The Fatal Clinical Outcome of Severe COVID-19 in Hospitalized Patients: Findings from a Prospective Cohort Study in Dhaka, Bangladesh
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Participants
2.2. Data and Sample Collection
2.3. SARS-CoV-2 RT-PCR and Viral Load from NPS
2.4. Immune Response Assessment
2.5. Statistical Analysis
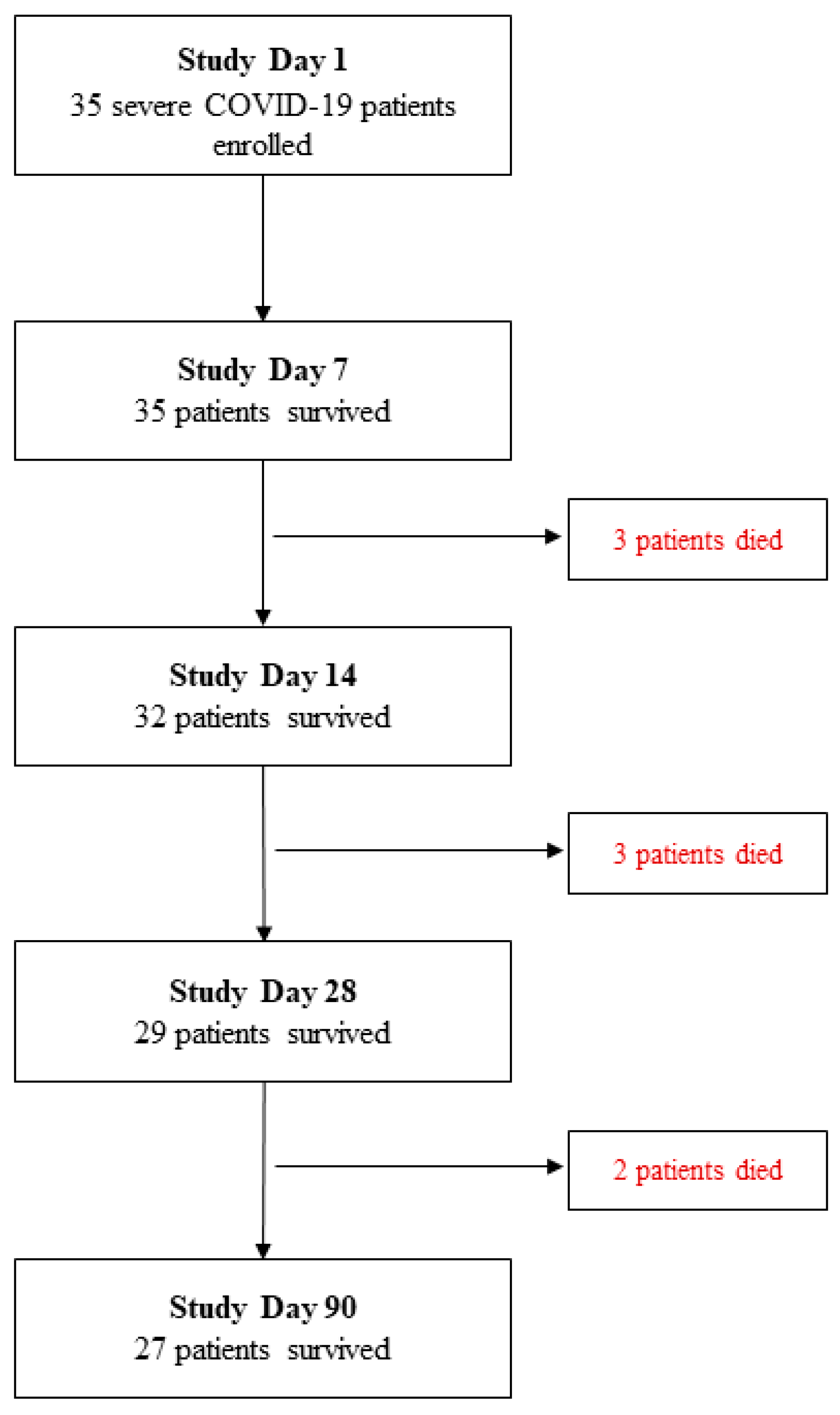
3. Results
3.1. Sociodemographic and Clinical Characteristics
3.2. Medication Received
3.3. Clinical Laboratory Findings
3.4. Viral Load from NPS
3.5. SARS-CoV-2 Specific IgG and IgM Antibody Responses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anwar, S.; Nasrullah, M.; Hosen, M.J. COVID-19 and Bangladesh: Challenges and How to Address Them. Front. Public Health 2020, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- COVID Live-Coronavirus Statistics-Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 30 December 2021).
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 20 May 2021).
- Bangladesh COVID-19 Hospital Information. Available online: https://dghs.gov.bd/images/docs/Notice/2020/corona/covid19_hospital_information.pdf (accessed on 17 August 2020).
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef] [PubMed]
- García, L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020, 11, 1441. [Google Scholar] [CrossRef]
- Kim, G.-U.; Kim, M.-J.; Ra, S.; Lee, J.; Bae, S.; Jung, J.; Kim, S.-H. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin. Microbiol. Infect. 2020, 26, 948.e1–948.e3. [Google Scholar] [CrossRef] [PubMed]
- AlJishi, J.M.; Alhajjaj, A.H.; Alkhabbaz, F.L.; AlAbduljabar, T.H.; Alsaif, A.; Alsaif, H.; Alomran, K.S.; Aljanobi, G.A.; Alghawi, Z.; Alsaif, M.; et al. Clinical characteristics of asymptomatic and symptomatic COVID-19 patients in the Eastern Province of Saudi Arabia. J. Infect. Public Health 2021, 14, 6–11. [Google Scholar] [CrossRef]
- Bansal Garg, I.; Srivastava, S.; Rai, C.; Kumar, V.; Hembrom, A.; Ghosh, N.; Kumari, B.; Bansal, A.; Kumar, B. Coronavirus (COVID-19): Prognostic risk associated with comorbidities and age. Int. J. Recent Sci. Res. 2020, 11, 37983–37986. [Google Scholar]
- Mejía, F.; Medina, C.; Cornejo, E.; Morello, E.; Vásquez, S.; Alave, J.; Schwalb, A.; Málaga, G. Oxygen saturation as a predictor of mortality in hospitalized adult patients with COVID-19 in a public hospital in Lima, Peru. PLoS ONE 2020, 15, e0244171. [Google Scholar] [CrossRef]
- Akter, A.; Ahmed, T.; Tauheed, I.; Akhtar, M.; Rahman, S.I.A.; Khaton, F.; Ahmmed, F.; Ferdous, J.; Afrad, M.H.; Kawser, Z.; et al. Disease characteristics and serological responses in patients with differing severity of COVID-19 infection: A longitudinal cohort study in Dhaka, Bangladesh. PLoS Neglected Trop. Dis. 2022, 16, e0010102. [Google Scholar] [CrossRef]
- Wang, B.; Li, R.; Lu, Z.; Huang, Y. Does comorbidity increase the risk of patients with COVID-19: Evidence from meta-analysis. Aging 2020, 12, 6049–6057. [Google Scholar] [CrossRef]
- WHO. Clinical Management of COVID-19: Interim Guidance, 27 May 2020; WHO IRIS; World Health Organization: Geneva, Switzerland, 2020; Available online: https://apps.who.int/iris/handle/10665/332196 (accessed on 25 March 2021).
- National Guidelines on Clinical Management of COVID-19 Version 8.0. Available online: https://covidlawlab.org/wp-content/uploads/2021/01/Bangladesh_2020.11.05_Guideline_National-Guidelines-on-Clinical-Management-of-COVID-19_EN.pdf (accessed on 23 August 2021).
- Wang, Y.; Zhao, J.; Yang, L.; Hu, J.; Yao, Y. Value of the Neutrophil-Lymphocyte Ratio in Predicting COVID-19 Severity: A Meta-analysis. Dis. Markers 2021, 2021, 2571912. [Google Scholar] [CrossRef]
- Imran, M.M.; Ahmad, U.; Usman, U.; Ali, M.; Shaukat, A.; Gul, N. Retracted: Neutrophil/lymphocyte ratio—A marker of COVID-19 pneumonia severity. Int. J. Clin. Pract. 2021, 75, e13698. [Google Scholar] [CrossRef] [PubMed]
- CDC Research Use Only 2019-Novel Coronavirus (2019-NCoV) Real-Time RT-PCR Primers and Probes. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 25 April 2021).
- Shirin, T.; Bhuiyan, T.R.; Charles, R.C.; Amin, S.; Bhuiyan, I.; Kawser, Z.; Rahat, A.; Alam, A.N.; Sultana, S.; Aleem, M.A.; et al. Antibody responses after COVID-19 infection in patients who are mildly symptomatic or asymptomatic in Bangladesh. Int. J. Infect. Dis. 2020, 101, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.S.; Jones, F.K.; Nodoushani, A.; Kelly, M.; Becker, M.; Slater, D.; Mills, R.; Teng, E.; Kamruzzaman, M.; Garcia-Beltran, W.F.; et al. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv 2020. [Google Scholar] [CrossRef]
- Bhuiyan, T.R.; Akhtar, M.; Akter, A.; Khaton, F.; Rahman, S.I.A.; Ferdous, J.; Nazneen, A.; Sumon, S.A.; Banik, K.C.; Bablu, A.R.; et al. Seroprevalence of SARS-CoV-2 Antibodies in Bangladesh Related to Novel Coronavirus Infection. IJID Reg. 2022, 2, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, T.; Weißbrich, B.; Wedekink, F.; Notz, Q.; Herrmann, J.; Krone, M.; Sitter, M.; Schmid, B.; Kredel, M.; Stumpner, J.; et al. Biodistribution and serologic response in SARS-CoV-2 induced ARDS: A cohort study. PLoS ONE 2020, 15, e0242917. [Google Scholar] [CrossRef]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W.; the Northwell COVID-19 Research Consortium. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized with COVID-19 in the New York City Area. JAMA 2020, 323, 2052–2059. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, J.; Liu, L.; Zhang, Y.; Liu, D.; Xiong, M.; Zhao, Y.; Chen, K.; Wang, Y.-M. Clinical Characteristics of Patients with Severe and Critical COVID-19 in Wuhan: A Single-Center, Retrospective Study. Infect. Dis. Ther. 2021, 10, 421–438. [Google Scholar] [CrossRef]
- Singer, A.J.; Morley, E.J.; Meyers, K.; Fernandes, R.; Rowe, A.L.; Viccellio, P.; Thode, H.C.; Bracey, A.; Henry, M.C. Cohort of Four Thousand Four Hundred Four Persons Under Investigation for COVID-19 in a New York Hospital and Predictors of ICU Care and Ventilation. Ann. Emerg. Med. 2020, 76, 394–404. [Google Scholar] [CrossRef]
- Chen, T.; Wu, D.; Chen, H.; Yan, W.; Yang, D.; Chen, G.; Ma, K.; Xu, D.; Yu, H.; Wang, H.; et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ 2020, 368, m1091. [Google Scholar] [CrossRef] [Green Version]
- Kremer, H.-J.; Thurner, W. Age Dependence in COVID-19 Mortality in Germany. Dtsch. Ärztebl. Int. 2020, 117, 432–433. [Google Scholar] [CrossRef]
- Kumar, V. Emerging Human Coronavirus Infections (SARS, MERS, and COVID-19): Where They Are Leading Us. Int. Rev. Immunol. 2021, 40, 5–53. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, K.; Wei, H.; Chen, W.; Wang, W.; Jia, L.; Liu, Q.; Zhang, J.; Shan, T.; Peng, Z.; et al. Dynamic relationship between D-dimer and COVID-19 severity. Br. J. Haematol. 2020, 190, e24–e27. [Google Scholar] [CrossRef] [PubMed]
- El Zein, S.; Chehab, O.; Kanj, A.; Akrawe, S.; Alkassis, S.; Mishra, T.; Shatta, M.; El-Hor, N.; Salimnia, H.; Chandrasekar, P. SARS-CoV-2 infection: Initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic. PLoS ONE 2021, 16, e0255981. [Google Scholar] [CrossRef] [PubMed]
- Pujadas, E.; Chaudhry, F.; McBride, R.; Richter, F.; Zhao, S.; Wajnberg, A.; Nadkarni, G.; Glicksberg, B.S.; Houldsworth, J.; Cordon-Cardo, C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020, 8, e70. [Google Scholar] [CrossRef]
- Longchamp, A.; Longchamp, J.; Croxatto, A.; Greub, G.; Sanchez, B.; Delaloye, J.; Whiting, L.; Jeanneret, S.; Coste, A.T.; Dumoulin, A.; et al. Serum antibody response in critically ill patients with COVID-19. Intensiv. Care Med. 2020, 46, 1921–1923. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Sang, L.; Ye, F.; Ruan, S.; Zhong, B.; Song, T.; Alshukairi, A.N.; Chen, R.; Zhang, Z.; et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Investig. 2020, 130, 5235–5244. [Google Scholar] [CrossRef]
- Alberca, R.W. Immune Response to COVID-19; IntechOpen: London, UK, 2021; ISBN 978-1-83968-217-9. [Google Scholar]
| Variables | All Patients n = 35 | Survivor (n = 27) | Death (n = 8) | p-Values * |
|---|---|---|---|---|
| Age, Median (IQR) | 55 (44, 62.5) | 50 (42.5, 62.5) | 61 (54.5, 62.75) | 0.215 |
| Sex, Male n (%) | 24 (68.57%) | 19 (70.37%) | 5 (62.5%) | 0.674 |
| Blood Group | ||||
| O Blood Group n (%) | 12 (34.29%) | 9 (33.33%) | 3 (37.5%) | 0.827 |
| Non-O Blood Group n (%) | 23 (65.71%) | 18 (66.67%) | 5 (62.5%) | |
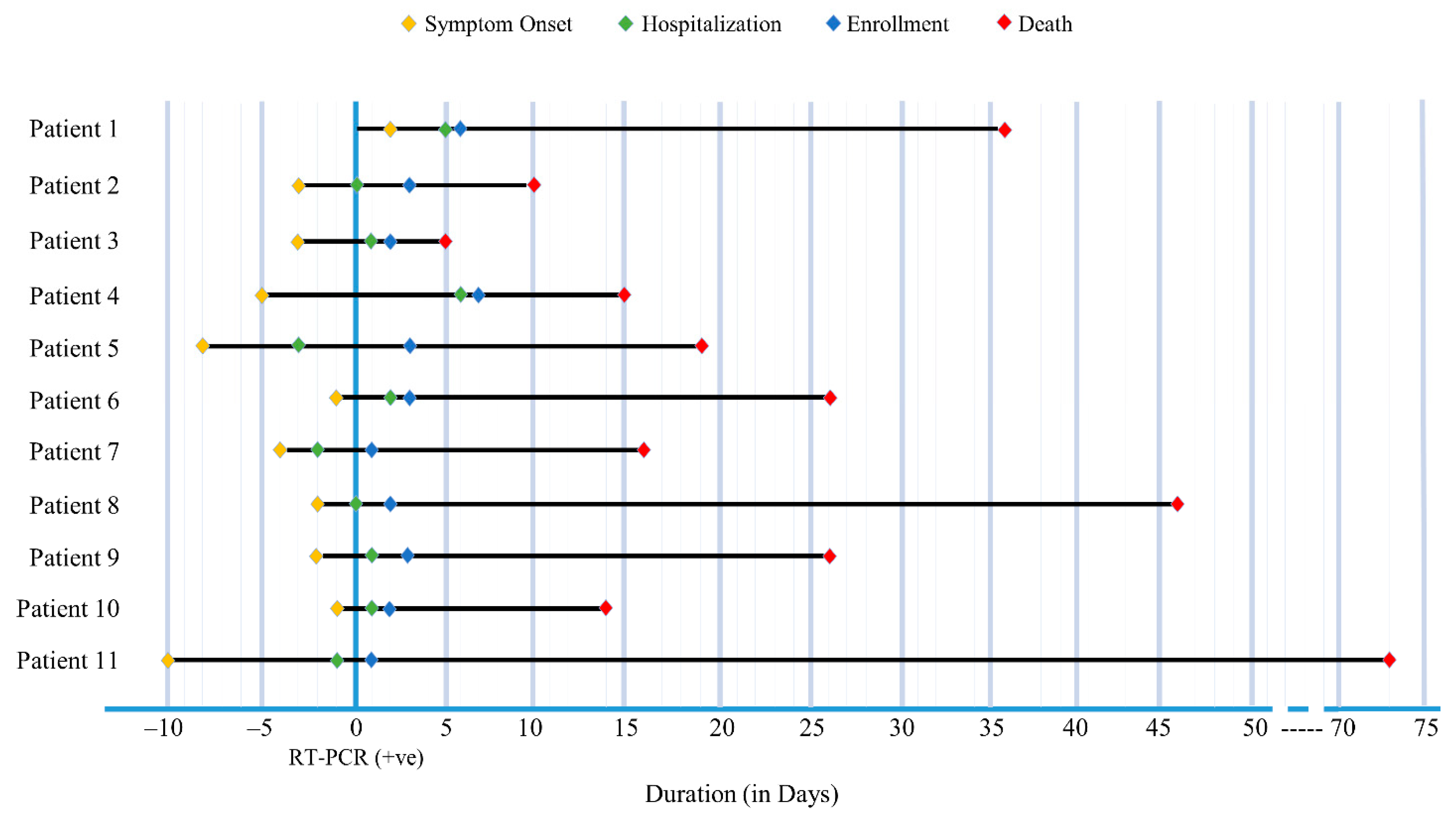
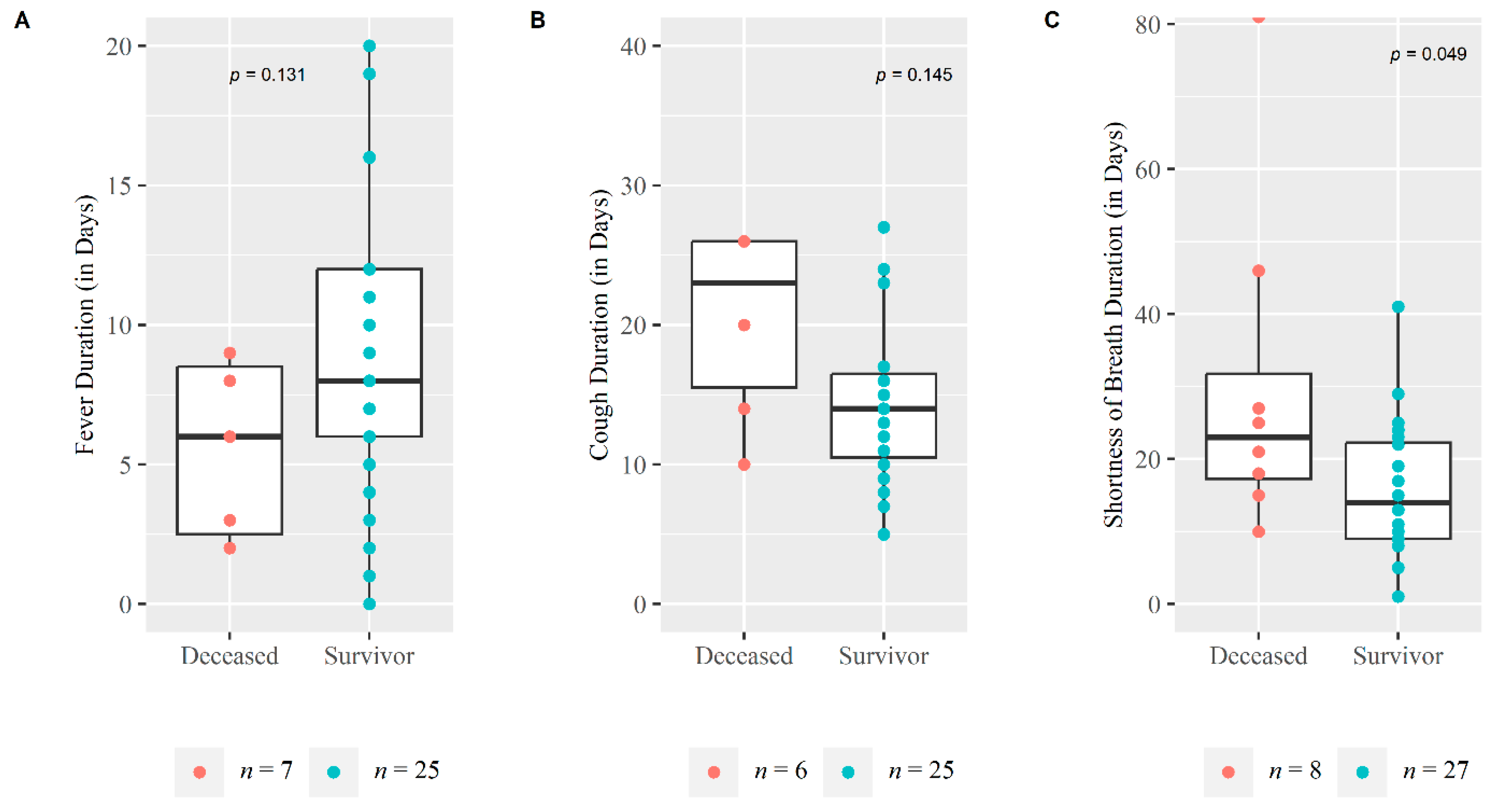
| Duration interval in days, median (IQR) a | ||||
| Disease Onset and Diagnosis | 5 (3, 7) | 6 (3, 7) | 3 (1.75, 5.75) | 0.206 |
| Disease Onset and Hospitalization | 7 (5, 9) | 8 (6, 7.5) | 3 (2, 6) | 0.046 |
| Duration of hospitalization | 14 (10, 22) | 13 (10, 19.5) | 19 (12.25, 28) | 0.245 |
| Disease Onset and Onset of Shortness of Breath | 4 (1.5, 6.5) | 4 (1.5, 7) | 2 (1.5, 3.75) | 0.303 |
| First symptom at disease onset, n (%) b | ||||
| Fever | 25 (71.43%) | 20 (74.07%) | 5 (62.5%) | 0.525 |
| Cough | 7 (20%) | 5 (18.52%) | 2 (25%) | 0.687 |
| Others (Loss of Taste, Runny Nose, Shortness of Breath) | 3 (8.57%) | 2 (7.41%) | 1 (12.5%) | 0.651 |
| Reported symptoms over the first 1 month, n (%) b | ||||
| Fever | 32 (91.43%) | 25 (92.59%) | 7 (87.5%) | 0.651 |
| Cough | 31 (88.57%) | 25 (92.59%) | 6 (75%) | 0.169 |
| Shortness of breath | 35 (100%) | 27 (100%) | 8 (100%) | - |
| Others ** | 21 (60%) | 16 (59.26%) | 5 (62.5%) | 0.869 |
| No. of symptoms reported over the first 1 month, n (%) b | ||||
| ≤3 symptoms | 16 (45.71%) | 13 (48.15%) | 3 (37.5%) | 0.595 |
| >3 symptoms | 19 (54.29%) | 14 (51.85%) | 5 (62.5%) | |
| Vital Signs, Mean (95% CI) c | ||||
| Systolic pressure (mmHg) | 125.51 (119.1, 131.9) | 128 (120, 136) | 117 (106, 128) | 0.079 |
| Diastolic pressure (mmHg) | 74.8 (70.86, 78.74) | 76.1 (71.4, 80.7) | 70.5 (62.3, 78.7) | 0.200 |
| Pulse rate (beats/min) | 84.49 (80.4, 88.6) | 85.5 (80.7, 90.4) | 81 (71.6, 90.4) | 0.347 |
| Respiratory rate (times/min) | 30.83 (28.5, 33.2) | 31.2 (28.7, 33.7) | 29.5 (22.5, 36.5) | 0.604 |
| Temperature (°F) | 97.60 (97.4, 97.8) | 97.6 (97.3, 97.8) | 97.7 (97.4, 98) | 0.477 |
| SpO2% (during hospital admission) | 83.51 (80.12, 86.91) | 83.93 (80.81, 87.04) | 82.13 (69.13, 95.12) | 0.760 |
| Oxygen flow at enrolment (L/min) | 8.21 (6.81, 9.6) | 8 (6.4, 9.6) | 8.88 (5.32, 12.43) | 0.616 |
| ICU required, n(%) b | 10 (28.57%) | 3 (11.11%) | 7 (87.5%) | 0.0003 |
| Ventilation required, n(%) b | 2 (5.71%) | 0 | 2 (25%) | 0.007 |
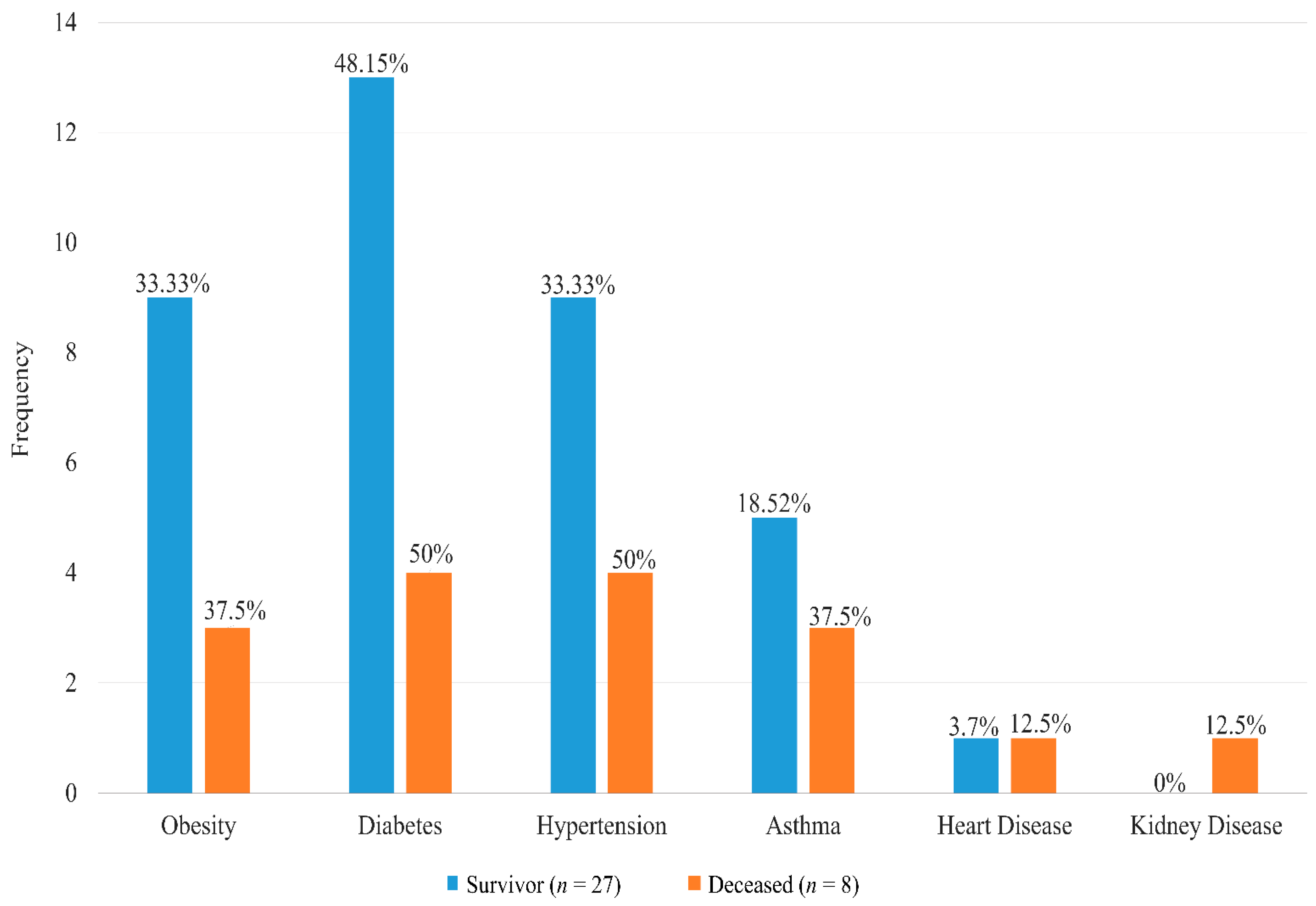
| Comorbidities, n(%) b | ||||
| Yes | 29 (82.86) | 22 (81.48) | 7 (87.5) | 0.692 |
| No. of Comorbidities b | ||||
| ≤1 | 17 (48.57%) | 14 (51.85%) | 3 (37.5%) | 0.476 |
| >1 | 18 (51.43%) | 13 (48.15%) | 5 (62.5%) | |
| Duration (in years) of Comorbidities, Median (IQR) a | ||||
| Diabetes (n = 17, nA = 13) | 7 (4, 10) | 8 (2,10) | 6 (4.75, 9) | 0.954 |
| Heart disease (n = 2, nA = 1) | 10 (7.5, 12.5) | 15 (15, 15) | 5 (5, 5) | - |
| Hypertension (n = 13, nA = 9) | 7 (2, 10) | 7 (2, 10) | 7 (4.5, 9.25) | 0.754 |
| Asthma (n = 8, nA = 5) | 10 (7, 20.75) | 10 (7, 20) | 10 (8.5, 17.5) | 0.763 |
| Kidney disease (n = 1, nA = 0) | 0.0822 (0.08, 0.08) | - | 0.0822 (0.08, 0.08) | - |
| Medication Received | All Patients n = 35 | Survivor n = 27 | Deceased n = 8 | p-Values |
|---|---|---|---|---|
| Antibiotic a, n (%) | 35 (100%) | 27 (100%) | 8 (100%) | - |
| Single | 6 (17.14%) | 4 (14.81%) | 2 (25%) | 0.502 |
| Multiple | 29 (82.86%) | 23 (85.19%) | 6 (75%) | |
| Interval between Hospitalization and antibiotic start, Median(IQR) | 0 (0, 4) | 0 (0, 4) | 1.5 (0, 5) | 0.289 |
| Anti-Coagulant b, n (%) | 35 (100%) | 27 (100%) | 8 (100%) | - |
| Single | 19 (54.29%) | 12 (44.44%) | 7 (87.5%) | 0.032 |
| Multiple | 16 (45.71%) | 15 (55.56%) | 1 (12.5%) | |
| Interval between Hospitalization and anti-Coagulant start, Median(IQR) | 0 (0, 9) | 0 (0, 10) | 0 (0, 1) | 0.402 |
| Anti-histamine c, n (%) | 28 (80%) | 22 (81.48%) | 6 (75%) | 0.687 |
| Single | 22 (78.57%) | 17 (77.27%) | 5 (83.33%) | 0.748 |
| Multiple | 6 (21.43%) | 5 (22.73%) | 1 (16.67%) | |
| Interval between Hospitalization and anti-histamine start, Median(IQR) | 0 (0, 1) | 0 (0, 1) | 0 (0, 1) | 0.783 |
| Anti-helminthic (Ivermectin), n (%) | 13 (37.14) | 10 (37.04) | 3 (37.5) | 0.981 |
| Interval between Hospitalization and Ivermectin start, Median(IQR) | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) | 1.00 |
| Anti-viral d, n (%) | 17 (48.57) | 15 (55.56) * | 2 (25) | 0.129 |
| Interval between Hospitalization and Anti-Viral start, Median(IQR) | 1 (0, 2) | 1 (0, 1.5) | 6.5 (6.25, 6.75) | 0.054 |
| Theophylline (Doxofylline), n (%) | 10 (28.57) | 9 (33.33) | 1 (12.5) | 0.252 |
| Interval between Hospitalization and theophylline start, Median(IQR) | 6 (2.25, 9.75) | 4 (2, 9) | 10 (10, 10) | 0.484 |
| Glucocorticoid steroid e, n (%) | 31 (88.57%) | 23 (85.19%) | 8 (100%) | 0.247 |
| Single | 26 (83.87%) | 18 (78.26%) | 8 (100%) | 0.569 |
| Multiple | 5 (16.13%) | 5 (21.74%) | 0 (0%) | |
| Duration between Hospitalization and steroid start, Median(IQR) | 1 (0, 9.25) | 1 (0, 10.25) | 0.5 (0, 1.50) | 0.324 |
| Treatment Initiation before Hospitalization n (%) * | 3 (8.57%) | 3 (11.11%) | 0 (0%) | 0.324 |
| Variables Normal Reference | All Patient n = 35 | Survivor (n = 27) | Death (n = 8) | p-Values * |
|---|---|---|---|---|
| TLC (109/L) 4.0–11.0 | 8.12 (6.06, 9.76) | 7.79 (6.06, 9.76) | 9.31 (7.34, 9.99) | 0.569 |
| Diff. Neu (%) 40–75 | 81.30 (77.40, 84.95) | 81.30 (77.25, 84.95) | 82.10 (80.90, 84.65) | 0.492 |
| Diff. Lymph (%) 20–45 | 11.80 (8.00, 15.20) | 11.80 (8.00, 15.20) | 11.50 (8.90, 13.93) | 0.922 |
| Diff. Mono (%) 2–10 | 5.50 (3.95, 8.65) | 6.80 (4.30, 8.95) | 4.50 (3.38, 5.90) | 0.074 |
| Diff. Eosino (%) 1–6 | 0.00 (0.00, 0.2) | 0.00 (0.00, 0.4) | 0.00 (0.00, 0.00) | 0.072 |
| Diff. Baso (%) 0.01–0.1 | 0.1 (0.1, 0.25) | 0.1 (0.1, 0.35) | 0.1 (0.1, 0.125) | 0.368 |
| Abs. Neut (109/L) 2.0–7.5 | 6.74 (4.96, 8.68) | 5.97 (4.96, 8.68) | 7.59 (6.04, 8.595) | 0.556 |
| Abs. Lymph (109/L) 1.5–4.0 | 0.83 (0.65, 1.195) | 0.83 (0.62, 1.16) | 0.9 (0.74, 1.24) | 0.492 |
| Abs. Mono (109/L) 0.2–0.8 | 0.44 (0.36, 0.63) | 0.44 (0.37, 0.64) | 0.46 (0.22, 0.52) | 0.346 |
| Abs. Eos (109/L) 0.04–0.45 | 0.00 (0.00, 0.015) | 0.00 (0.00, 0.025) | 0.00 (0.00, 0.00) | 0.0589 |
| Abs. Bas (109/L) 0.01–0.1 | 0.01 (0.01, 0.02) | 0.01 (0.01, 0.02) | 0.01 (0.01, 0.01) | 0.406 |
| Plt (109/L) 150–450 | 266 (208, 365.5) | 282 (215.5, 371.5) | 207 (155.2, 298) | 0.107 |
| RBS (mmol/L) 4.20–7.80 | 10.71 (7.29, 14.24) | 10.71 (7.69, 13.28) | 11.47 (7.32, 14.95) | 0.738 |
| Cr (µmol/L) 64–104 | 79.80 (64.82, 100.01) | 78.01 (63.48, 97.57) | 85.05 (74.19, 111.25) | 0.492 |
| ALT (U/L) <50 | 43 (25.34, 101.50) | 43 (25.34, 86.50) | 68.50 (27.50, 127.40) | 0.709 |
| AST (U/L) <50 | 47 (29, 61.50) | 42 (29, 57.8) | 64 (41.23, 101.50) | 0.121 |
| LDH (U/L) <248 | 401.2 (306.1, 485.5) | 401.2 (318, 485.5) | 373.1 (271.7, 491.5) | 0.768 |
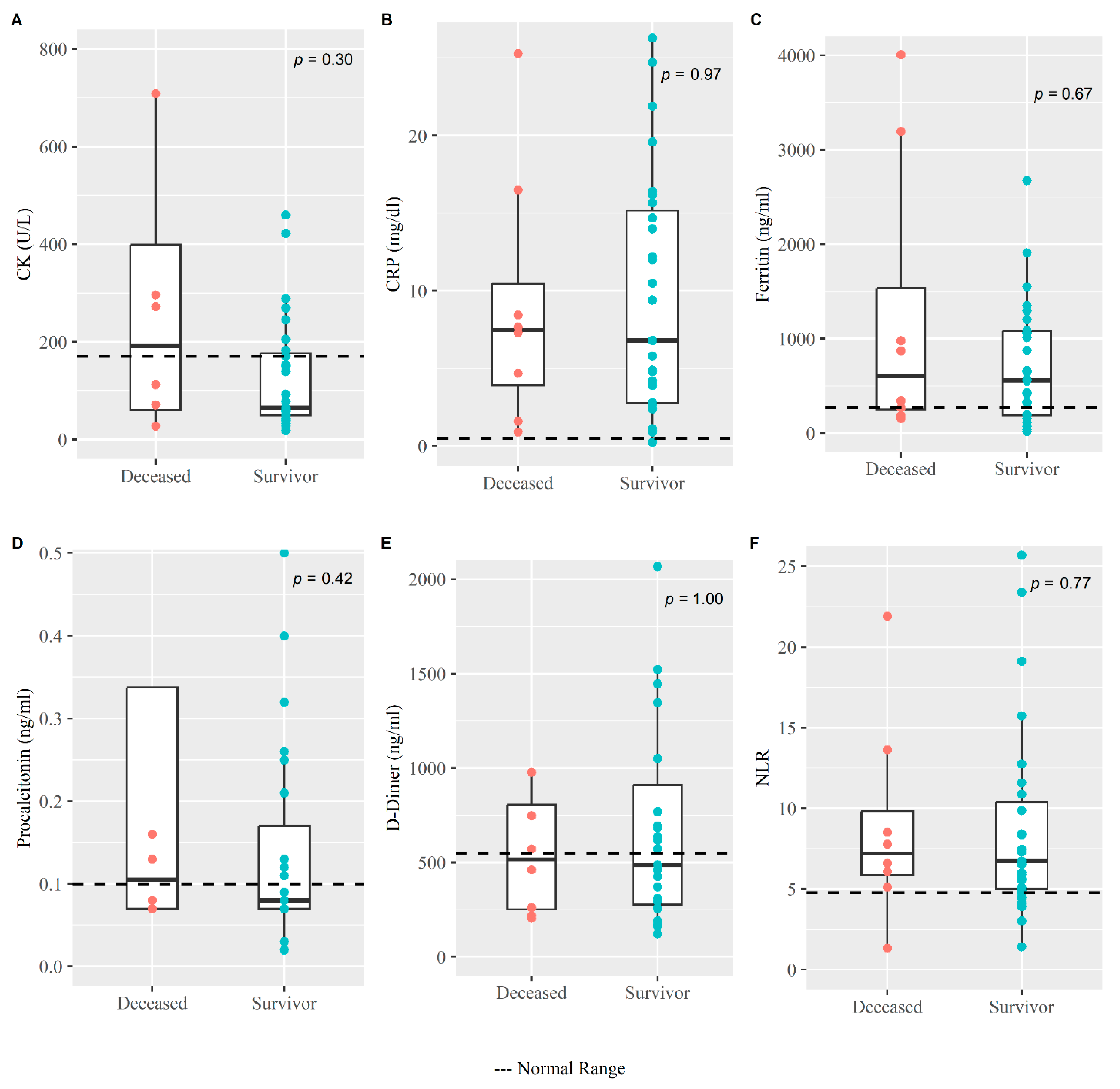
| CK (U/L) <171 | 77.03 (50.05, 225.65) | 65.42 (50.05, 176.87) | 192.54 (60.33, 399.1) | 0.298 |
| CRP (mg/dL) <0.5 | 7.30 (2.75, 15.185) | 6.80 (2.75, 15.19) | 7.485 (3.91, 10.46) | 0.969 |
| Ferritin (ng/mL) ≤274.66 | 561.9 (190.8, 1082.8) | 561.9 (190, 1082.8) | 608.6 (251, 1534) | 0.670 |
| PCT (ng/mL) <0.1 | 0.08 (0.07, 0.185) | 0.08 (0.07, 0.17) | 0.105 (0.07, 0.338) | 0.417 |
| D-Dimer (ng/mL) <550 | 488 (260.5, 873.5) | 488 (276.5, 910) | 516.7 (251.2, 805.7) | 1.000 |
| Variables | All Patients n = 35 | Survivor (nS = 27) | Deceased (nD = 8) | p-Values a |
|---|---|---|---|---|
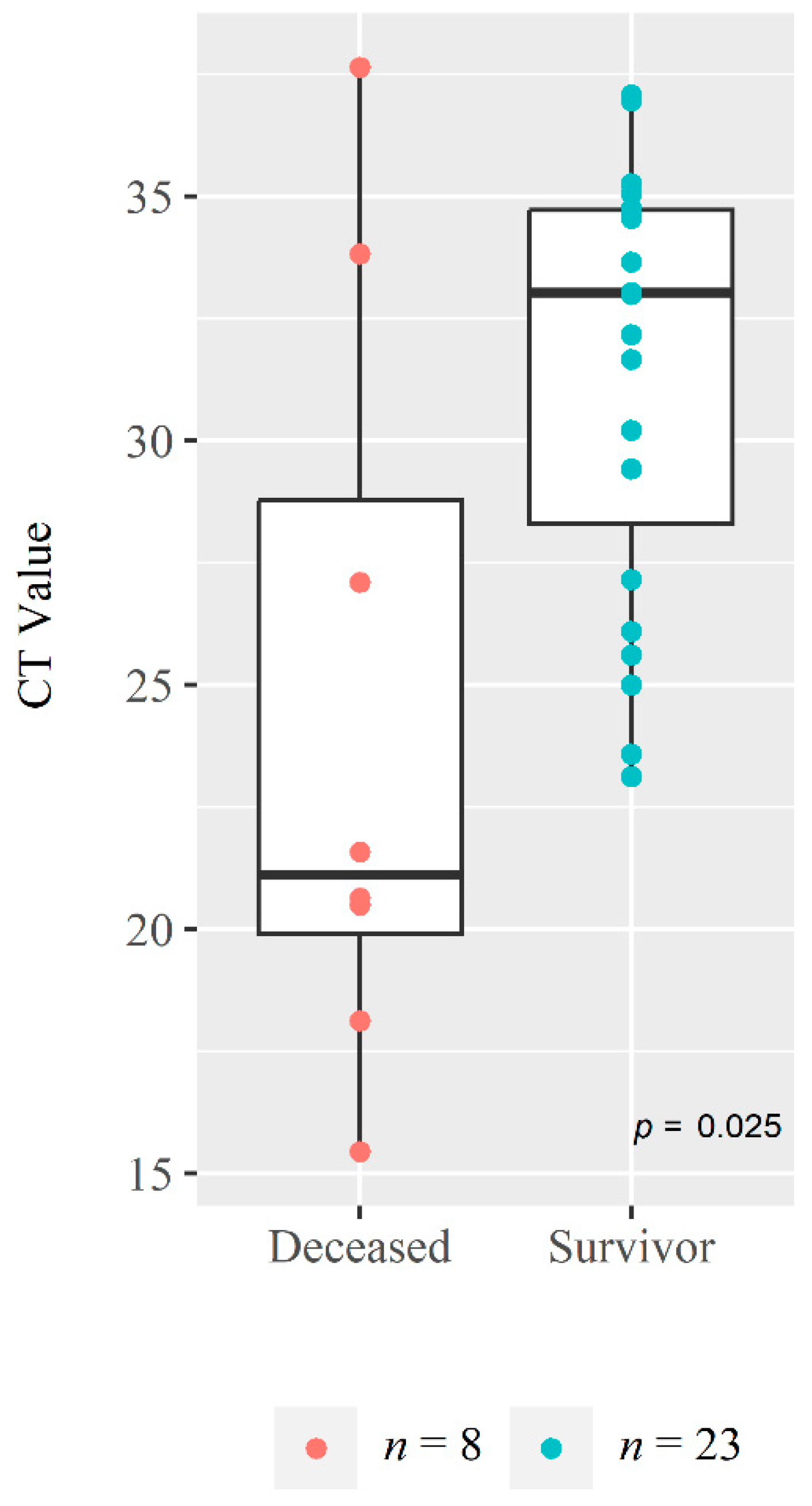
| Viral Load (n = 24), GM (95% CI) * | 10.63 × 104 (25.62 × 103, 44.08 × 104) | 59.77 × 103 (15.40 × 103, 23.19 × 104) | 94.64 × 104 (27.58 × 102, 32.47 × 107) | 0.191 |
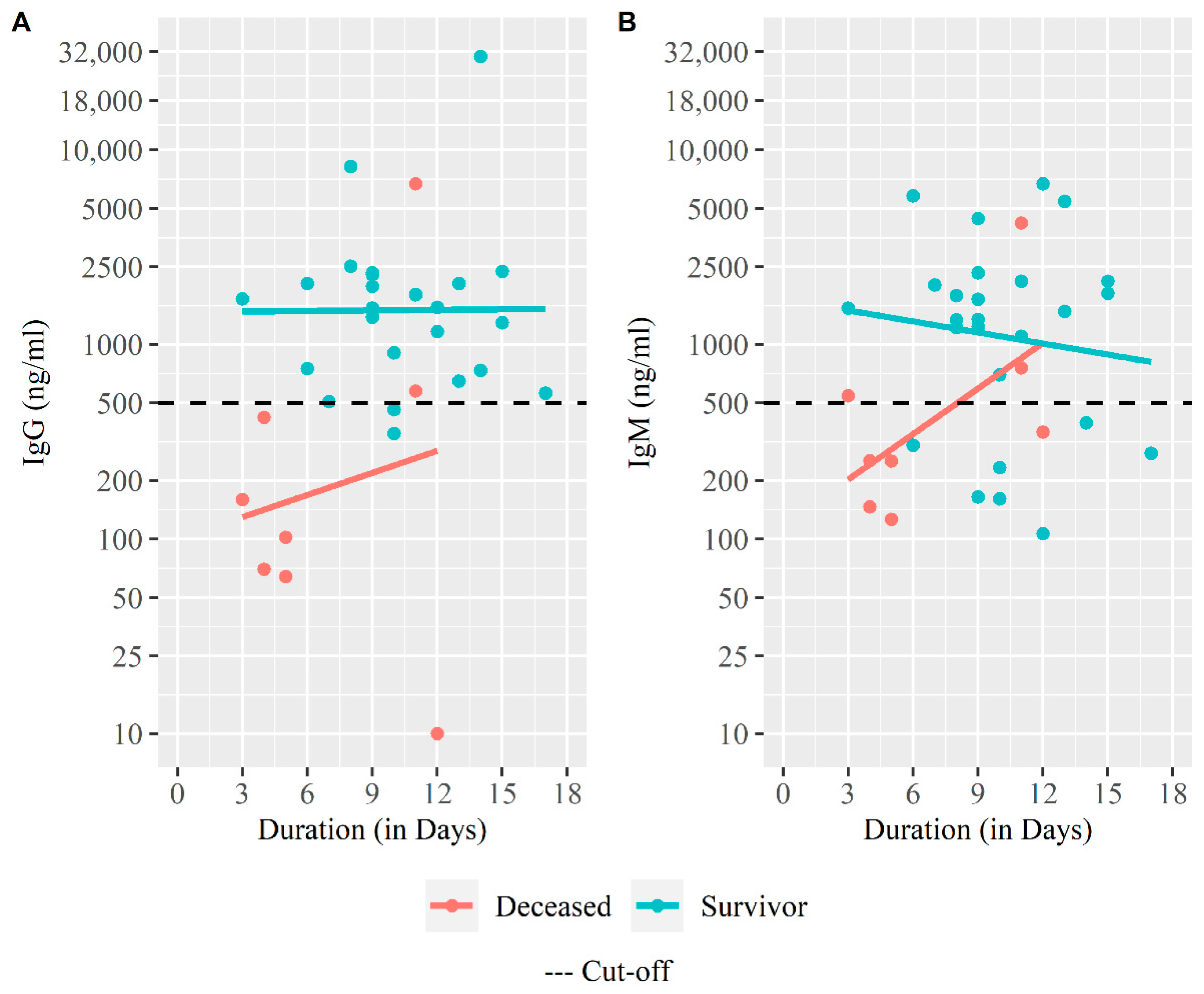
| IgG (ng/mL) | ||||
| ≥500, n (%) | 27 (77.14%) | 25 (92.59%) | 2 (25%) | <0.001 |
| GM (95% CI) | 1013.38 (585.98, 1752.51) | 1685.59 (1101.55, 2579.29) | 181.96 (36.55, 905.84) | 0.013 |
| IgM (ng/mL) | ||||
| ≥500, n (%) | 23 (65.71%) | 20 (74.07%) | 3 (37.5%) | 0.056 |
| GM (95% CI) | 999.58 (607.79, 1643.93) | 1305.58 (737.98, 2309.72) | 405.85 (158.78, 1037.40) | 0.030 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, T.; Akter, A.; Tauheed, I.; Akhtar, M.; Rahman, S.I.A.; Khaton, F.; Ahmmed, F.; Firoj, M.G.; Ferdous, J.; Afrad, M.H.; et al. The Fatal Clinical Outcome of Severe COVID-19 in Hospitalized Patients: Findings from a Prospective Cohort Study in Dhaka, Bangladesh. Medicina 2023, 59, 1280. https://doi.org/10.3390/medicina59071280
Ahmed T, Akter A, Tauheed I, Akhtar M, Rahman SIA, Khaton F, Ahmmed F, Firoj MG, Ferdous J, Afrad MH, et al. The Fatal Clinical Outcome of Severe COVID-19 in Hospitalized Patients: Findings from a Prospective Cohort Study in Dhaka, Bangladesh. Medicina. 2023; 59(7):1280. https://doi.org/10.3390/medicina59071280
Chicago/Turabian StyleAhmed, Tasnuva, Afroza Akter, Imam Tauheed, Marjahan Akhtar, Sadia Isfat Ara Rahman, Fatema Khaton, Faisal Ahmmed, Md. Golam Firoj, Jannatul Ferdous, Mokibul Hassan Afrad, and et al. 2023. "The Fatal Clinical Outcome of Severe COVID-19 in Hospitalized Patients: Findings from a Prospective Cohort Study in Dhaka, Bangladesh" Medicina 59, no. 7: 1280. https://doi.org/10.3390/medicina59071280






