Sphingolipid Metabolism as a New Predictive Target Correlated with Aging and AD: A Transcriptomic Analysis
Abstract
1. Introduction
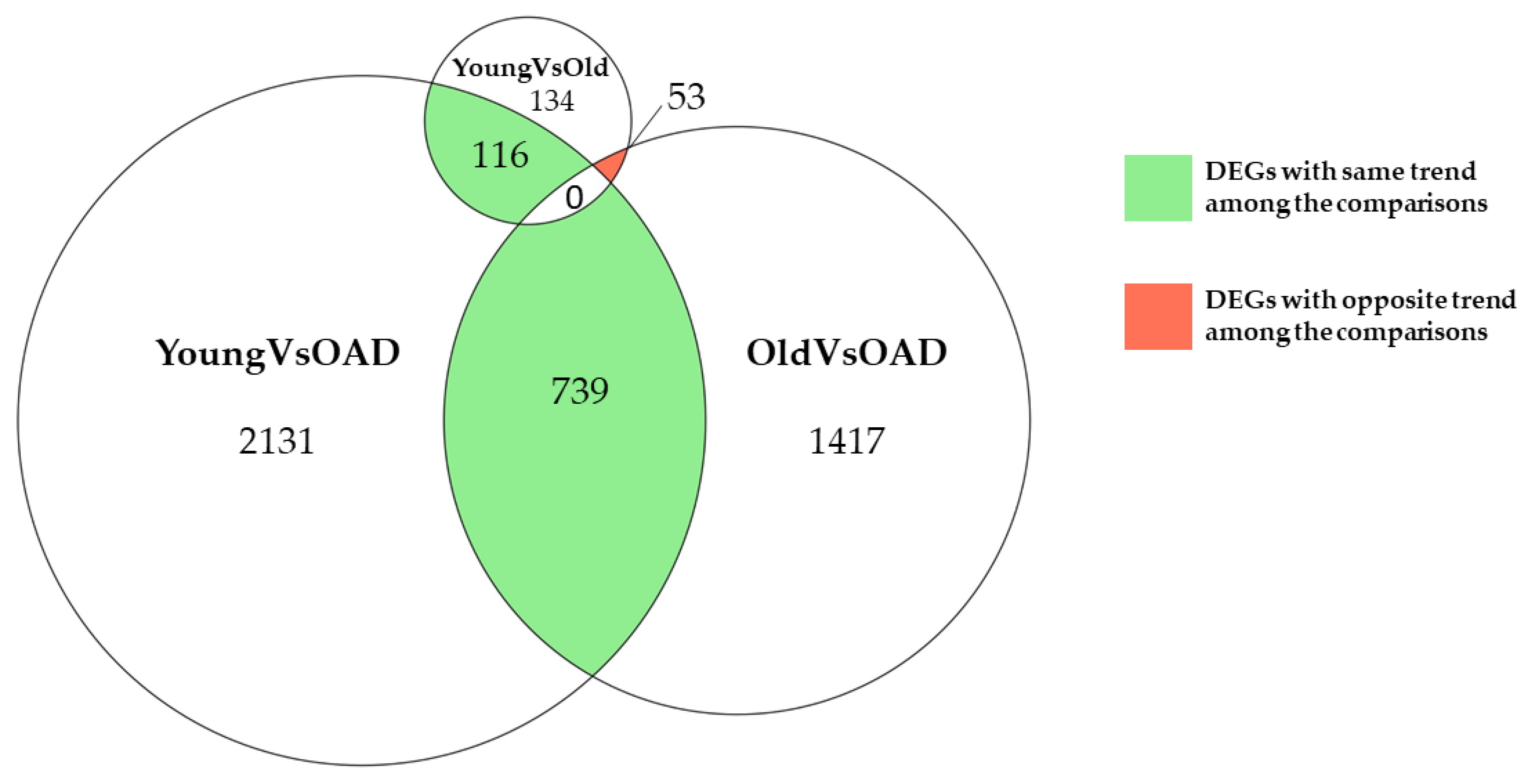
2. Results
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 1 December 2021).
- Kalaria, R.N.; Maestre, G.E.; Arizaga, R.; Friedland, R.P.; Galasko, D.; Hall, K.; Luchsinger, J.A.; Ogunniyi, A.; Perry, E.K.; Potocnik, F.; et al. Alzheimer’s disease and vascular dementia in developing countries: Prevalence, management, and risk factors. Lancet Neurol. 2008, 7, 812–826. [Google Scholar] [CrossRef]
- Schachter, A.S.; Davis, K.L. Alzheimer’s disease. Dialogues Clin. Neurosci. 2000, 2, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, O.; Coleman, M. Alzheimer’s Disease: Etiology, Neuropathology and Pathogenesis. In Alzheimer’s Disease: Drug Discovery; Huang, X., Ed.; Exon Publications: Brisbane, Australia, 2020. [Google Scholar]
- Tiwari, S.; Atluri, V.; Kaushik, A.; Yndart, A.; Nair, M. Alzheimer’s disease: Pathogenesis, diagnostics, and therapeutics. Int. J. Nanomed. 2019, 14, 5541–5554. [Google Scholar] [CrossRef] [PubMed]
- Apostolova, L.G. Alzheimer Disease. Continuum 2016, 22, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Govaerts, L.; Schoenen, J.; Bouhy, D. Pathogenesis of Alzheimer’s disease: Molecular and cellular mechanisms. Rev. Med. Liege 2007, 62, 209–216. [Google Scholar] [PubMed]
- Goate, A.; Chartier-Harlin, M.C.; Mullan, M.; Brown, J.; Crawford, F.; Fidani, L.; Giuffra, L.; Haynes, A.; Irving, N.; James, L.; et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 1991, 349, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Levy-Lahad, E.; Wasco, W.; Poorkaj, P.; Romano, D.M.; Oshima, J.; Pettingell, W.H.; Yu, C.E.; Jondro, P.D.; Schmidt, S.D.; Wang, K.; et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 1995, 269, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Sherrington, R.; Rogaev, E.I.; Liang, Y.; Rogaeva, E.A.; Levesque, G.; Ikeda, M.; Chi, H.; Lin, C.; Li, G.; Holman, K.; et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 1995, 375, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Fraering, P.C.; Ye, W.; Strub, J.M.; Dolios, G.; LaVoie, M.J.; Ostaszewski, B.L.; van Dorsselaer, A.; Wang, R.; Selkoe, D.J.; Wolfe, M.S. Purification and characterization of the human gamma-secretase complex. Biochemistry 2004, 43, 9774–9789. [Google Scholar] [CrossRef] [PubMed]
- Heinzen, E.L.; Need, A.C.; Hayden, K.M.; Chiba-Falek, O.; Roses, A.D.; Strittmatter, W.J.; Burke, J.R.; Hulette, C.M.; Welsh-Bohmer, K.A.; Goldstein, D.B. Genome-wide scan of copy number variation in late-onset Alzheimer’s disease. J. Alzheimer’s Dis. 2010, 19, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, R.; Bras, J.; Hardy, J. SnapShot: Genetics of Alzheimer’s disease. Cell 2013, 155, 968–968.e1. [Google Scholar] [CrossRef] [PubMed]
- Annese, A.; Manzari, C.; Lionetti, C.; Picardi, E.; Horner, D.S.; Chiara, M.; Caratozzolo, M.F.; Tullo, A.; Fosso, B.; Pesole, G.; et al. Whole transcriptome profiling of Late-Onset Alzheimer’s Disease patients provides insights into the molecular changes involved in the disease. Sci. Rep. 2018, 8, 4282. [Google Scholar] [CrossRef] [PubMed]
- Magistri, M.; Velmeshev, D.; Makhmutova, M.; Faghihi, M.A. Transcriptomics Profiling of Alzheimer’s Disease Reveal Neurovascular Defects, Altered Amyloid-beta Homeostasis, and Deregulated Expression of Long Noncoding RNAs. J. Alzheimer’s Dis. 2015, 48, 647–665. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Zeng, L.; Haure-Mirande, J.V.; Wang, M.; Huffman, D.M.; Haroutunian, V.; Ehrlich, M.E.; Zhang, B.; Tu, Z. Transcriptomic Changes Highly Similar to Alzheimer’s Disease Are Observed in a Subpopulation of Individuals During Normal Brain Aging. Front. Aging Neurosci. 2021, 13, 711524. [Google Scholar] [CrossRef]
- Xia, X.; Jiang, Q.; McDermott, J.; Han, J.J. Aging and Alzheimer’s disease: Comparison and associations from molecular to system level. Aging Cell 2018, 17, e12802. [Google Scholar] [CrossRef]
- de Wit, N.M.; Mol, K.; Rodriguez-Lorenzo, S.; de Vries, H.E.; Kooij, G. The Role of Sphingolipids and Specialized Pro-Resolving Mediators in Alzheimer’s Disease. Front. Immunol. 2020, 11, 620348. [Google Scholar] [CrossRef]
- Czubowicz, K.; Jesko, H.; Wencel, P.; Lukiw, W.J.; Strosznajder, R.P. The Role of Ceramide and Sphingosine-1-Phosphate in Alzheimer’s Disease and Other Neurodegenerative Disorders. Mol. Neurobiol. 2019, 56, 5436–5455. [Google Scholar] [CrossRef]
- Maceyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef]
- Mattson, M.P.; Duan, W.; Maswood, N. How does the brain control lifespan? Ageing Res. Rev. 2002, 1, 155–165. [Google Scholar] [CrossRef]
- Cutler, R.G.; Mattson, M.P. Sphingomyelin and ceramide as regulators of development and lifespan. Mech. Ageing Dev. 2001, 122, 895–908. [Google Scholar] [CrossRef]
- Kolter, T.; Sandhoff, K. Sphingolipid metabolism diseases. Biochim. Biophys. Acta 2006, 1758, 2057–2079. [Google Scholar] [CrossRef] [PubMed]
- Jove, M.; Naudi, A.; Gambini, J.; Borras, C.; Cabre, R.; Portero-Otin, M.; Vina, J.; Pamplona, R. A Stress-Resistant Lipidomic Signature Confers Extreme Longevity to Humans. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2017, 72, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Mielke, M.M.; Haughey, N.J. Could plasma sphingolipids be diagnostic or prognostic biomarkers for Alzheimer’s disease? Clin. Lipidol. 2012, 7, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Satoi, H.; Tomimoto, H.; Ohtani, R.; Kitano, T.; Kondo, T.; Watanabe, M.; Oka, N.; Akiguchi, I.; Furuya, S.; Hirabayashi, Y.; et al. Astroglial expression of ceramide in Alzheimer’s disease brains: A role during neuronal apoptosis. Neuroscience 2005, 130, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Greeve, I.; Hermans-Borgmeyer, I.; Brellinger, C.; Kasper, D.; Gomez-Isla, T.; Behl, C.; Levkau, B.; Nitsch, R.M. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer’s disease-associated neurodegeneration and oxidative stress. J. Neurosci. 2000, 20, 7345–7352. [Google Scholar] [CrossRef]
- Pandit, J.; Danley, D.E.; Schulte, G.K.; Mazzalupo, S.; Pauly, T.A.; Hayward, C.M.; Hamanaka, E.S.; Thompson, J.F.; Harwood, H.J., Jr. Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis. J. Biol. Chem. 2000, 275, 30610–30617. [Google Scholar] [CrossRef]
- Crameri, A.; Biondi, E.; Kuehnle, K.; Lutjohann, D.; Thelen, K.M.; Perga, S.; Dotti, C.G.; Nitsch, R.M.; Ledesma, M.D.; Mohajeri, M.H. The role of seladin-1/DHCR24 in cholesterol biosynthesis, APP processing and Abeta generation in vivo. EMBO J. 2006, 25, 432–443. [Google Scholar] [CrossRef]
- Laviad, E.L.; Albee, L.; Pankova-Kholmyansky, I.; Epstein, S.; Park, H.; Merrill, A.H., Jr.; Futerman, A.H. Characterization of ceramide synthase 2: Tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 2008, 283, 5677–5684. [Google Scholar] [CrossRef]
- Kapitonov, D.; Yu, R.K. Cloning, characterization, and expression of human ceramide galactosyltransferase cDNA. Biochem. Biophys. Res. Commun. 1997, 232, 449–453. [Google Scholar] [CrossRef]
- Busnelli, M.; Manzini, S.; Parolini, C.; Escalante-Alcalde, D.; Chiesa, G. Lipid phosphate phosphatase 3 in vascular pathophysiology. Atherosclerosis 2018, 271, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.; Sciorra, V.A.; Morris, A.J. Human type 2 phosphatidic acid phosphohydrolases. Substrate specificity of the type 2a, 2b, and 2c enzymes and cell surface activity of the 2a isoform. J. Biol. Chem. 1998, 273, 22059–22067. [Google Scholar] [CrossRef] [PubMed]
- Nativio, R.; Donahue, G.; Berson, A.; Lan, Y.; Amlie-Wolf, A.; Tuzer, F.; Toledo, J.B.; Gosai, S.J.; Gregory, B.D.; Torres, C.; et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci. 2018, 21, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Pike, C.J. Sex and the development of Alzheimer’s disease. J. Neurosci. Res. 2017, 95, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Puglielli, L.; Ellis, B.C.; Saunders, A.J.; Kovacs, D.M. Ceramide stabilizes beta-site amyloid precursor protein-cleaving enzyme 1 and promotes amyloid beta-peptide biogenesis. J. Biol. Chem. 2003, 278, 19777–19783. [Google Scholar] [CrossRef]
- He, X.; Huang, Y.; Li, B.; Gong, C.X.; Schuchman, E.H. Deregulation of sphingolipid metabolism in Alzheimer’s disease. Neurobiol. Aging 2010, 31, 398–408. [Google Scholar] [CrossRef]
- Mencarelli, C.; Martinez-Martinez, P. Ceramide function in the brain: When a slight tilt is enough. Cell. Mol. Life Sci. CMLS 2013, 70, 181–203. [Google Scholar] [CrossRef]
- Brann, A.B.; Scott, R.; Neuberger, Y.; Abulafia, D.; Boldin, S.; Fainzilber, M.; Futerman, A.H. Ceramide signaling downstream of the p75 neurotrophin receptor mediates the effects of nerve growth factor on outgrowth of cultured hippocampal neurons. J. Neurosci. 1999, 19, 8199–8206. [Google Scholar] [CrossRef]
- Schwarz, A.; Futerman, A.H. Distinct roles for ceramide and glucosylceramide at different stages of neuronal growth. J. Neurosci. 1997, 17, 2929–2938. [Google Scholar] [CrossRef]
- Goodman, Y.; Mattson, M.P. Ceramide protects hippocampal neurons against excitotoxic and oxidative insults, and amyloid beta-peptide toxicity. J. Neurochem. 1996, 66, 869–872. [Google Scholar] [CrossRef]
- Schimizzi, G.V.; Longmore, G.D. Ajuba proteins. Curr. Biol. 2015, 25, R445–R446. [Google Scholar] [CrossRef] [PubMed]
- Das Thakur, M.; Feng, Y.; Jagannathan, R.; Seppa, M.J.; Skeath, J.B.; Longmore, G.D. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 2010, 20, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Heng, B.C.; Zhang, X.; Aubel, D.; Bai, Y.; Li, X.; Wei, Y.; Fussenegger, M.; Deng, X. An overview of signaling pathways regulating YAP/TAZ activity. Cell. Mol. Life Sci. 2021, 78, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Fallahi, E.; O’Driscoll, N.A.; Matallanas, D. The MST/Hippo Pathway and Cell Death: A Non-Canonical Affair. Genes 2016, 7, 28. [Google Scholar] [CrossRef]
- Sahu, M.R.; Mondal, A.C. The emerging role of Hippo signaling in neurodegeneration. J. Neurosci. Res. 2020, 98, 796–814. [Google Scholar] [CrossRef]
- Bruno, L.; Karagil, S.; Mahmood, A.; Elbediwy, A.; Stolinski, M.; Mackenzie, F.E. Mechanosensing and the Hippo Pathway in Microglia: A Potential Link to Alzheimer’s Disease Pathogenesis? Cells 2021, 10, 3144. [Google Scholar] [CrossRef]
- Meotti, F.C.; Figueiredo, C.P.; Manjavachi, M.; Calixto, J.B. The transient receptor potential ankyrin-1 mediates mechanical hyperalgesia induced by the activation of B1 receptor in mice. Biochem. Pharmacol. 2017, 125, 75–83. [Google Scholar] [CrossRef]
- Zhu, D.; Kosik, K.S.; Meigs, T.E.; Yanamadala, V.; Denker, B.M. Galpha12 directly interacts with PP2A: Evidence FOR Galpha12-stimulated PP2A phosphatase activity and dephosphorylation of microtubule-associated protein, tau. J. Biol. Chem. 2004, 279, 54983–54986. [Google Scholar] [CrossRef]
- Torrent, L.; Ferrer, I. PP2A and Alzheimer disease. Curr. Alzheimer Res. 2012, 9, 248–256. [Google Scholar] [CrossRef]
- Laurent, C.; Buee, L.; Blum, D. Tau and neuroinflammation: What impact for Alzheimer’s Disease and Tauopathies? Biomed. J. 2018, 41, 21–33. [Google Scholar] [CrossRef]
- Swerdlow, R.H. Mitochondria and Mitochondrial Cascades in Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 62, 1403–1416. [Google Scholar] [CrossRef] [PubMed]
- Gueli, M.C.; Taibi, G. Alzheimer’s disease: Amino acid levels and brain metabolic status. Neurol. Sci. 2013, 34, 1575–1579. [Google Scholar] [CrossRef] [PubMed]
- Svetlov, V.; Nudler, E. Basic mechanism of transcription by RNA polymerase II. Biochim. Biophys. Acta 2013, 1829, 20–28. [Google Scholar] [CrossRef][Green Version]
- Kershnar, E.; Wu, S.Y.; Chiang, C.M. Immunoaffinity purification and functional characterization of human transcription factor IIH and RNA polymerase II from clonal cell lines that conditionally express epitope-tagged subunits of the multiprotein complexes. J. Biol. Chem. 1998, 273, 34444–34453. [Google Scholar] [CrossRef] [PubMed]
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J.Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019, 570, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]

| Group Comparisons | Enriched Pathway | Involved Gene | Log Fold Change | ||
|---|---|---|---|---|---|
| YoungVsOld | OldVsOAD | YoungVsOAD | |||
| YoungVsOAD & YoungVsOld | Steroid biosynthesis | DHCR24 | −0.67 | N.S. | −0.85 |
| FDFT1 | −0.84 | N.S. | −0.60 | ||
| SQLE | −1.00 | N.S. | -1.03 | ||
| YoungVsOld | OldVsOAD | YoungVsOAD | |||
| YoungVsOld & OldVsOAD | Sphingolipid metabolism | CERS2 | −1.50 | 0.68 | N.S. |
| UGT8 | −2.10 | 1.22 | N.S. | ||
| PLPP2 | −1.76 | 1.23 | N.S. | ||
| Gene | Pathway | Biological Function | Reference |
|---|---|---|---|
| DHCR24 | Steroid biosynthesis | Metabolism of desmosterol and impedes the formation of amyloid-β | [28] |
| FDFT1 | Synthesis of squalene | [29] | |
| SQLE | Cholesterol synthesis | [30] | |
| CERS2 | Sphingolipid Metabolism | Synthesis of ceramide | [31] |
| UGT8 | Transfer of galactose to ceramide | [32] | |
| PLPP2 | Catalyzes the dephosphorylation of sphingosine and ceramide | [33,34] |
| Genes | Log Fold Change | Betweenness Centrality | Degree | |||
|---|---|---|---|---|---|---|
| YoungVsOAD | OldVsOAD | YoungVsOAD | OldVsOAD | YoungVsOAD | OldVsOAD | |
| AJUBA | 0.91 | 0.76 | 0.02 | 0.04 | 39 | 21 |
| ATP5F1A | −0.72 | −0.61 | 0.02 | 0.04 | 24 | 19 |
| GNB2 | −0.36 | −0.36 | 0.02 | 0.09 | 51 | 45 |
| POLR2A | −0.80 | −0.46 | 0.03 | 0.08 | 126 | 50 |
| POLR2E | −0.28 | −0.25 | 0.02 | 0.05 | 140 | 53 |
| PPP2R1A | −0.54 | −0.55 | 0.03 | 0.05 | 74 | 55 |
| YWHAZ | −0.84 | −0.58 | 0.04 | 0.09 | 27 | 22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Angiolini, S.; Chiricosta, L.; Mazzon, E. Sphingolipid Metabolism as a New Predictive Target Correlated with Aging and AD: A Transcriptomic Analysis. Medicina 2022, 58, 493. https://doi.org/10.3390/medicina58040493
D’Angiolini S, Chiricosta L, Mazzon E. Sphingolipid Metabolism as a New Predictive Target Correlated with Aging and AD: A Transcriptomic Analysis. Medicina. 2022; 58(4):493. https://doi.org/10.3390/medicina58040493
Chicago/Turabian StyleD’Angiolini, Simone, Luigi Chiricosta, and Emanuela Mazzon. 2022. "Sphingolipid Metabolism as a New Predictive Target Correlated with Aging and AD: A Transcriptomic Analysis" Medicina 58, no. 4: 493. https://doi.org/10.3390/medicina58040493
APA StyleD’Angiolini, S., Chiricosta, L., & Mazzon, E. (2022). Sphingolipid Metabolism as a New Predictive Target Correlated with Aging and AD: A Transcriptomic Analysis. Medicina, 58(4), 493. https://doi.org/10.3390/medicina58040493






