Endoplasmic Reticulum Stress and Impairment of Ribosome Biogenesis Mediate the Apoptosis Induced by Ocimum x africanum Essential Oil in a Human Gastric Cancer Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Essential Oil Preparation
2.2. AGS Cell Culture
2.3. AGS Cell Viability by MTT Assay
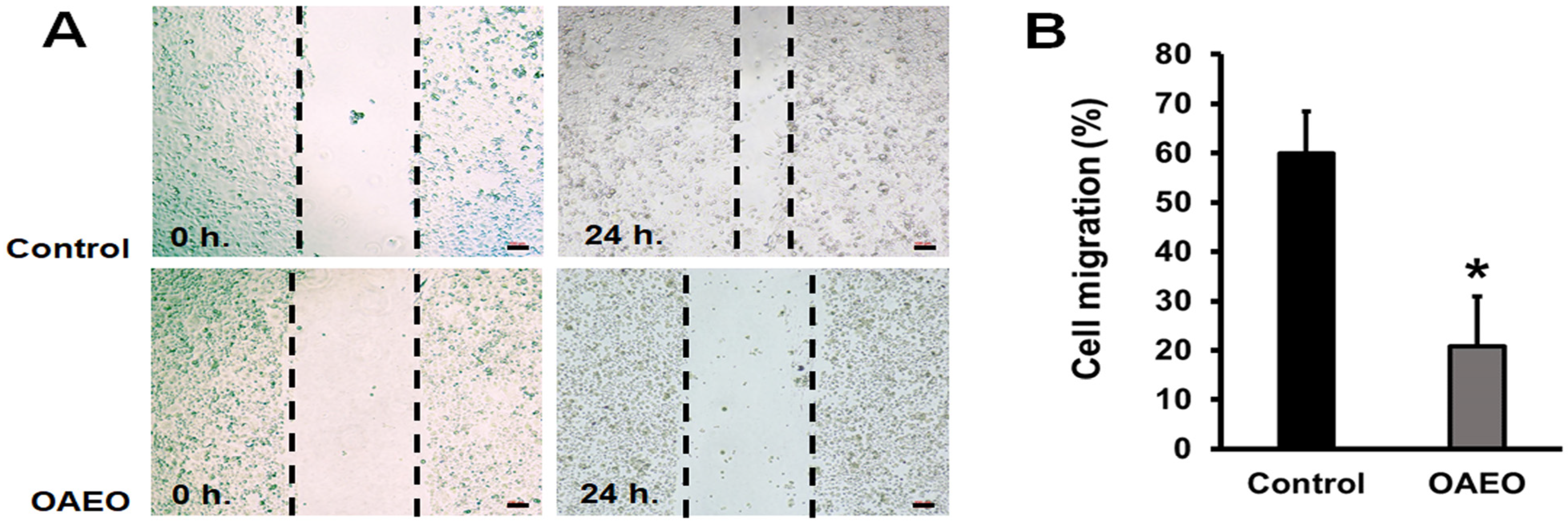
2.4. Cell Migration Assay
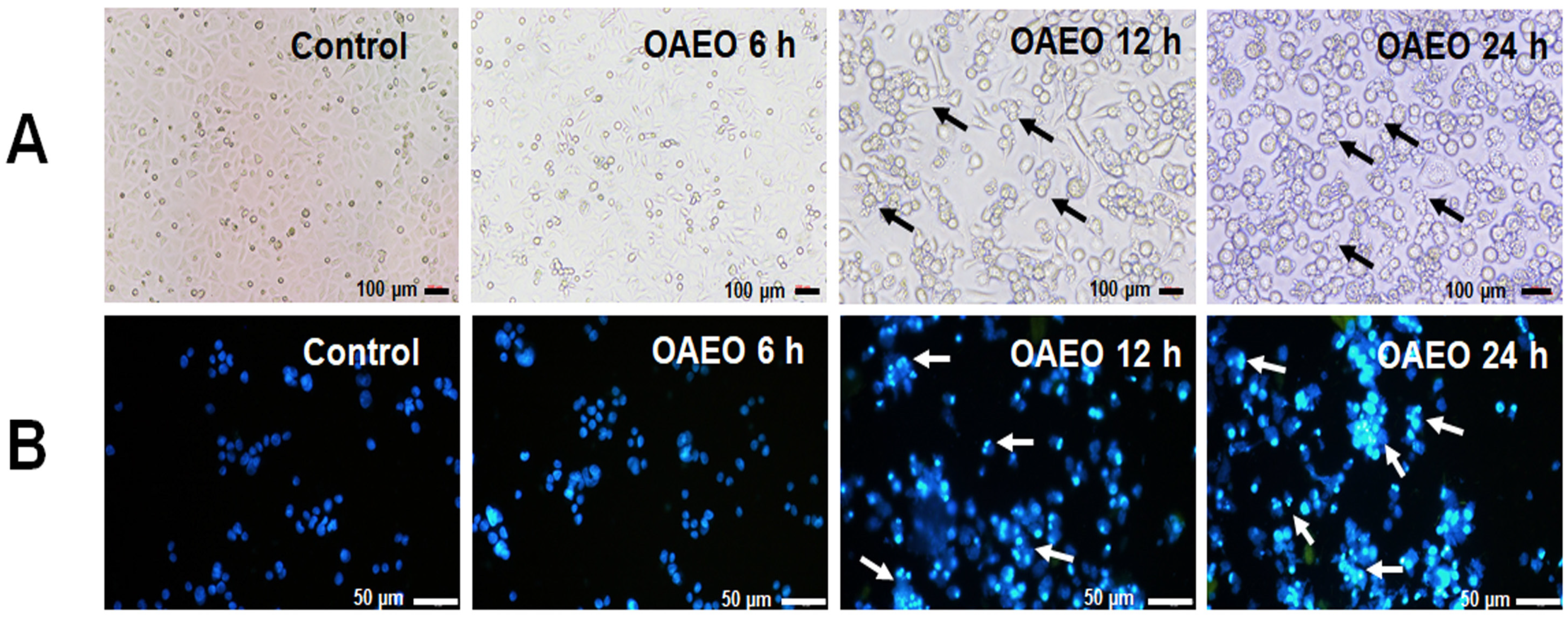
2.5. AGS Nuclear Morphology by DAPI Staining
2.6. Expression Levels of Apoptosis-Related Genes by qRT–PCR
2.7. Differential Protein Analysis by LC–MS/MS
2.8. Analysis of OAEO Chemical Constituents by GC–MS
2.9. Statistical Analysis
3. Results
3.1. Inhibition of AGS Cell Viability by OAEO
3.2. Inhibition of AGS Cell Migration by OAEO
3.3. Morphological Features of Cell Death
3.4. Expression Levels of Apoptosis-Related Genes in OAEO-Treated AGS Cells
3.5. Differential Protein Expression of OAEO-Treated AGS Cells by LC–MS/MS
3.6. Protein–Protein Interaction Network of the Identified Proteins
3.7. GC–MS Analysis of OAEO
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moss, S.F. The Clinical Evidence Linking Helicobacter pylori to Gastric Cancer. Cell Mol. Gastroenterol. Hepatol. 2017, 3, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Van Cutsem, E.; Sagaert, X.; Topal, B.; Haustermans, K.; Prenen, H. Gastric cancer. Lancet 2016, 388, 2654–2664. [Google Scholar] [CrossRef]
- Ohnishi, S.; Takeda, H. Herbal medicines for the treatment of cancer chemotherapy-induced side effects. Front. Pharmacol. 2015, 6, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, T.; Liu, W.; Tao, K.; Wu, C. A Review of Research Progress in Multidrug-Resistance Mechanisms in Gastric Cancer. Onco Targets Ther. 2020, 13, 1797–1807. [Google Scholar] [CrossRef] [Green Version]
- Martucciello, S.; Masullo, M.; Cerulli, A.; Piacente, S. Natural Products Targeting ER Stress, and the Functional Link to Mitochondria. Int. J. Mol. Sci. 2020, 21, 1905. [Google Scholar] [CrossRef] [Green Version]
- Cimino, C.; Maurel, O.M.; Musumeci, T.; Bonaccorso, A.; Drago, F.; Souto, E.M.B.; Pignatello, R.; Carbone, C. Essential Oils: Pharmaceutical Applications and Encapsulation Strategies into Lipid-Based Delivery Systems. Pharmaceutics 2021, 13, 327. [Google Scholar] [CrossRef]
- Gautam, N.; Mantha, A.K.; Mittal, S. Essential oils and their constituents as anticancer agents: A mechanistic view. BioMed. Res. Int. 2014, 2014, 154106. [Google Scholar] [CrossRef] [Green Version]
- Makri, O.; Kintzios, S. Ocimum sp. (Basil): Botany, Cultivation, Pharmaceutical Properties, and Biotechnology. J. Herbs Spices Med. Plants 2008, 13, 123–150. [Google Scholar]
- Pandey, A.K.; Singh, H.; Tripathi, N.N. Chemistry and bioactivities of essential oils of some Ocimum species: An overview. Asian Pac. J. Trop. Biomed. 2014, 4, 682–694. [Google Scholar] [CrossRef] [Green Version]
- Pisutthanan, N.; Pisutthanan, S. Variability of essential oil constituents of Ocimum africanum. Naresuan Univ. J. 2009, 17, 269–274. [Google Scholar]
- Thaweboon, S.; Thaweboon, B. In vitro antimicrobial activity of Ocimum americanum L. essential oil against oral microorganisms. Southeast Asian J. Trop. Med. Public Health 2009, 40, 1025–1033. [Google Scholar] [PubMed]
- Boonyanugomol, W.; Rukseree, K.; Prapatpong, P.; Reamtong, O.; Baik, S.C.; Jung, M.; Shin, M.K.; Kang, H.L.; Lee, W.K. An In Vitro Anti-Cancer Activity of Ocimum tenuiflorum Essential Oil by Inducing Apoptosis in Human Gastric Cancer Cell Line. Medicina 2021, 57, 784. [Google Scholar] [CrossRef] [PubMed]
- Mesri, M. Advances in Proteomic Technologies and Its Contribution to the Field of Cancer. Adv. Med. 2014, 2014, 238045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpievitch, Y.V.; Polpitiya, A.D.; Anderson, G.A.; Smith, R.D.; Dabney, A.R. Liquid Chromatography Mass Spectrometry-Based Proteomics: Biological and Technological Aspects. Ann. Appl. Stat. 2010, 4, 1797–1823. [Google Scholar] [CrossRef]
- Namwat, N.; Amimanan, P.; Loilome, W.; Jearanaikoon, P.; Sripa, B.; Bhudhisawasdi, V.; Tassaneeyakul, W. Characterization of 5-fluorouracil-resistant cholangiocarcinoma cell lines. Chemotherapy 2008, 54, 343–351. [Google Scholar] [CrossRef]
- Jafari, N.; Zargar, S.J.; Yassa, N.; Delnavazi, M.R. Induction of Apoptosis and Cell Cycle Arrest by Dorema Glabrum Root Extracts in a Gastric Adenocarcinoma (AGS) Cell Line. Asian Pac. J. Cancer Prev. 2016, 17, 5189–5193. [Google Scholar] [CrossRef]
- Wang, G.; Dai, L.; Luo, L.; Xu, W.; Zhang, C.; Zhu, Y.; Chen, Z.; Hu, W.; Xu, X.; Pan, W. Non-essential amino acids attenuate apoptosis of gastric cancer cells induced by glucose starvation. Oncol. Rep. 2014, 32, 332–340. [Google Scholar] [CrossRef] [Green Version]
- Soutto, M.; Chen, Z.; Saleh, M.A.; Katsha, A.; Zhu, S.; Zaika, A.; Belkhiri, A.; El-Rifai, W. TFF1 activates p53 through down-regulation of miR-504 in gastric cancer. Oncotarget 2014, 5, 5663–5673. [Google Scholar] [CrossRef] [Green Version]
- Karaliotas, G.I.; Mavridis, K.; Scorilas, A.; Babis, G.C. Quantitative analysis of the mRNA expression levels of BCL2 and BAX genes in human osteoarthritis and normal articular cartilage: An investigation into their differential expression. Mol. Med. Rep. 2015, 12, 4514–4521. [Google Scholar] [CrossRef] [Green Version]
- Borhani, N.; Manoochehri, M.; Gargari, S.S.; Novin, M.G.; Mansouri, A.; Omrani, M.D. Decreased Expression of Proapoptotic Genes Caspase-8- and BCL2-Associated Agonist of Cell Death (BAD) in Ovarian Cancer. Clin. Ovarian Other Gynecologic. Cancer 2014, 7, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Mane, S.D.; Thoh, M.; Sharma, D.; Sandur, S.K.; Naidu, K.A. Ascorbyl Stearate Promotes Apoptosis Through Intrinsic Mitochondrial Pathway in HeLa Cancer Cells. Anticancer Res. 2016, 36, 6409–6417. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Qi, L.; Chen, K.; Li, R.; Song, S.; Zhou, C.; Zhai, W. Metformin induces TPC-1 cell apoptosis through endoplasmic reticulum stress-associated pathways in vitro and in vivo. Int. J. Oncol. 2019, 55, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Jan, R.; Chaudhry, G.E. Understanding Apoptosis and Apoptotic Pathways Targeted Cancer Therapeutics. Adv. Pharm. Bull. 2019, 9, 205–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haupt, S.; Berger, M.; Goldberg, Z.; Haupt, Y. Apoptosis—the p53 network. J. Cell Sci. 2003, 116, 4077–4085. [Google Scholar] [CrossRef] [Green Version]
- Wong, R.S. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Kim, B. Anti-Cancer Natural Products and Their Bioactive Compounds Inducing ER Stress-Mediated Apoptosis: A Review. Nutrients 2018, 10, 1021. [Google Scholar] [CrossRef] [Green Version]
- Rao, R.V.; Ellerby, H.M.; Bredesen, D.E. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004, 11, 372–380. [Google Scholar] [CrossRef] [Green Version]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Wihadmadyatami, H.; Karnati, S.; Hening, P.; Tjahjono, Y.; Rizal; Maharjanti, F.; Kusindarta, D.L.; Triyono, T.; Supriatno. Ethanolic extract Ocimum sanctum Linn. induces an apoptosis in human lung adenocarcinoma (A549) cells. Heliyon 2019, 5, e02772. [Google Scholar] [CrossRef]
- Dhandayuthapani, S.; Azad, H.; Rathinavelu, A. Apoptosis Induction by Ocimum sanctum Extract in LNCaP Prostate Cancer Cells. J. Med. Food 2015, 18, 776–785. [Google Scholar] [CrossRef]
- Manaharan, T.; Thirugnanasampandan, R.; Jayakumar, R.; Kanthimathi, M.S.; Ramya, G.; Ramnath, M.G. Purified Essential Oil from Ocimum sanctum Linn. Triggers the Apoptotic Mechanism in Human Breast Cancer Cells. Pharm. Mag. 2016, 12, S327–S331. [Google Scholar] [CrossRef]
- Kathirvel, P.; Ravi, S. Chemical composition of the essential oil from basil (Ocimum basilicum Linn.) and its in vitro cytotoxicity against HeLa and HEp-2 human cancer cell lines and NIH 3T3 mouse embryonic fibroblasts. Nat. Prod. Res. 2012, 26, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wey, S.; Zhang, Y.; Ye, R.; Lee, A.S. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid Redox Signal. 2009, 11, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Casas, C. GRP78 at the Centre of the Stage in Cancer and Neuroprotection. Front. Neurosci. 2017, 11, 177. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The Unfolded Protein Response and Cell Fate Control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; Hu, Y.; Ruan, L.; Ji, Y.; Lou, Z. Role of endoplasmic reticulum stress in depression (Review). Mol. Med. Rep. 2019, 20, 4774–4780. [Google Scholar] [CrossRef] [Green Version]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chang, A. Heat shock response relieves ER stress. EMBO J. 2008, 27, 1049–1059. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.H.; Li, H.; Yasumura, D.; Cohen, H.R.; Zhang, C.; Panning, B.; Shokat, K.M.; Lavail, M.M.; Walter, P. IRE1 signaling affects cell fate during the unfolded protein response. Science 2007, 318, 944–949. [Google Scholar] [CrossRef] [Green Version]
- Golomb, L.; Volarevic, S.; Oren, M. p53 and ribosome biogenesis stress: The essentials. FEBS Lett. 2014, 588, 2571–2579. [Google Scholar] [CrossRef]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, E.; Rossi, A.; Trere, D. Treating hematological malignancies with drugs inhibiting ribosome biogenesis: When and why. J. Hematol. Oncol. 2018, 11, 75. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, T.; Farrar, S.; Ji, L.; Liu, T.; Ma, X. Antioxidants Maintain Cellular Redox Homeostasis by Elimination of Reactive Oxygen Species. Cell. Physiol. Biochem. 2017, 44, 532–553. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Jiang, J.; Lei, Y.; Zhou, S.; Wei, Y.; Huang, C. Targeting Metabolic-Redox Circuits for Cancer Therapy. Trends Biochem. Sci. 2019, 44, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, H.; Wang, D.; Zhang, L.; Ma, T. Peroxiredoxin2 (Prdx2) Reduces Oxidative Stress and Apoptosis of Myocardial Cells Induced by Acute Myocardial Infarction by Inhibiting the TLR4/Nuclear Factor kappa B (NF-kappaB) Signaling Pathway. Med. Sci. Monit. 2020, 26, e926281. [Google Scholar] [CrossRef]
- Lehtonen, S.T.; Svensk, A.M.; Soini, Y.; Paakko, P.; Hirvikoski, P.; Kang, S.W.; Saily, M.; Kinnula, V.L. Peroxiredoxins, a novel protein family in lung cancer. Int. J. Cancer 2004, 111, 514–521. [Google Scholar] [CrossRef]
- Park, Y.H.; Kim, S.U.; Kwon, T.H.; Kim, J.M.; Song, I.S.; Shin, H.J.; Lee, B.K.; Bang, D.H.; Lee, S.J.; Lee, D.S.; et al. Peroxiredoxin II promotes hepatic tumorigenesis through cooperation with Ras/Forkhead box M1 signaling pathway. Oncogene 2016, 35, 3503–3513. [Google Scholar] [CrossRef]
- Soini, Y.; Kallio, J.P.; Hirvikoski, P.; Helin, H.; Kellokumpu-Lehtinen, P.; Kang, S.W.; Tammela, T.L.; Peltoniemi, M.; Martikainen, P.M.; Kinnula, V.L. Oxidative/nitrosative stress and peroxiredoxin 2 are associated with grade and prognosis of human renal carcinoma. APMIS 2006, 114, 329–337. [Google Scholar] [CrossRef]
- Niu, L.; Liu, A.; Xu, W.; Yang, L.; Zhu, W.; Gu, Y. Downregulation of peroxiredoxin II suppresses the proliferation and metastasis of gastric cancer cells. Oncol. Lett. 2018, 16, 4551–4560. [Google Scholar] [CrossRef]
- Jing, X.; Du, L.; Niu, A.; Wang, Y.; Wang, Y.; Wang, C. Silencing of PRDX2 Inhibits the Proliferation and Invasion of Non-Small Cell Lung Cancer Cells. Biomed. Res. Int. 2020, 2020, 1276328. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Wyckoff, J.; Condeelis, J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005, 17, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Dvorakova, M.; Nenutil, R.; Bouchal, P. Transgelins, cytoskeletal proteins implicated in different aspects of cancer development. Expert Rev. Proteom. 2014, 11, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.; Liu, L.; Hao, R.; Chen, S.; Dong, Y. Transgelin-2: A potential oncogenic factor. Tumour Biol. 2017, 39, 1010428317702650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhifi, W.; Bellili, S.; Jazi, S.; Bahloul, N.; Mnif, W. Essential Oils’ Chemical Characterization and Investigation of Some Biological Activities: A Critical Review. Medicines 2016, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Sharifi-Rad, J.; Sureda, A.; Tenore, G.C.; Daglia, M.; Sharifi-Rad, M.; Valussi, M.; Tundis, R.; Sharifi-Rad, M.; Loizzo, M.R.; Ademiluyi, A.O.; et al. Biological Activities of Essential Oils: From Plant Chemoecology to Traditional Healing Systems. Molecules 2017, 22, 70. [Google Scholar] [CrossRef]
- Barra, A. Factors affecting chemical variability of essential oils: A review of recent developments. Nat. Prod. Commun. 2009, 4, 1147–1154. [Google Scholar] [CrossRef] [Green Version]




| Gene | Primer Sequences | Annealing (°C) | Ref |
|---|---|---|---|
| GAPDH | 5′-TCATCAGCAATGCCTCCTGCA-3′ | 55 | [15] |
| 5′-TGGGTGGCAGTGATGGCA-3′ | |||
| BCL-2 | 5′-CAGGATAACGGAGGCTGGGATG-3′ | 60 | [16] |
| 5′-AGAAATCAAACAGAGGCCGCA-3′ | |||
| BCL-xL | 5′-ACCCCAGGGACAGCATATCA-3′ | 60 | [17] |
| 5′-TGCGATCCGACTCACCAATA-3′ | |||
| TP53 | 5′-TAACAGTTCCTGCATGGGCGGC-3′ | 55 | [18] |
| 5′-AGGACAGGCACAAACACGCACC-3′ | |||
| BAX | 5′-TGGCAGCTGACATGTTTTCTGAC-3′ | 60 | [19] |
| 5′-TCACCCAACCACCCTGGTCTT-3′ | |||
| CASP8 | 5′-AGAGTCTGTGCCCAAATCAAC-3′ | 60 | [20] |
| 5′-GCTGCTTCTCTCTTTGCTGAA-3′ | |||
| CASP9 | 5′-CGAACTAACAGGCAAGCAGC-3′ | 60 | [21] |
| 5′-ACCTCACCAAATCCTCCAGAAC-3′ | |||
| CASP12 | 5′-GCTCAGGAAATGGAAACAGC-3′ | 60 | [22] |
| 5′-AGTGCTTGGTCCCACAGATT-3′ | |||
| CASP3 | 5′-GCGGTTGTAGAAGAGTTTCGTG-3′ | 60 | [16] |
| 5′-CTCACGGCCTGGGATTTCAA-3′ |
| Protein Accession | Protein Description | Fold Change | pI | % Coverage | Peptides Matched |
|---|---|---|---|---|---|
| GRP78_HUMAN | 78 kDa glucose-regulated protein | +26.6 | 5.07 | 44.3 | 24 |
| CH60_HUMAN | 60 kDa heat shock protein, mitochondrial | +15.6 | 5.70 | 34.9 | 14 |
| HSP71_HUMAN | Heat shock 70 kDa protein 1A/1B | +10.8 | 5.48 | 29 | 17 |
| FLNB_HUMAN | Filamin-B | +10.00 | 5.49 | 23.5 | 45 |
| HSP76_HUMAN | Heat shock 70 kDa protein 6 | +8.8 | 5.81 | 20.5 | 12 |
| RS5_HUMAN | 40S ribosomal protein S5 | −6.53 | 9.73 | 26.5 | 6 |
| TAGL2_HUMAN | Transgelin-2 | −5.97 | 8.41 | 82.4 | 16 |
| RL31_HUMAN | 60S ribosomal protein L31 | −5.54 | 10.54 | 43.2 | 4 |
| PRDX2_HUMAN | Peroxiredoxin-2 | −5.13 | 5.66 | 25.3 | 5 |
| TPIS_HUMAN | Triosephosphate isomerase | −3.90 | 6.45 | 56.2 | 13 |
| Retention Time | Compound Name | CAS no. | Area (%) |
|---|---|---|---|
| 2.37 | Oxirane, tetramethyl- | 5076-20-0 | 0.81 |
| 3.56 | 7-methyl-1,6-octadiene | 42152-47-6 | 0.99 |
| 4.51 | Pulegone | 89-82-7 | 0.84 |
| 5.42 | α-pinene | 80-56-8 | 5.99 |
| 6.53 | 6-Methyl-5-hepten-2-one | 110-93-0 | 21.02 |
| 6.75 | 2,3-dehydro-1,8-cineole | 92760-25-3 | 1.46 |
| 7.05 | 3-hexen-1-ol, acetate, (e)- | 3681-82-1 | 0.55 |
| 7.81 | D-limonene | 5989-27-5 | 0.76 |
| 7.91 | Eucalyptol | 470-82-6 | 2.28 |
| 9.01 | Trans-linalool oxide (furanoid) | 34995-77-2 | 0.54 |
| 9.56 | L-fenchone | 7787-20-4 | 2.14 |
| 9.89 | Linalool | 78-70-6 | 17.66 |
| 11.08 | Photocitral A | 55253-28-6 | 0.80 |
| 11.41 | Trans-chrysanthemal | 20104-05-6 | 0.89 |
| 12.92 | α-terpineol | 98-55-5 | 1.15 |
| 13.00 | Estragole | 140-67-0 | 1.95 |
| 13.87 | β-myrcene | 123-35-3 | 0.87 |
| 14.35 | Neral | 106-26-3 | 17.67 |
| 14.84 | 3-cyclohexen-1-one, 2-isopropyl-5-methyl | 900155-47-0 | 0.50 |
| 15.32 | Citral | 5392-40-5 | 19.20 |
| 15.53 | β-pinene | 127-91-3 | 0.38 |
| 18.15 | Camphene | 79-92-5 | 0.14 |
| 18.77 | Copaene | 3856-25-5 | 0.29 |
| 20.16 | Caryophyllene | 87-44-5 | 0.56 |
| 20.55 | Trans-α-bergamotene | 13474-59-4 | 0.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boonyanugomol, W.; Rukseree, K.; Prapatpong, P.; Reamtong, O.; Baik, S.-C.; Jung, M.; Shin, M.-K.; Kang, H.-L.; Lee, W.-K. Endoplasmic Reticulum Stress and Impairment of Ribosome Biogenesis Mediate the Apoptosis Induced by Ocimum x africanum Essential Oil in a Human Gastric Cancer Cell Line. Medicina 2022, 58, 799. https://doi.org/10.3390/medicina58060799
Boonyanugomol W, Rukseree K, Prapatpong P, Reamtong O, Baik S-C, Jung M, Shin M-K, Kang H-L, Lee W-K. Endoplasmic Reticulum Stress and Impairment of Ribosome Biogenesis Mediate the Apoptosis Induced by Ocimum x africanum Essential Oil in a Human Gastric Cancer Cell Line. Medicina. 2022; 58(6):799. https://doi.org/10.3390/medicina58060799
Chicago/Turabian StyleBoonyanugomol, Wongwarut, Kamolchanok Rukseree, Pornpan Prapatpong, Onrapak Reamtong, Seung-Chul Baik, Myunghwan Jung, Min-Kyoung Shin, Hyung-Lyun Kang, and Woo-Kon Lee. 2022. "Endoplasmic Reticulum Stress and Impairment of Ribosome Biogenesis Mediate the Apoptosis Induced by Ocimum x africanum Essential Oil in a Human Gastric Cancer Cell Line" Medicina 58, no. 6: 799. https://doi.org/10.3390/medicina58060799
APA StyleBoonyanugomol, W., Rukseree, K., Prapatpong, P., Reamtong, O., Baik, S.-C., Jung, M., Shin, M.-K., Kang, H.-L., & Lee, W.-K. (2022). Endoplasmic Reticulum Stress and Impairment of Ribosome Biogenesis Mediate the Apoptosis Induced by Ocimum x africanum Essential Oil in a Human Gastric Cancer Cell Line. Medicina, 58(6), 799. https://doi.org/10.3390/medicina58060799






