Early Transcriptional Changes of Adipose-Derived Stem Cells (ADSCs) in Cell Culture
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Sample Acquisition
2.2. Cell Isolation and Culture Conditions
2.3. mRNA-Sequencing and Bioinformatics
3. Results
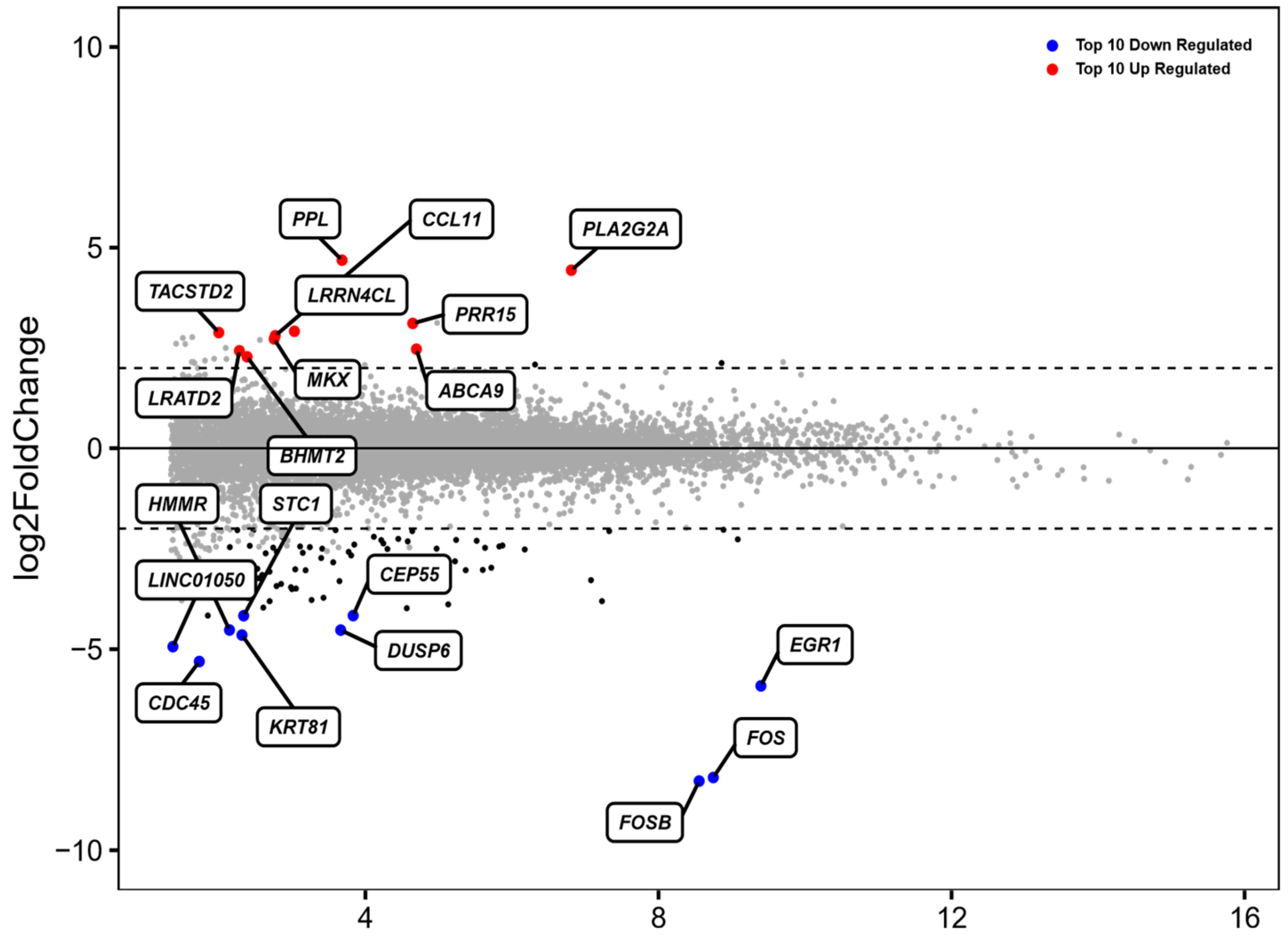
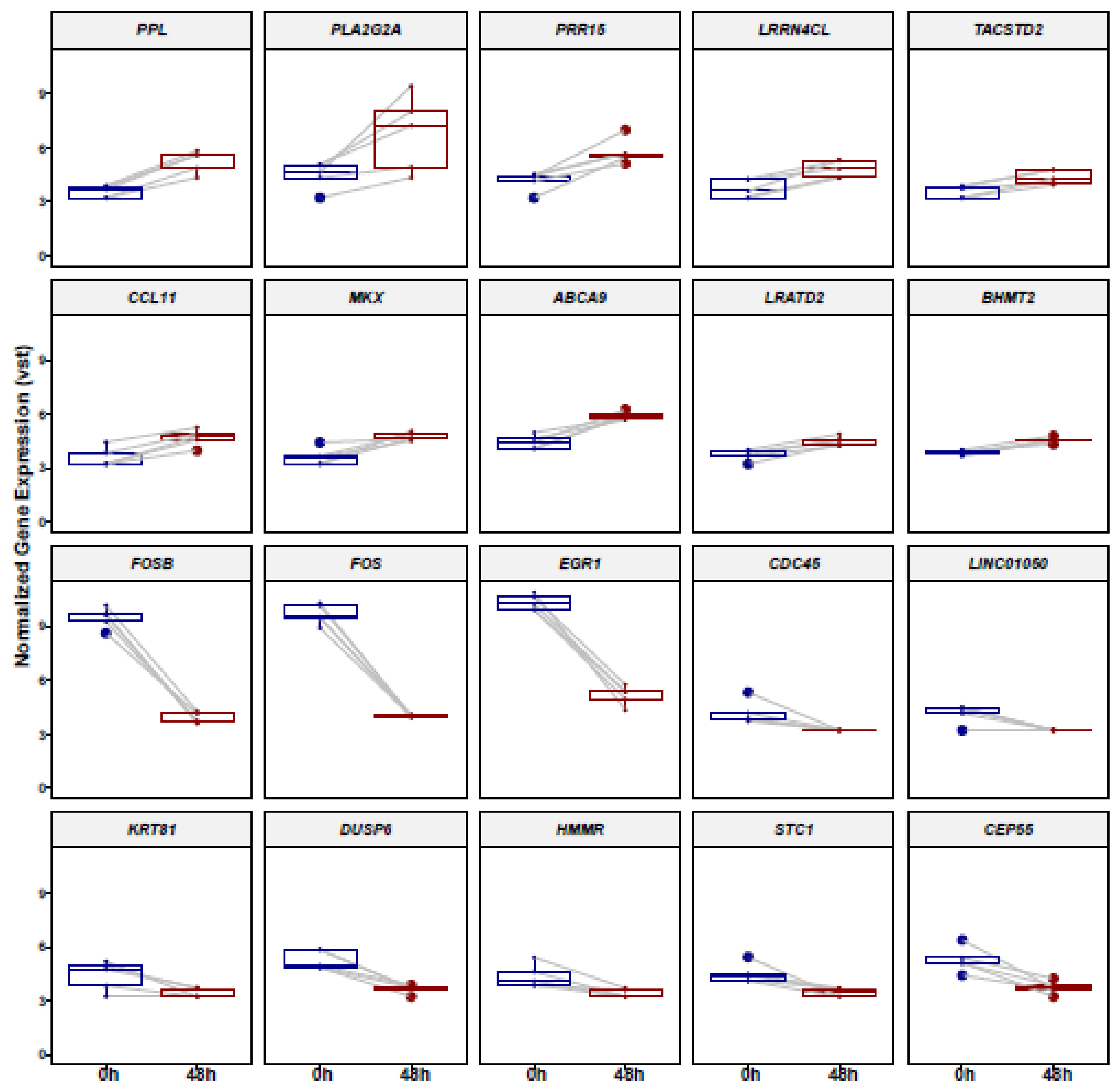
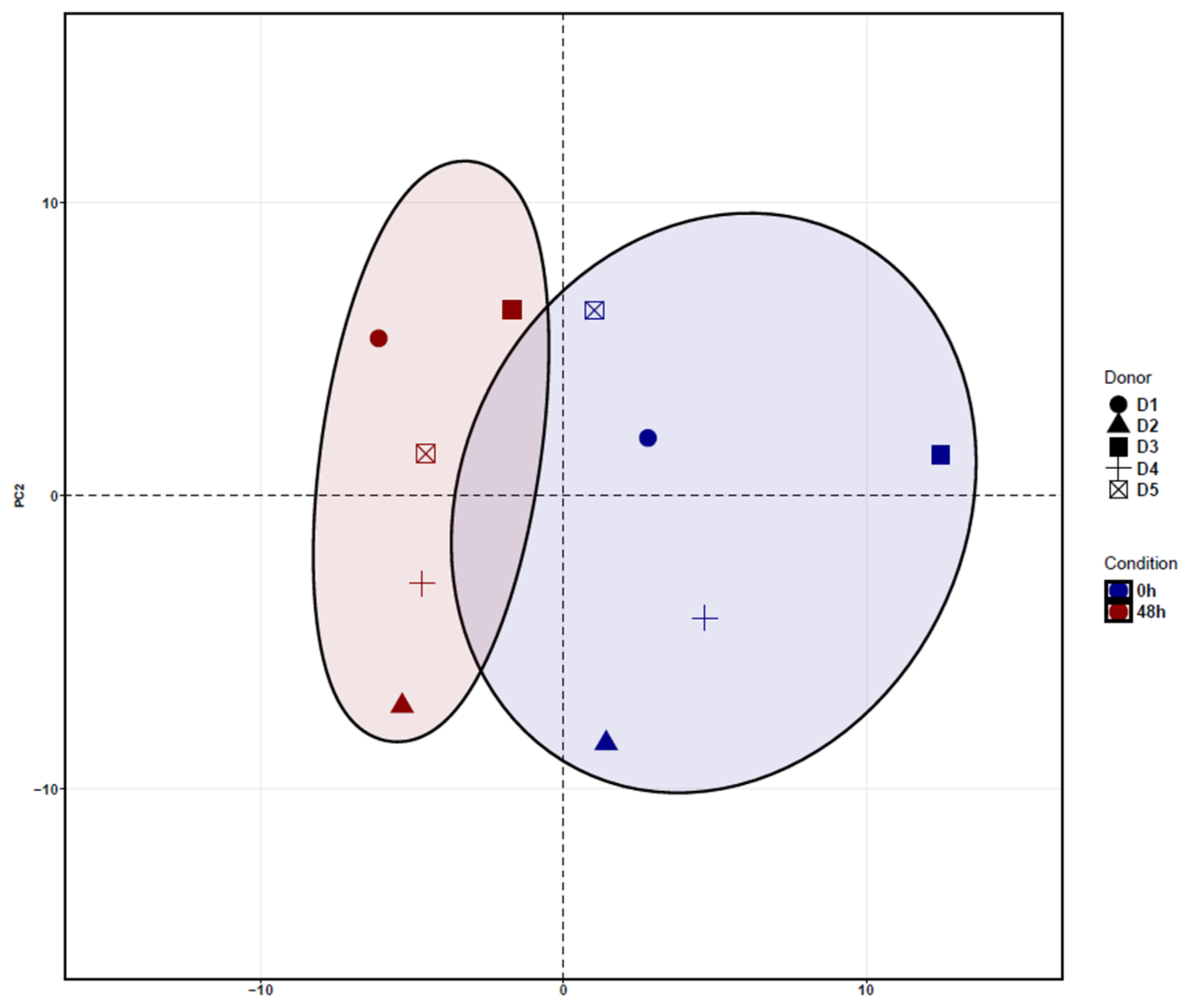
3.1. Identification of Differentially Expressed Genes (DEGs) in ADSCs after Cell Cultivation for 48 h
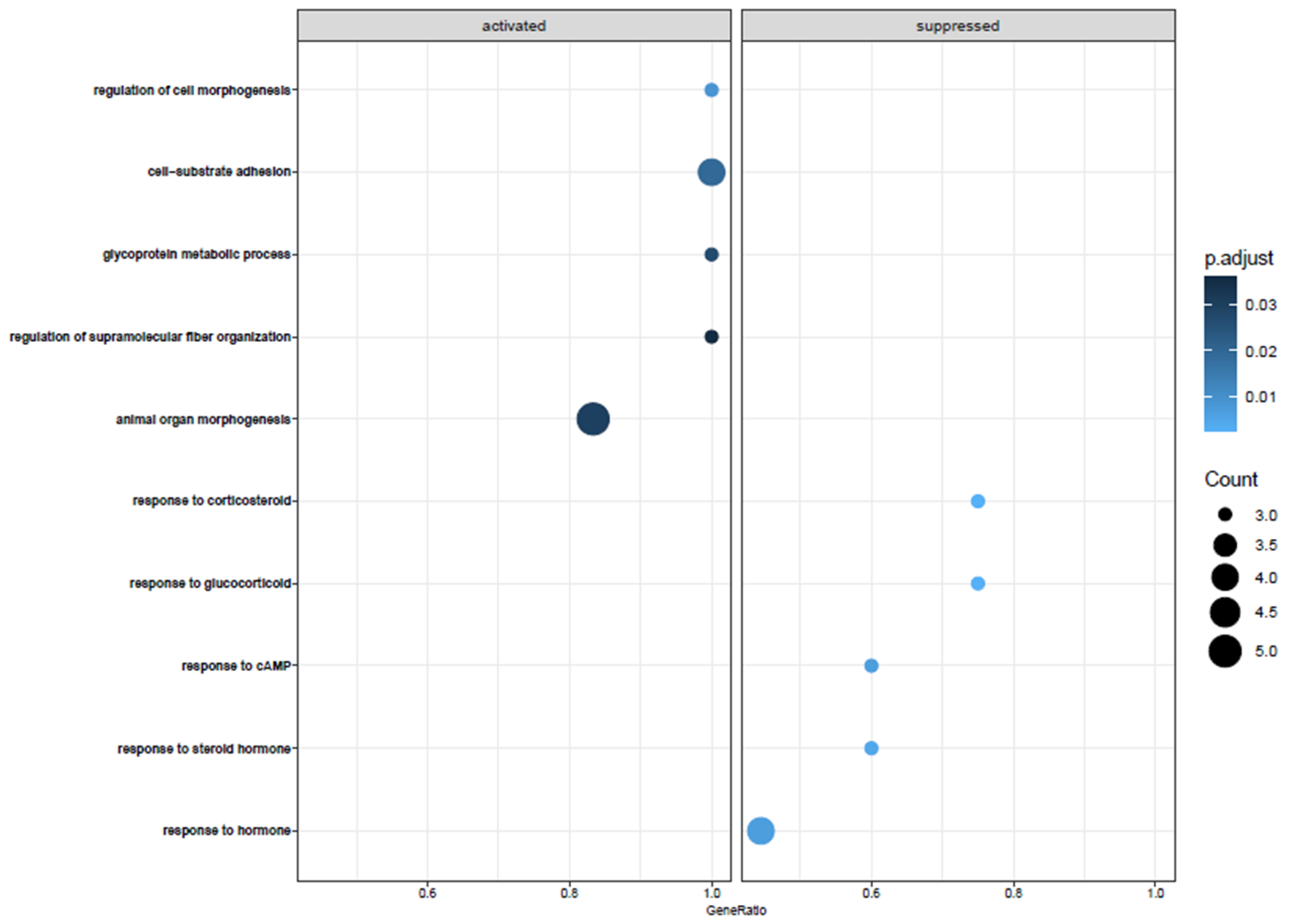
3.2. Biological Pathway Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Witkowska-Zimny, M.; Walenko, K. Stem cells from adipose tissue. Cell. Mol. Biol. Lett. 2011, 16, 236–257. [Google Scholar] [CrossRef] [PubMed]
- Gimble, J.M.; Bunnell, B.A.; Frazier, T.; Rowan, B.; Shah, F.; Thomas-Porch, C.; Wu, X. Adipose-derived stromal/stem cells. Organogenesis 2013, 9, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, H.J.; Yeo, J.E.; Choi, Y.J.; Koh, Y.G. Isolation and Characterization of Human Mesenchymal Stem Cells Derived From Synovial Fluid in Patients With Osteochondral Lesion of the Talus. Am. J. Sports Med. 2014, 43, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Butler, P.E.; Seifalian, A.M. Adipose-derived stem cells for clinical applications: A review. Cell Prolif. 2010, 44, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Uysal, A.C. Tendon Regeneration and Repair with Adipose Derived Stem Cells. Curr. Stem Cell Res. Ther. 2010, 5, 161–167. [Google Scholar] [CrossRef]
- Jiang, L.-B.; Lee, S.; Wang, Y.; Xu, Q.-T.; Meng, D.-H.; Zhang, J. Adipose-derived stem cells induce autophagic activation and inhibit catabolic response to pro-inflammatory cytokines in rat chondrocytes. Osteoarthr. Cartil. 2016, 24, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Cui, W.; Song, F.; Zhai, C.; Hu, H.; Zuo, Q.; Fanglong, S. Effects of mesenchymal stem cells on interleukin-1β-treated chondrocytes and cartilage in a rat osteoarthritic model. Mol. Med. Rep. 2015, 12, 1753–1760. [Google Scholar] [CrossRef]
- Lopatina, T.; Kalinina, N.; Karagyaur, M.; Stambolsky, D.; Rubina, K.; Revischin, A.; Pavlova, G.; Parfyonova, Y.; Tkachuk, V. Adipose-Derived Stem Cells Stimulate Regeneration of Peripheral Nerves: BDNF Secreted by These Cells Promotes Nerve Healing and Axon Growth De Novo. PLoS ONE 2011, 6, e17899. [Google Scholar] [CrossRef]
- Yun, I.S.; Jeon, Y.R.; Lee, W.J.; Lee, J.W.; Rah, D.K.; Tark, K.C.; Lew, D.H. Effect of Human Adipose Derived Stem Cells on Scar Formation and Remodeling in a Pig Model: A Pilot Study. Dermatol. Surg. 2012, 38, 1678–1688. [Google Scholar] [CrossRef]
- Taha, S.; Saller, M.M.; Haas, E.; Farkas, Z.; Aszodi, A.; Giunta, R.; Volkmer, E. Adipose-derived stem/progenitor cells from lipoaspirates: A comparison between the Lipivage200-5 liposuction system and the Body-Jet liposuction system. J. Plast. Reconstr. Aesthetic Surg. 2019, 73, 166–175. [Google Scholar] [CrossRef] [Green Version]
- Dudas, J.R.; Marra, K.G.; Cooper, G.M.; Penascino, V.M.; Mooney, M.P.; Jiang, S.; Rubin, J.P.; Losee, J.E. The Osteogenic Potential of Adipose-Derived Stem Cells for the Repair of Rabbit Calvarial Defects. Ann. Plast. Surg. 2006, 56, 543–548. [Google Scholar] [CrossRef]
- Yoon, E.; Dhar, S.; Chun, D.E.; Gharibjanian, N.A.; Evans, G.R. In Vivo Osteogenic Potential of Human Adipose-Derived Stem Cells/Poly Lactide-Co-Glycolic Acid Constructs for Bone Regeneration in a Rat Critical-Sized Calvarial Defect Model. Tissue Eng. 2007, 13, 619–627. [Google Scholar] [CrossRef]
- Guilak, H.; Awad, F.; Fermor, B.; Leddy, H.; Gimble, J.M. Adipose-derived adult stem cells for cartilage tissue engineering. Biorheology 2004, 41, 389–399. [Google Scholar] [PubMed]
- Moutos, F.T.; Glass, K.A.; Compton, S.A.; Ross, A.K.; Gersbach, C.A.; Guilak, F.; Estes, B.T. Anatomically shaped tissue-engineered cartilage with tunable and inducible anticytokine delivery for biological joint resurfacing. Proc. Natl. Acad. Sci. USA 2016, 113, E4513–E4522. [Google Scholar] [CrossRef] [PubMed]
- Miranville, A.; Heeschen, C.; Sengenès, C.; Curat, C.; Busse, R.; Bouloumié, A. Improvement of Postnatal Neovascularization by Human Adipose Tissue-Derived Stem Cells. Circulation 2004, 110, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Baer, P.C.; Geiger, H. Adipose-Derived Mesenchymal Stromal/Stem Cells: Tissue Localization, Characterization, and Heterogeneity. Stem Cells Int. 2012, 2012, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bowen, R.E. Stromal Vascular Fraction from Lipoaspirate Infranatant: Comparison Between Suction-Assisted Liposuction and Nutational Infrasonic Liposuction. Aesthetic Plast. Surg. 2016, 40, 367–371. [Google Scholar] [CrossRef]
- Yin, S.; Luan, J.; Fu, S.; Wang, Q.; Zhuang, Q. Does Water-Jet Force Make a Difference in Fat Grafting? In Vitro and In Vivo Evidence of Improved Lipoaspirate Viability and Fat Graft Survival. Plast. Reconstr. Surg. 2015, 135, 127–138. [Google Scholar] [CrossRef]
- Duscher, D.; Luan, A.; Rennert, R.C.; Atashroo, D.; Maan, Z.N.; Brett, E.A.; Whittam, A.J.; Ho, N.; Lin, M.; Hu, M.S.; et al. Suction assisted liposuction does not impair the regenerative potential of adipose derived stem cells. J. Transl. Med. 2016, 14, 126. [Google Scholar] [CrossRef]
- Yoshimura, K.; Shigeura, T.; Matsumoto, D.; Sato, T.; Takaki, Y.; Aiba-Kojima, E.; Sato, K.; Inoue, K.; Nagase, T.; Koshima, I.; et al. Characterization of freshly isolated and cultured cells derived from the fatty and fluid portions of liposuction aspirates. J. Cell. Physiol. 2006, 208, 64–76. [Google Scholar] [CrossRef]
- Folgiero, V.; Migliano, E.; Tedesco, M.; Iacovelli, S.; Bon, G.; Torre, M.L.; Sacchi, A.; Marazzi, M.; Bucher, S.; Falcioni, R. Purification and Characterization of Adipose-Derived Stem Cells from Patients with Lipoaspirate Transplant. Cell Transplant. 2010, 19, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.B.; McIntosh, K.; Zvonic, S.; Garrett, S.; Floyd, Z.E.; Kloster, A.; Di Halvorsen, Y.; Storms, R.W.; Goh, B.; Kilroy, G.; et al. Immunophenotype of Human Adipose-Derived Cells: Temporal Changes in Stromal-Associated and Stem Cell–Associated Markers. Stem Cells 2005, 24, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Roobrouck, V.D.; Ulloa-Montoya, F.; Verfaillie, C.M. Self-renewal and differentiation capacity of young and aged stem cells. Exp. Cell Res. 2008, 314, 1937–1944. [Google Scholar] [CrossRef] [PubMed]
- Czapla, J.; Matuszczak, S.; Kulik, K.; Wiśniewska, E.; Pilny, E.; Jarosz-Biej, M.; Smolarczyk, R.; Sirek, T.; Zembala, M.O.; Zembala, M.; et al. The effect of culture media on large-scale expansion and characteristic of adipose tissue-derived mesenchymal stromal cells. Stem Cell Res. Ther. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Lee, B.-Y.; Li, Q.; Song, W.-J.; Chae, H.-K.; Kweon, K.; Ahn, J.-O.; Youn, H.-Y. Altered properties of feline adipose-derived mesenchymal stem cells during continuous in vitro cultivation. J. Veter- Med Sci. 2018, 80, 930–938. [Google Scholar] [CrossRef]
- Simon, M.; Green, H. Participation of membrane-associated proteins in the formation of the cross-linked envelope of the keratinocyte. Cell 1984, 36, 827–834. [Google Scholar] [CrossRef]
- Ruhrberg, C.; Hajibagheri, M.N.; Parry, D.A.; Watt, F. Periplakin, a Novel Component of Cornified Envelopes and Desmosomes That Belongs to the Plakin Family and Forms Complexes with Envoplakin. J. Cell Biol. 1997, 139, 1835–1849. [Google Scholar] [CrossRef]
- Adamska, A.; Falasca, M. ATP-binding cassette transporters in progression and clinical outcome of pancreatic cancer: What is the way forward? World J. Gastroenterol. 2018, 24, 3222–3238. [Google Scholar] [CrossRef]
- Nobili, S.; Lapucci, A.; Landini, I.; Coronnello, M.; Roviello, G.; Mini, E. Role of ATP-binding cassette transporters in cancer initiation and progression. Semin. Cancer Biol. 2020, 60, 72–95. [Google Scholar] [CrossRef] [PubMed]
- Hedditch, E.L.; Gao, B.; Russell, A.J.; Lu, Y.; Emmanuel, C.; Beesley, J.; Johnatty, S.E.; Chen, X.; Harnett, P.; George, J.; et al. ABCA Transporter Gene Expression and Poor Outcome in Epithelial Ovarian Cancer. JNCI: J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Piehler, A.; E Kaminski, W.; Wenzel, J.J.; Langmann, T.; Schmitz, G. Molecular structure of a novel cholesterol-responsive A subclass ABC transporter, ABCA9. Biochem. Biophys. Res. Commun. 2002, 295, 408–416. [Google Scholar] [CrossRef]
- Chen, J.; Qian, X.; He, Y.; Han, X.; Pan, Y. Novel key genes in triple-negative breast cancer identified by weighted gene co-expression network analysis. J. Cell. Biochem. 2019, 120, 16900–16912. [Google Scholar] [CrossRef] [PubMed]
- Meunier, D.; Patra, K.; Smits, R.; Hägebarth, A.; Lüttges, A.; Jaussi, R.; Wieduwilt, M.J.; Quintanilla-Fend, L.; Himmelbauer, H.; Fodde, R.; et al. Expression analysis of proline rich 15 (Prr15) in mouse and human gastrointestinal tumors. Mol. Carcinog. 2010, 50, 8–15. [Google Scholar] [CrossRef]
- Bult, C.J.; Eppig, J.T.; Kadin, J.A.; Richardson, J.E.; Blake, J.A.; the Mouse Genome Database Group. The Mouse Genome Database (MGD): Mouse biology and model systems. Nucleic Acids Res. 2008, 36, D724–D728. [Google Scholar] [CrossRef]
- Jose, P.J.; A Griffiths-Johnson, D.; Collins, P.D.; Walsh, D.T.; Moqbel, R.; Totty, N.F.; Truong, O.; Hsuan, J.J.; Williams, T.J. Eotaxin: A potent eosinophil chemoattractant cytokine detected in a guinea pig model of allergic airways inflammation. J. Exp. Med. 1994, 179, 881–887. [Google Scholar] [CrossRef]
- Kindstedt, E.; Holm, C.K.; Sulniute, R.; Martinez-Carrasco, I.; Lundmark, R.; Lundberg, P. CCL11, a novel mediator of inflammatory bone resorption. Sci. Rep. 2017, 7, 5334. [Google Scholar] [CrossRef]
- Xiao, B.; Wang, G.; Li, W. Weighted gene correlation network analysis reveals novel biomarkers associated with mesenchymal stromal cell differentiation in early phase. PeerJ 2020, 8, e8907. [Google Scholar] [CrossRef]
- Schneider, A.K.; Cama, G.; Ghuman, M.; Hughes, F.J.; Gharibi, B. Sprouty 2, an Early Response Gene Regulator of FosB and Mesenchymal Stem Cell Proliferation During Mechanical Loading and Osteogenic Differentiation. J. Cell. Biochem. 2017, 118, 2606–2614. [Google Scholar] [CrossRef] [PubMed]
- Mikell, I.; Crawford, L.B.; Hancock, M.H.; Mitchell, J.; Buehler, J.; Goodrum, F.; Nelson, J.A. HCMV miR-US22 down-regulation of EGR-1 regulates CD34+ hematopoietic progenitor cell proliferation and viral reactivation. PLOS Pathog. 2019, 15, e1007854. [Google Scholar] [CrossRef] [PubMed]
- Monard, H.; Grimaud, J.A. Matrix metalloproteinases. A review. Cell. Mol. Biol. 1990, 36, 131–153. [Google Scholar]
- Morita-Fujimura, Y.; Fujimura, M.; Gasche, Y.; Copin, J.; Chan, P.H. Overexpression of copper and zinc superoxide dismutase in transgenic mice prevents the induction and activation of matrix metalloproteinases after cold injury-induced brain trauma. J. Cereb. Blood Flow Metab. 2000, 20, 130–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Patient Overview | Sex | Age | Harvesting Site | BMI | Surgery Type |
|---|---|---|---|---|---|
| donor 1 | female | 56 | abdomen | 24.7 | aesthetic |
| donor 2 | female | 76 | thighs | 35.5 | aesthetic |
| donor 3 | male | 25 | abdomen | 23.2 | aesthetic |
| donor 4 | female | 26 | thighs | 21 | aesthetic |
| donor 5 | male | 38 | abdomen | 27.8 | aesthetic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taha, S.; Akova, E.; Saller, M.M.; Giunta, R.E.; Haas-Lützenberger, E.M. Early Transcriptional Changes of Adipose-Derived Stem Cells (ADSCs) in Cell Culture. Medicina 2022, 58, 1249. https://doi.org/10.3390/medicina58091249
Taha S, Akova E, Saller MM, Giunta RE, Haas-Lützenberger EM. Early Transcriptional Changes of Adipose-Derived Stem Cells (ADSCs) in Cell Culture. Medicina. 2022; 58(9):1249. https://doi.org/10.3390/medicina58091249
Chicago/Turabian StyleTaha, Sara, Elif Akova, Maximilian Michael Saller, Riccardo Enzo Giunta, and Elisabeth Maria Haas-Lützenberger. 2022. "Early Transcriptional Changes of Adipose-Derived Stem Cells (ADSCs) in Cell Culture" Medicina 58, no. 9: 1249. https://doi.org/10.3390/medicina58091249







