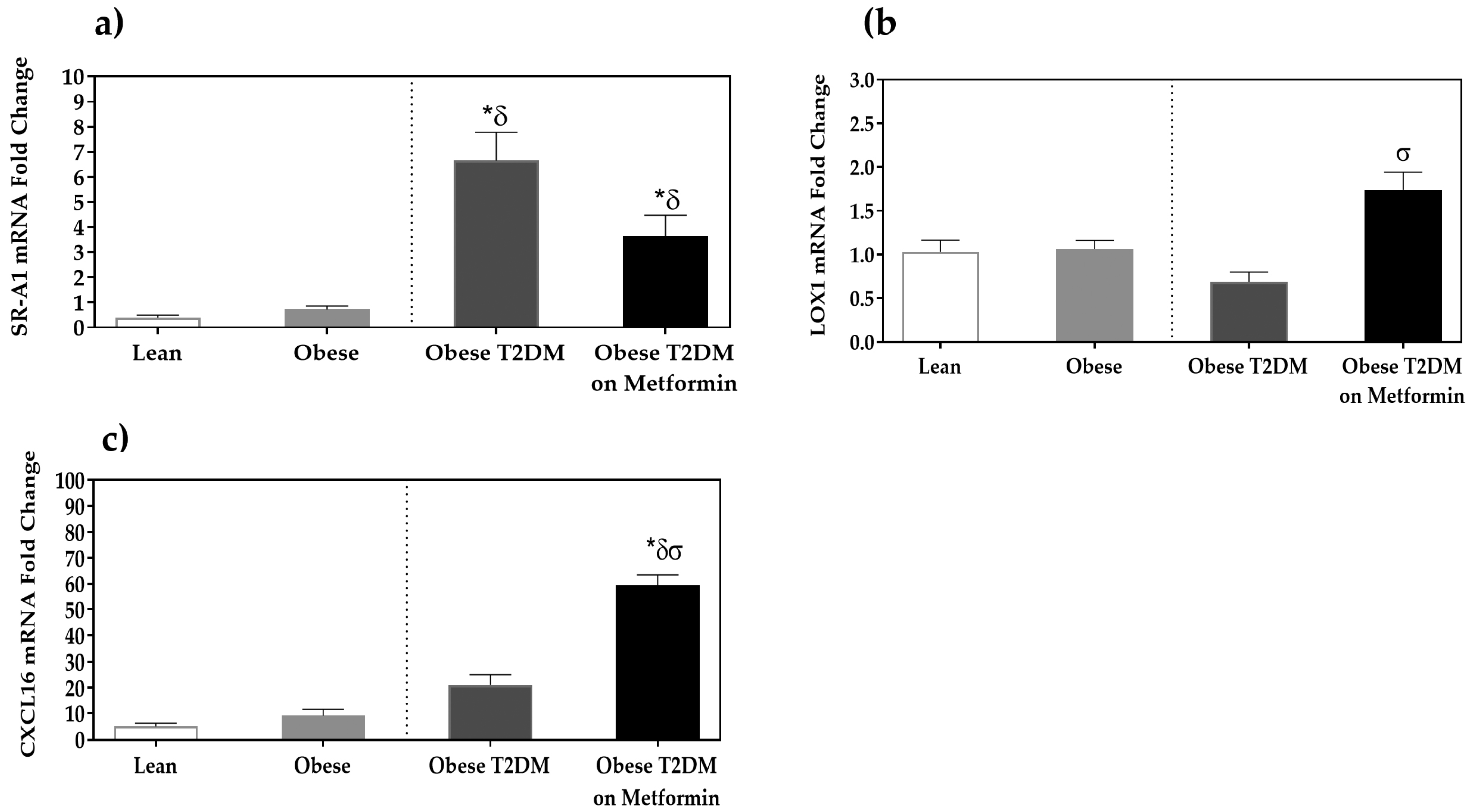
3.4. Expression Analysis of Scavenger Receptors
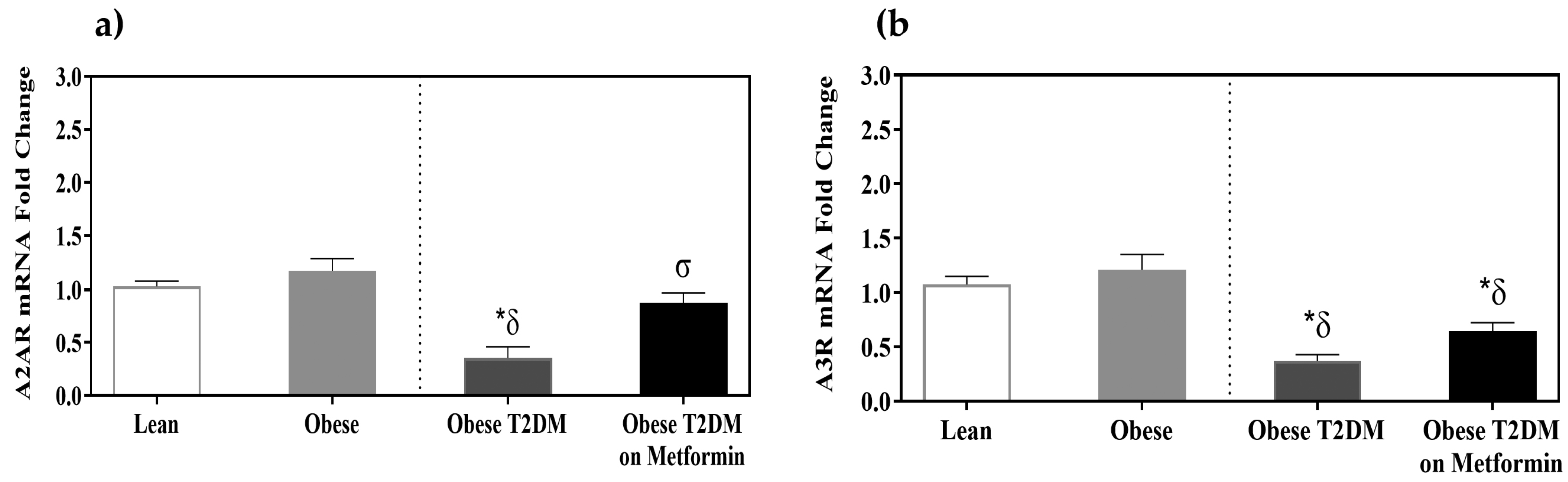
3.5. Adenosine Receptors A2AR and A3R Expression
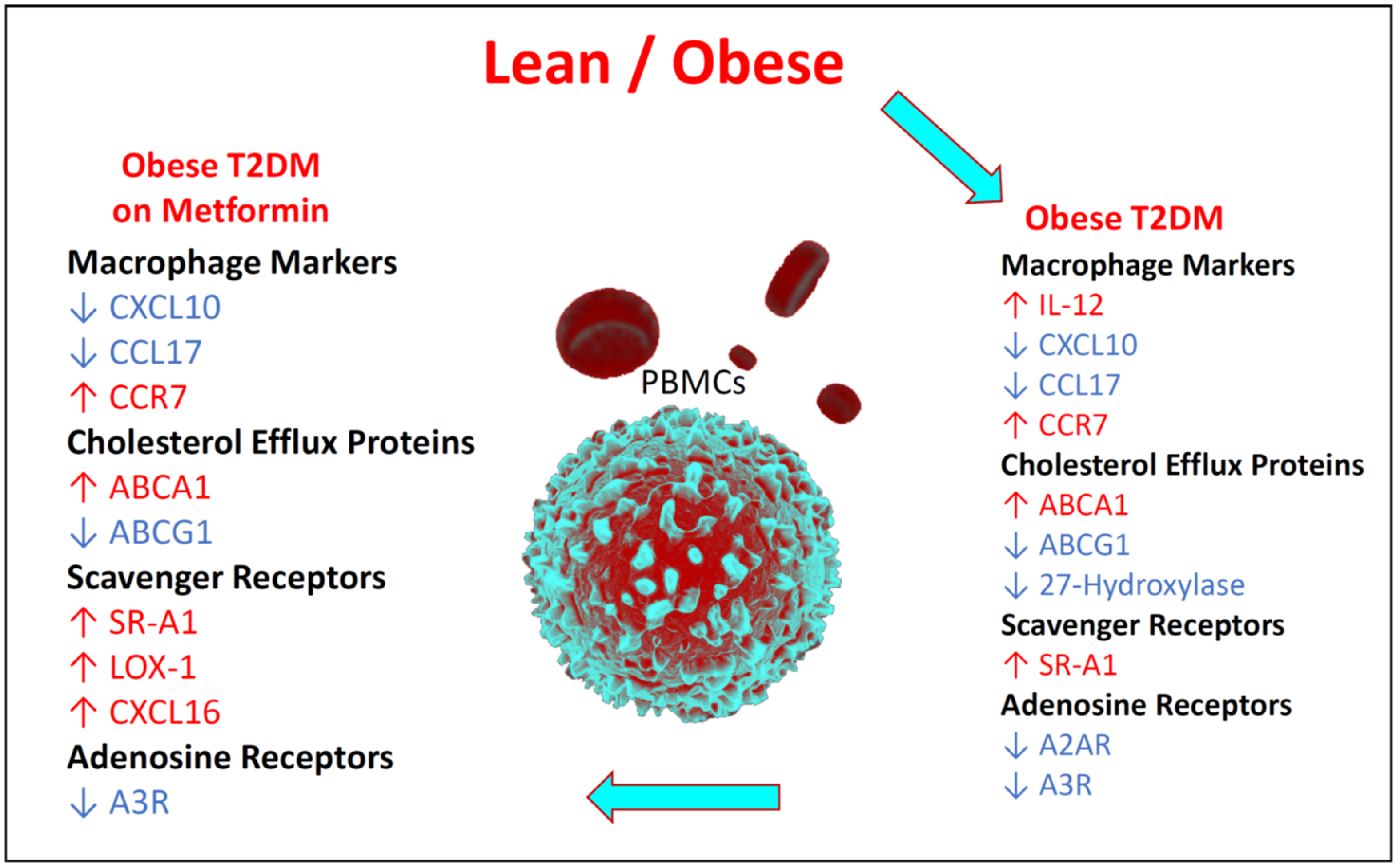
3.6. Summary of the Changes in the Relative Expression of the Studied Biomarkers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PBMCs | Peripheral blood mononuclear cells |
| MMe | Metabolically activated macrophages |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| NF-κB | Nuclear factor κB |
| LOX-1 | Lectin-like oxidized LDL receptor-1 |
| oxLDL | Oxidized LDL |
| S.E.M. | Standard error of the mean |
References
- Oguntibeju, O.O. Type 2 diabetes mellitus, oxidative stress and inflammation: Examining the links. Int. J. Physiol. Pathophysiol. Pharmacol. 2019, 11, 45–63. [Google Scholar]
- Fujisaka, S. The role of adipose tissue M1/M2 macrophages in type 2 diabetes mellitus. Diabetol. Int. 2021, 12, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ballantyne, C.M. Metabolic Inflammation and Insulin Resistance in Obesity. Circ. Res. 2020, 126, 1549–1564. [Google Scholar] [CrossRef] [PubMed]
- Donath, M.Y.; Shoelson, S.E. Type 2 diabetes as an inflammatory disease. Nat. Rev. Immunol. 2011, 11, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Valtierra-Alvarado, M.A.; Castaneda Delgado, J.E.; Ramirez-Talavera, S.I.; Lugo-Villarino, G.; Duenas-Arteaga, F.; Lugo-Sanchez, A.; Adame-Villalpando, M.S.; Rivas-Santiago, B.; Enciso-Moreno, J.; Serrano, C.J. Type 2 diabetes mellitus metabolic control correlates with the phenotype of human monocytes and monocyte-derived macrophages. J. Diabetes Complicat. 2020, 34, 107708. [Google Scholar] [CrossRef]
- Kratz, M.; Coats, B.R.; Hisert, K.B.; Hagman, D.; Mutskov, V.; Peris, E.; Schoenfelt, K.Q.; Kuzma, J.N.; Larson, I.; Billing, P.S.; et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014, 20, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Al Dubayee, M.S.; Alayed, H.; Almansour, R.; Alqaoud, N.; Alnamlah, R.; Obeid, D.; Alshahrani, A.; Zahra, M.M.; Nasr, A.; Al-Bawab, A.; et al. Differential Expression of Human Peripheral Mononuclear Cells Phenotype Markers in Type 2 Diabetic Patients and Type 2 Diabetic Patients on Metformin. Front. Endocrinol. 2018, 9, 537. [Google Scholar] [CrossRef] [Green Version]
- Alexovic, M.; Lindner, J.R.; Bober, P.; Longuespee, R.; Sabo, J.; Davalieva, K. Human peripheral blood mononuclear cells: A review of recent proteomic applications. Proteomics 2022, 22, e2200026. [Google Scholar] [CrossRef]
- Rempel, J.D.; Packiasamy, J.; Dean, H.J.; McGavock, J.; Janke, A.; Collister, M.; Wicklow, B.; Sellers, E.A. Preliminary analysis of immune activation in early onset type 2 diabetes. Int. J. Circumpolar. Health 2013, 72, 21190. [Google Scholar] [CrossRef]
- Ghanim, H.; Aljada, A.; Hofmeyer, D.; Syed, T.; Mohanty, P.; Dandona, P. Circulating mononuclear cells in the obese are in a proinflammatory state. Circulation 2004, 110, 1564–1571. [Google Scholar] [CrossRef] [Green Version]
- Dandona, P.; Aljada, A.; Bandyopadhyay, A. Inflammation: The link between insulin resistance, obesity and diabetes. Trends Immunol. 2004, 25, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Dentan, C.; Lesnik, P.; Chapman, M.J.; Ninio, E. Phagocytic activation induces formation of platelet-activating factor in human monocyte-derived macrophages and in macrophage-derived foam cells. Relevance to the inflammatory reaction in atherogenesis. Eur. J. Biochem. 1996, 236, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Fredholm, B.B. Adenosine, an endogenous distress signal, modulates tissue damage and repair. Cell Death Differ. 2007, 14, 1315–1323. [Google Scholar] [CrossRef] [Green Version]
- Cauwels, A.; Rogge, E.; Vandendriessche, B.; Shiva, S.; Brouckaert, P. Extracellular ATP drives systemic inflammation, tissue damage and mortality. Cell Death Dis. 2014, 5, e1102. [Google Scholar] [CrossRef] [Green Version]
- Baharav, E.; Bar-Yehuda, S.; Madi, L.; Silberman, D.; Rath-Wolfson, L.; Halpren, M.; Ochaion, A.; Weinberger, A.; Fishman, P. Antiinflammatory effect of A3 adenosine receptor agonists in murine autoimmune arthritis models. J. Rheumatol. 2005, 32, 469–476. [Google Scholar] [PubMed]
- Li, S.; Geiger, N.H.; Soliman, M.L.; Hui, L.; Geiger, J.D.; Chen, X. Caffeine, Through Adenosine A3 Receptor-Mediated Actions, Suppresses Amyloid-beta Protein Precursor Internalization and Amyloid-beta Generation. J. Alzheimer’s Dis. 2015, 47, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Mazziotta, C.; Rotondo, J.C.; Lanzillotti, C.; Campione, G.; Martini, F.; Tognon, M. Cancer biology and molecular genetics of A(3) adenosine receptor. Oncogene 2022, 41, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Patten, D.A.; Shetty, S. More Than Just a Removal Service: Scavenger Receptors in Leukocyte Trafficking. Front. Immunol. 2018, 9, 2904. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Yao, K.; Huang, D.; Sun, A.; Zou, Y.; Qian, J.; Ge, J. High glucose induces upregulation of scavenger receptors and promotes maturation of dendritic cells. Cardiovasc. Diabetol. 2013, 12, 80. [Google Scholar] [CrossRef] [Green Version]
- Rasouli, N.; Yao-Borengasser, A.; Varma, V.; Spencer, H.J.; McGehee, R.E., Jr.; Peterson, C.A.; Mehta, J.L.; Kern, P.A. Association of scavenger receptors in adipose tissue with insulin resistance in nondiabetic humans. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 1328–1335. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.Y.; Nicholson, A.C.; Hajjar, D.P. Inhibition of macrophage scavenger receptor activity by tumor necrosis factor-alpha is transcriptionally and post-transcriptionally regulated. J. Biol. Chem. 1996, 271, 7767–7773. [Google Scholar] [CrossRef] [PubMed]
- Groenen, A.G.; Halmos, B.; Tall, A.R.; Westerterp, M. Cholesterol efflux pathways, inflammation, and atherosclerosis. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Bonora, E.; Formentini, G.; Calcaterra, F.; Lombardi, S.; Marini, F.; Zenari, L.; Saggiani, F.; Poli, M.; Perbellini, S.; Raffaelli, A.; et al. HOMA-estimated insulin resistance is an independent predictor of cardiovascular disease in type 2 diabetic subjects: Prospective data from the Verona Diabetes Complications Study. Diabetes Care 2002, 25, 1135–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Littlefield, M.; Teboul, I.; Voloshyna, I.; Reiss, A.; Reiss, A. Polarization of human THP-1 macrophages: Link between Adenosine Receptors, Inflammation and Lipid accumulation. Int. J. Immunol. Immunother. 2014, 1, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Katakura, T.; Miyazaki, M.; Kobayashi, M.; Herndon, D.N.; Suzuki, F. CCL17 and IL-10 as effectors that enable alternatively activated macrophages to inhibit the generation of classically activated macrophages. J. Immunol. 2004, 172, 1407–1413. [Google Scholar] [CrossRef] [Green Version]
- Kochumon, S.; Madhoun, A.A.; Al-Rashed, F.; Azim, R.; Al-Ozairi, E.; Al-Mulla, F.; Ahmad, R. Adipose tissue gene expression of CXCL10 and CXCL11 modulates inflammatory markers in obesity: Implications for metabolic inflammation and insulin resistance. Ther. Adv. Endocrinol. Metab. 2020, 11, 2042018820930902. [Google Scholar] [CrossRef]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Pei, Y.; Li, H.; Cai, Y.; Zhou, J.; Luo, X.; Ma, L.; McDaniel, K.; Zeng, T.; Chen, Y.; Qian, X.; et al. Regulation of adipose tissue inflammation by adenosine 2A receptor in obese mice. J. Endocrinol. 2018, 239, 365–376. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jhun, B.S.; Oh, Y.T.; Lee, J.H.; Choe, W.; Baik, H.H.; Ha, J.; Yoon, K.S.; Kim, S.S.; Kang, I. Activation of adenosine A3 receptor suppresses lipopolysaccharide-induced TNF-alpha production through inhibition of PI 3-kinase/Akt and NF-kappaB activation in murine BV2 microglial cells. Neurosci. Lett. 2006, 396, 1–6. [Google Scholar] [CrossRef]
- Bingham, T.C.; Parathath, S.; Tian, H.; Reiss, A.; Chan, E.; Fisher, E.A.; Cronstein, B.N. Cholesterol 27-hydroxylase but not apolipoprotein apoE contributes to A2A adenosine receptor stimulated reverse cholesterol transport. Inflammation 2012, 35, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Escher, G.; Krozowski, Z.; Croft, K.D.; Sviridov, D. Expression of sterol 27-hydroxylase (CYP27A1) enhances cholesterol efflux. J. Biol. Chem. 2003, 278, 11015–11019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, A.J.; Yvan-Charvet, L. Adipose modulation of ABCG1 uncovers an intimate link between sphingomyelin and triglyceride storage. Diabetes 2015, 64, 689–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldo, S.W.; Li, Y.; Buono, C.; Zhao, B.; Billings, E.M.; Chang, J.; Kruth, H.S. Heterogeneity of human macrophages in culture and in atherosclerotic plaques. Am. J. Pathol. 2008, 172, 1112–1126. [Google Scholar] [CrossRef] [Green Version]
- Tavoosi, Z.; Moradi-Sardareh, H.; Saidijam, M.; Yadegarazari, R.; Borzuei, S.; Soltanian, A.; Goodarzi, M.T. Cholesterol Transporters ABCA1 and ABCG1 Gene Expression in Peripheral Blood Mononuclear Cells in Patients with Metabolic Syndrome. Cholesterol 2015, 2015, 682904. [Google Scholar] [CrossRef] [Green Version]
- Frisdal, E.; Le Goff, W. Adipose ABCG1: A potential therapeutic target in obesity? Adipocyte 2015, 4, 315–318. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Tarling, E.J.; McMillen, T.S.; Tang, C.; LeBoeuf, R.C. ABCG1 regulates mouse adipose tissue macrophage cholesterol levels and ratio of M1 to M2 cells in obesity and caloric restriction. J. Lipid Res. 2015, 56, 2337–2347. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.; Liu, Y.; Yang, W.; Storey, C.; McMillen, T.S.; Houston, B.A.; Heinecke, J.W.; LeBoeuf, R.C. Hematopoietic ABCA1 deletion promotes monocytosis and worsens diet-induced insulin resistance in mice. J. Lipid Res. 2016, 57, 100–108. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Scavini, M.; Orlando, R.A.; Murata, G.H.; Servilla, K.S.; Tzamaloukas, A.H.; Schrader, R.; Bedrick, E.J.; Burge, M.R.; Abumrad, N.A.; et al. Increased CD36 expression signals monocyte activation among patients with type 2 diabetes. Diabetes Care 2010, 33, 2065–2067. [Google Scholar] [CrossRef] [Green Version]
- Cai, L.; Wang, Z.; Ji, A.; Meyer, J.M.; van der Westhuyzen, D.R. Scavenger receptor CD36 expression contributes to adipose tissue inflammation and cell death in diet-induced obesity. PLoS ONE 2012, 7, e36785. [Google Scholar] [CrossRef] [Green Version]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Marquie, G. Metformin action on lipid metabolism in lesions of experimental aortic atherosclerosis of rabbits. Atherosclerosis 1983, 47, 7–17. [Google Scholar] [CrossRef]
- Bouissou, H.; Thiers, J.C.; Douste-Blazy, L.; Pieraggi, M.T.; Julian, M. Chronic lathyrism and atheromatosis in the rat. Protective effect of metformin. Gerontology 1980, 26, 188–199. [Google Scholar] [CrossRef]
- Mamputu, J.C.; Wiernsperger, N.F.; Renier, G. Antiatherogenic properties of metformin: The experimental evidence. Diabetes Metab. 2003, 29, S71–S76. [Google Scholar] [CrossRef] [PubMed]
- Montanari, G.; Bondioli, A.; Rizzato, G.; Puttini, M.; Tremoli, E.; Mussoni, L.; Mannucci, L.; Pazzucconi, F.; Sirtori, C.R. Treatment with low dose metformin in patients with peripheral vascular disease. Pharmacol. Res. 1992, 25, 63–73. [Google Scholar] [CrossRef] [PubMed]
- UK Prospective Diabetes Study (UKPDS) Group. Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (UKPDS 34). Lancet 1998, 352, 854–865. [Google Scholar] [CrossRef]
- Aslanian, A.M.; Charo, I.F. Targeted disruption of the scavenger receptor and chemokine CXCL16 accelerates atherosclerosis. Circulation 2006, 114, 583–590. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, M.C.; Andersen, M.N.; Moller, H.J. Monocyte isolation techniques significantly impact the phenotype of both isolated monocytes and derived macrophages in vitro. Immunology 2020, 159, 63–74. [Google Scholar] [CrossRef]
- Klimcakova, E.; Roussel, B.; Kovacova, Z.; Kovacikova, M.; Siklova-Vitkova, M.; Combes, M.; Hejnova, J.; Decaunes, P.; Maoret, J.J.; Vedral, T.; et al. Macrophage gene expression is related to obesity and the metabolic syndrome in human subcutaneous fat as well as in visceral fat. Diabetologia 2011, 54, 876–887. [Google Scholar] [CrossRef] [Green Version]
- Njemini, R.; Onyema, O.O.; Renmans, W.; Bautmans, I.; De Waele, M.; Mets, T. Shortcomings in the application of multicolour flow cytometry in lymphocyte subsets enumeration. Scand. J. Immunol. 2014, 79, 75–89. [Google Scholar] [CrossRef]
- Drescher, H.; Weiskirchen, S.; Weiskirchen, R. Flow Cytometry: A Blessing and a Curse. Biomedicines 2021, 9, 1613. [Google Scholar] [CrossRef] [PubMed]
| Primer | Sense (5′→3′) | Anti-Sense (5′→3′) | Accession Number |
|---|---|---|---|
| IL-12 (IL12RB2) | AAAGGACATCTGCGAGGAAAGTTC | CGAGGTGAGGTGCGTTTATGC | NM_001559.2 |
| CXCL10 | GAAAGCAGTTAGCAAGGAAAGGTC | ATGTAGGGAAGTGATGGGAGAGG | NM_001565.4 |
| CCL17 | CGGGACTACCTGGGACCTC | CCTCACTGTGGCTCTTCTTCG | BC112068.1 |
| CCR7 | TGGTGGTGGCTCTCCTTGTC | TGTGGTGTTGTCTCCGATGTAATC | NM_001838.4 |
| ABCA1 | GAAGTACATCAGAACATGGGC | GATCAAAGCCATGGCTGTAG | NM_005502.4 |
| ABCG1 | CAGGAAGATTAGACACTGTGG | GAAAGGGGAATGGAGAGAAG | NM_016818.2 |
| CYP27A | AAGCGATACCTGGATGGTTG | TGTTGGATGTCGTGTCCACT | NM_000784.4 |
| SR-A1 | CTCGTGTTTGCAGTTCTCA | CCATGTTGCTCATGTGTTCC | NM_138715.3 |
| LOX-1 | TTACTCTCCATGGTGGTGCC | AGCTTCTTCTGCTTGTTGCC | NM_002543.4 |
| CXCL16 | ACTACACGACGTTCCAGCTCC | CTTTGTCCGAGGACAGTGATC | NM_022059.4 |
| A2AR | CGAATTCAACCTGCAGAACGTCACC | TCGAATTCGCGGTCAATGGCGATG | NM_001278497.1 |
| A3R | ACCACTCACAGAAGAATATG | ACTTAGCCGTCTTGAACTCC | L22607.1 |
| Ubiquitin C | ACTACAACATCCAGAAAGAGTCCA | CCAGTCAGGGTCTTCACGAAG | NM_021009.6 |
| RPL13 | AACAAGTTGAAGTACCTGGCTTTC | TGGTTTTGTGGGGCAGCATA | NM_000977.3 |
| Cyclophilin A | CCCACCGTGTTCTTCGACAT | TTTCTGCTGTCTTTGGGACCTT | NM_021130.4 |
| Group | Gender | Age | BMI | Glucose | Insulin | HOMA-IR | LDL | HDL | Triglycerides | HbA1c |
|---|---|---|---|---|---|---|---|---|---|---|
| (Years) | kg/m2 | (mmol/L) | µIU/mL | (mmol/L) | (mmol/L) | (mmol/L) | (%) | |||
| Normal weight | N = 30, 18 Males, 12 Females | 25.7 ± 1.1 | 23.0 ± 0.3 | 5.1 ± 0.1 | 4.8 ± 0.60 | 1.1 ± 0.1 | 2.54 ± 0.26 | 1.35 ± 0.04 | 0.83 ± 0.09 | 5.8 ± 0.10 |
| Obese | N = 30, 11 Males, 19 Females | 35.1 ± 2.3 | 39.1 ± 1.7 * | 5.4 ± 0.1 | 10.0 ± 1.54 * | 2.1 ± 0.3 * | 3.04 ± 0.16 | 1.18 ± 0.05 | 1.22 ± 0.12 | 5.6 ± 0.25 |
| Obese with T2D | N = 20, 15 Males, 5 Females | 48.4 ± 3.0 *δ | 32.5 ± 1.9 * | 10.0 ± 1.1 *δ | 7.4 ± 2.57 * | 2.8 ± 0.8 * | 3.53 ± 0.19 * | 1.02 ± 0.05 *δ | 1.97 ± 0.23 *δ | 8.0 ± 0.62 *δ |
| T2DM on Metformin | N = 30, 12 Males, 18 Females | 47.1 ± 2.0 *δ | 40.5 ± 1.5 * | 10.0 ± 0.8 *δ | 7.8 ± 1.52 * | 4.1 ± 1.3 * | 2.60 ± 0.17 σ | 0.99 ± 0.04 *δ | 1.56 ± 0.16 * | 8.7 ± 0.35 *δ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fakhoury, H.M.A.; Elahi, M.A.; Al Sarheed, S.; Al Dubayee, M.; Alshahrani, A.; Zhra, M.; Almassri, A.; Aljada, A. Gene Expression Profiling of Peripheral Blood Mononuclear Cells in Type 2 Diabetes: An Exploratory Study. Medicina 2022, 58, 1829. https://doi.org/10.3390/medicina58121829
Fakhoury HMA, Elahi MA, Al Sarheed S, Al Dubayee M, Alshahrani A, Zhra M, Almassri A, Aljada A. Gene Expression Profiling of Peripheral Blood Mononuclear Cells in Type 2 Diabetes: An Exploratory Study. Medicina. 2022; 58(12):1829. https://doi.org/10.3390/medicina58121829
Chicago/Turabian StyleFakhoury, Hana M. A., Muhammad Affan Elahi, Saud Al Sarheed, Mohammed Al Dubayee, Awad Alshahrani, Mahmoud Zhra, Arwa Almassri, and Ahmad Aljada. 2022. "Gene Expression Profiling of Peripheral Blood Mononuclear Cells in Type 2 Diabetes: An Exploratory Study" Medicina 58, no. 12: 1829. https://doi.org/10.3390/medicina58121829
APA StyleFakhoury, H. M. A., Elahi, M. A., Al Sarheed, S., Al Dubayee, M., Alshahrani, A., Zhra, M., Almassri, A., & Aljada, A. (2022). Gene Expression Profiling of Peripheral Blood Mononuclear Cells in Type 2 Diabetes: An Exploratory Study. Medicina, 58(12), 1829. https://doi.org/10.3390/medicina58121829






