MicroRNAs as a Potential Biomarker in the Diagnosis of Cardiovascular Diseases
Abstract
1. Introduction
2. Cardiovascular Diseases (CVD)
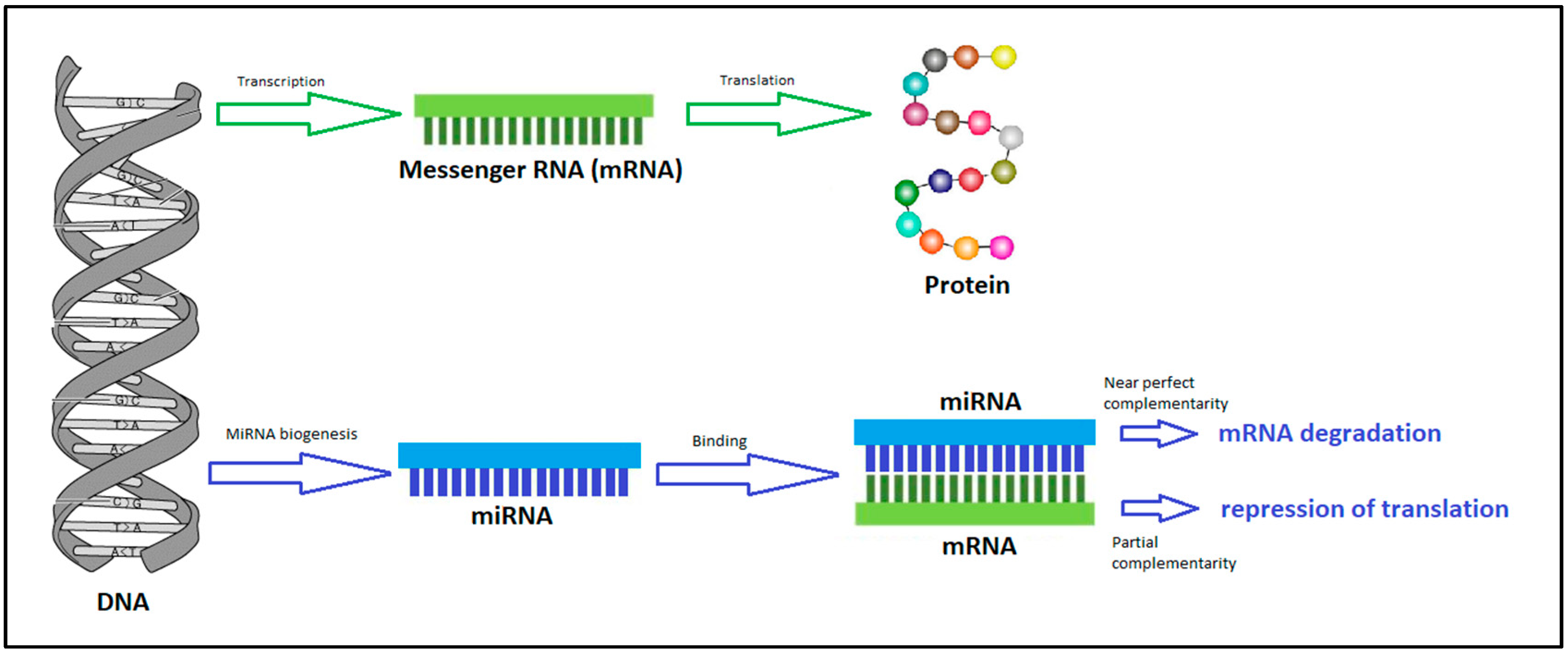
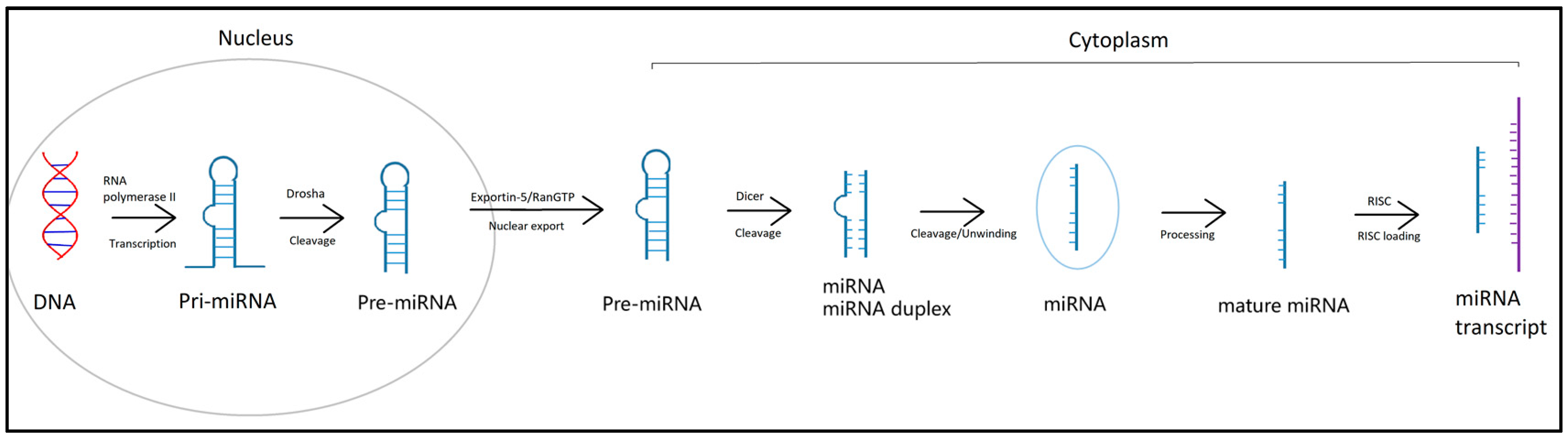
3. MicroRNA
MicroRNA as a Potential Biomarker
4. Changes at The MicroRNA Level in Cardiovascular Diseases
4.1. Atherosclerosis and Coronary Heart Diseases (CHD)
4.2. Acute Coronary Syndrome—Myocardial Infarction (MI) and Unstable Angina Pectoris (AP)
4.3. Heart Failure (HF)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. “Cardiovascular Diseases”. 2023. Available online: https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1 (accessed on 30 June 2023).
- World Health Organization. “The Top 10 Causes of Death”. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 30 June 2023).
- Visseren, F.; Mach, F.; Smulders, Y.; Carballo, D.; Koskinas, K.; Bäck, M.; Benetos, A.; Biffi, A.; Boavida, J.; Capodanno, D.; et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice. Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar] [CrossRef] [PubMed]
- Cífková, R.; Vaverková, H.; Filipovský, J.; Aschermann, M. Summary of the European Guidelines on cardiovascular disease prevention in clinical practice (version 2012): Prepared by the Czech Society of Cardiology: Prepared by the Czech Society of Cardiology. Cor et Vasa 2014, 56, e169–e189. [Google Scholar] [CrossRef]
- Çakmak, H.; Demir, M. Microrna and cardiovascular diseases. Balk. Med. J. 2020, 37, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Rochette, L. Emerging New Biomarkers for Cardiovascular Disease. Int. J. Mol. Sci. 2022, 23, 3274. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Grant, B. The safe-neighborhood hypothesis of junk DNA. J. Theor. Biol. 1981, 90, 149–150. [Google Scholar] [CrossRef]
- Bartel, D. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lytle, J.; Yario, T.; Steitz, J. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Batkai, S.; Dangwal, S.; Gupta, S.; Foinquinos, A.; Holzmann, A.; Just, A.; Remke, J.; Zimmer, K.; Zeug, A.; et al. Cardiac fibroblast–derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Investig. 2014, 124, 2136–2146. [Google Scholar] [CrossRef]
- Colpaert, R.; Calore, M. Epigenetics and microRNAs in cardiovascular diseases. Genomics 2021, 113, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D. MicroRNAs: Target Recognition and Regulatory Functions: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Siasos, G.; Bletsa, E.; Stampouloglou, P.; Oikonomou, E.; Tsigkou, V.; Paschou, S.; Vlasis, K.; Marinos, G.; Vavuranakis, M.; Stefanadis, C.; et al. MicroRNAs in cardiovascular disease. Hell. J. Cardiol. 2020, 61, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Szelenberger, R.; Kacprzak, M.; Saluk-Bijak, J.; Zielinska, M.; Bijak, M. Plasma MicroRNA as a novel diagnostic. Clin. Chim. Acta 2019, 499, 98–107. [Google Scholar] [CrossRef]
- Schulte, C.; Karakas, M.; Zeller, T. microRNAs in cardiovascular disease—Clinical application. Clin. Chem. Lab. Med. 2017, 55, 687–704. [Google Scholar] [CrossRef]
- Fazmin, I.; Achercouk, Z.; Edling, C.; Said, A.; Jeevaratnam, K. Circulating microRNA as a Biomarker for Coronary Artery Disease. Biomolecules 2020, 10, 1354. [Google Scholar] [CrossRef]
- Cavarretta, E.; Frati, G. MicroRNAs in Coronary Heart Disease: Ready to Enter the Clinical Arena? BioMed Res. Int. 2016, 2016, 2150763. [Google Scholar] [CrossRef]
- Jensen, R.; Hjortbak, M.; Bøtker, H. Ischemic Heart Disease: An Update. Semin. Nucl. Med. 2020, 50, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Churov, A.; Summerhill, V.; Grechko, A.; Orekhova, V.; Orekhov, A. MicroRNAs as Potential Biomarkers in Atherosclerosis. Int. J. Mol. Sci. 2019, 20, 5547. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.D.B. Coronary Heart Disease: From Diagnosis to Treatment; Addicus Books, Inc.: Omaha, NE, USA, 2019; ISBN 978-1-943886-85-2. [Google Scholar]
- Fichtlscherer, S.; Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.; Röxe, T.; Müller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Conroy, R.; Pyörälä, K.; Fitzgerald, A.; Sans, S.; Menotti, A.; Backer, G.; Bacquer, D.; Ducimetière, P.; Jousilahti, P.; Keil, U.; et al. Estimation of ten-year risk of fatal cardiovascular disease in Europe: The SCORE project: The SCORE project. Eur. Heart J. 2003, 24, 987–1003. [Google Scholar] [CrossRef] [PubMed]
- Cifkova, R.; Byma, S.; Ceska, R.; Horky, K.; Karen, I.; Kunesova, M.; Kralikova, E.; Rosolova, H.; Roztocil, K.; Soska, V.; et al. Prevence kardiovaskularnich onemocneni v dospelem veku. Spolecne doporuceni ceskych odbornych spolecnosti. Vnitr. Lek. 2005, 51, 1021–1036. [Google Scholar]
- Najafi-Shoushtari, S.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.; Gerszten, R.; Näär, A. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef]
- Allen, R.; Marquart, T.; Albert, C.; Suchy, F.; Wang, D.; Ananthanarayanan, M.; Ford, D.; Baldán, Á. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol. Med. 2012, 4, 882–895. [Google Scholar] [CrossRef]
- Vickers, K.; Shoucri, B.; Levin, M.; Wu, H.; Pearson, D.; Osei-Hwedieh, D.; Collins, F.; Remaley, A.; Sethupathy, P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 2013, 57, 533–542. [Google Scholar] [CrossRef]
- Goedeke, L.; Rotllan, N.; Canfrán-Duque, A.; Aranda, J.; Ramírez, C.; Araldi, E.; Lin, C.; Anderson, N.; Wagschal, A.; de Cabo, R.; et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015, 21, 1280–1289. [Google Scholar] [CrossRef]
- Vickers, K.; Landstreet, S.; Levin, M.; Shoucri, B.; Toth, C.; Taylor, R.; Palmisano, B.; Tabet, F.; Cui, H.; Rye, K.; et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef]
- Rayner, K.; Sheedy, F.; Esau, C.; Hussain, F.; Temel, R.; Parathath, S.; van Gils, J.; Rayner, A.; Chang, A.; Suarez, Y.; et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 2011, 121, 2921–2931. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, X.; Liu, Y.; Meng, Z.; Wang, D.; Yang, F.; Shi, Q. Plasma MicroRNA-126-5p is Associated with the Complexity and Severity of Coronary Artery Disease in Patients with Stable Angina Pectoris. Cell. Physiol. Biochem. 2016, 39, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, T.; Lo, H.; Huang, P.; Lin, C.; Chang, S.; Liao, K.; Tsai, C.; Chan, C.; Tsai, C.; et al. Deficiency of the MicroRNA-31-MicroRNA-720 pathway in the plasma and endothelial progenitor cells from patients with coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shao, G.; Chen, X.; Yang, X.; Huang, X.; Peng, P.; Ba, Y.; Zhang, L.; Jehangir, T.; Bu, S.; et al. miRNA 206 and miRNA 574-5p are highly expression in coronary artery disease. Biosci. Rep. 2016, 36, 295. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.; Xia, K.; Li, F.; Deng, X.; Salma, U.; Li, T.; Deng, H.; Yang, D.; Haoyang, Z.; Yang, T.; et al. The diagnostic value of circulating microRNAs for middle-aged (40–60-year-old) coronary artery disease patients. Clinics 2015, 70, 257–263. [Google Scholar] [CrossRef]
- Han, H.; Qu, G.; Han, C.; Wang, Y.; Sun, T.; Li, F.; Wang, J.; Luo, S. MiR-34a, miR-21 and miR-23a as potential biomarkers for coronary artery disease: A pilot microarray study and confirmation in a 32 patient cohort: A pilot microarray study and confirmation in a 32 patient cohort. Exp. Mol. Med. 2015, 47, e138. [Google Scholar] [CrossRef]
- Du, Y.; Yang, S.H.; Li, S.; Cui, C.J.; Zhang, Y.; Zhu, C.G.; Guo, Y.L.; Wu, N.Q.; Gao, Y.; Sun, J.; et al. Circulating MicroRNAs as Novel Diagnostic Biomarkers for Very Early-onset (≤40 years) Coronary Artery Disease. Biomed. Environ. Sci. 2016, 29, 545–554. [Google Scholar] [CrossRef]
- Wang, F.; Long, G.; Zhao, C.; Li, H.; Chaugai, S.; Wang, Y.; Chen, C.; Wang, D. Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J. Transl. Med. 2013, 11, 222. [Google Scholar] [CrossRef]
- Faccini, J.; Ruidavets, J.; Cordelier, P.; Martins, F.; Maoret, J.; Bongard, V.; Ferrières, J.; Roncalli, J.; Elbaz, M.; Vindis, C. Circulating MIR-155, MIR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci. Rep. 2017, 7, srep42916. [Google Scholar] [CrossRef]
- Ibanez, B.; James, S.; Agewall, S.; Antunes, M.; Bucciarelli-Ducci, C.; Bueno, H.; Caforio, A.; Crea, F.; Goudevenos, J.; Halvorsen, S.; et al. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation. Eur. Heart J. 2018, 39, 119–177. [Google Scholar] [CrossRef]
- Panteghini, P. Recommendations on Use of Biochemical Markers in Acute Coronary Syndrome: IFCC Proposals: IFCC Proposals. EJIFCC 2003, 14, 104. [Google Scholar] [PubMed]
- Dekker, M.; Mosterd, A.; Van’t Hof, A.; Hoes, A. Novel biochemical markers in suspected acute coronary syndrome: Systematic review and critical appraisal: Systematic review and critical appraisal. Heart 2010, 96, 1001. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.; Khan, M. Cardiac Biomarkers: What Is and What Can Be. Indian J. Cardiovasc. Dis. Women WINCARS 2019, 3, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.; Orekhov, A.; Bobryshev, Y. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J. Mol. Cell. Cardiol. 2016, 94, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhu, J.; Zhang, J.; Li, Q.; Li, Y.; He, J.; Qin, Y.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666. [Google Scholar] [CrossRef]
- Navickas, R.; Gal, D.; Laucevičius, A.; Taparauskaite, A.; Zdanyte, M.; Holvoet, P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: A systematic review: A systematic review. Cardiovasc. Res. 2016, 111, 322–337. [Google Scholar] [CrossRef]
- Li, S.; Lee, C.; Song, J.; Lu, C.; Liu, J.; Cui, Y.; Liang, H.; Cao, C.; Zhang, F.; Chen, H. Circulating microRNAs as potential biomarkers for coronary plaque rupture. Oncotarget 2017, 8, 48145–48156. [Google Scholar] [CrossRef]
- Zhong, J.; He, Y.; Chen, W.; Shui, X.; Chen, C.; Lei, W. Circulating microRNA-19a as a potential novel biomarker for diagnosis of acute myocardial infarction. Int. J. Mol. Sci. 2014, 15, 20355–20364. [Google Scholar] [CrossRef]
- Zeller, T.; Tanja, T.; Ojeda, F.; Reichlin, T.; Twerenbold, R.; Tzikas, S.; Wild, P.; Reiter, M.; Czyz, E.; Lackner, K.; et al. Assessment of microRNAs in patients with unstable angina pectoris. Eur. Heart J. 2014, 35, 2106–2114. [Google Scholar] [CrossRef]
- Gidlöf, O.; Andersson, P.; Van Der Pals, J.; Götberg, M.; Erlinge, D. Cardiospecific microRNA Plasma Levels Correlate with Troponin and Cardiac Function in Patients with ST Elevation Myocardial Infarction, Are Selectively Dependent on Renal Elimination, and Can Be Detected in Urine Samples. Cardiology 2011, 118, 217–226. [Google Scholar] [CrossRef]
- Oerlemans, M.; Mosterd, A.; Dekker, M.; de Vrey, E.; van Mil, A.; Pasterkamp, G.; Doevendans, P.; Hoes, A.; Sluijter, J. Early assessment of acute coronary syndromes in the emergency department: The potential diagnostic value of circulating microRNAs: The potential diagnostic value of circulating microRNAs. EMBO Mol. Med. 2012, 4, 1176. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Mueller, M.; Haaf, P.; Goretti, E.; Twerenbold, R.; Zangrando, J.; Vausort, M.; Reichlin, T.; Wildi, K.; Moehring, B.; et al. Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J. Intern. Med. 2015, 277, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Widera, C.; Gupta, S.; Lorenzen, J.; Bang, C.; Bauersachs, J.; Bethmann, K.; Kempf, T.; Wollert, K.; Thum, T. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J. Mol. Cell. Cardiol. 2011, 51, 872–875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, X.; Su, T.; Li, H.; Huang, Q.; Wu, D.; Yang, C.; Han, Z. Circulating miR-499 are novel and sensitive biomarker of acute myocardial infarction. J. Thorac. Dis. 2015, 7, 303. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Wagner, D.; Staessen, J.; Heymans, S.; Schroen, B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 2010, 3, 499–506. [Google Scholar] [CrossRef]
- Gacon, J.; Kablak-Ziembicka, A.; Stepien, E.; Enguita, F.; Karch, I.; Derlaga, B.; Zmudka, K.; Przewlocki, T. Decision-making microRNAs (miR-124, -133a/b, -34a and -134) in patients with occluded target vessel in acute coronary syndrome. Kardiol. Pol. 2016, 74, 280–288. [Google Scholar] [CrossRef]
- Gao, H.; Guddeti, R.; Matsuzawa, Y.; Liu, L.; Su, L.; Guo, D.; Nie, S.; Du, J.; Zhang, M. Plasma Levels of microRNA-145 Are Associated with Severity of Coronary Artery Disease. PLoS ONE 2015, 10, e0123477. [Google Scholar] [CrossRef]
- Matsumoto, S.; Sakata, Y.; Nakatani, D.; Suna, S.; Mizuno, H.; Shimizu, M.; Usami, M.; Sasaki, T.; Sato, H.; Kawahara, Y.; et al. A subset of circulating microRNAs are predictive for cardiac death after discharge for acute myocardial infarction. Biochem. Biophys. Res. Commun. 2012, 427, 280–284. [Google Scholar] [CrossRef]
- Devaux, Y.; Vausort, M.; McCann, G.; Kelly, D.; Collignon, O.; Ng, L.; Wagner, D.; Squire, I. A Panel of 4 microRNAs Facilitates the Prediction of Left Ventricular Contractility after Acute Myocardial Infarction. PLoS ONE 2013, 8, e70644. [Google Scholar] [CrossRef]
- Bye, A.; Røsjø, H.; Nauman, J.; Silva, G.; Follestad, T.; Omland, T.; Wisløff, U. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals—The HUNT study. J. Mol. Cell. Cardiol. 2016, 97, 162–168. [Google Scholar] [CrossRef]
- Karakas, M.; Schulte, C.; Appelbaum, S.; Ojeda, F.; Lackner, K.; Münzel, T.; Schnabel, R.; Blankenberg, S.; Zeller, T. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease—Results from the large AtheroGene study. Eur. Heart J. 2017, 38, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Willeit, P.; Tilling, L.; Drozdov, I.; Prokopi, M.; Renard, J.; Mayr, A.; Weger, S.; Schett, G.; Shah, A.; et al. Prospective Study on Circulating MicroRNAs and Risk of Myocardial Infarction. J. Am. Coll. Cardiol. 2012, 60, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Molz, S.; Appelbaum, S.; Karakas, M.; Ojeda, F.; Lau, D.; Hartmann, T.; Lackner, K.; Westermann, D.; Schnabel, R.; et al. miRNA-197 and miRNA-223 Predict Cardiovascular Death in a Cohort of Patients with Symptomatic Coronary Artery Disease. PLoS ONE 2015, 10, e0145930. [Google Scholar] [CrossRef]
- Tsutsui, H.; Isobe, M.; Ito, H.; Ito, H.; Okumura, K.; Ono, M.; Kitakaze, M.; Kinugawa, K.; Kihara, Y.; Goto, Y.; et al. JCS 2017/JHFS 2017 Guideline on Diagnosis and Treatment of Acute and Chronic Heart Failure—Digest Version. Circ. J. 2019, 83, 2084–2184. [Google Scholar] [CrossRef]
- McDonagh, T.; Metra, M.; Adamo, M.; Gardner, R.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.; Wang, J.; Liew, O.; Richards, A.; Chen, Y. MicroRNA and Heart Failure. Int. J. Mol. Sci. 2016, 17, 502. [Google Scholar] [CrossRef]
- Duan, Q.; Chen, C.; Yang, L.; Li, N.; Gong, W.; Li, S.; Wang, D. MicroRNA regulation of unfolded protein response transcription factor XBP1 in the progression of cardiac hypertrophy and heart failure in vivo. J. Transl. Med. 2015, 13, 363. [Google Scholar] [CrossRef] [PubMed]
- Da Costa Martins, P.; De Windt, L. MicroRNAs in control of cardiac hypertrophy. Cardiovasc. Res. 2012, 93, 563–572. [Google Scholar] [CrossRef]
- Pan, Z.; Sun, X.; Ren, J.; Li, X.; Gao, X.; Lu, C.; Zhang, Y.; Sun, H.; Wang, Y.; Wang, H.; et al. miR-1 Exacerbates Cardiac Ischemia-Reperfusion Injury in Mouse Models. PLoS ONE 2012, 7, e50515. [Google Scholar] [CrossRef]
- Pan, W.; Zhong, Y.; Cheng, C.; Liu, B.; Wang, L.; Li, A.; Xiong, L.; Liu, S. MiR-30-Regulated Autophagy Mediates Angiotensin II-Induced Myocardial Hypertrophy. PLoS ONE 2013, 8, e53950. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, Y.; Wang, Y.; Li, J.; Schiller, P.; Peng, T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc. Res. 2011, 92, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jia, Z.; Zhang, C.; Sun, M.; Wang, W.; Chen, P.; Ma, K.; Zhang, Y.; Li, X.; Zhou, C. miR-499 protects cardiomyocytes from H2O2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol. 2014, 11, 339. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, M.; Li, C.; Zhou, J.; Li, H.; Zhu, D.; Wang, Z.; Chen, A.; Zhao, Q. MicroRNA-92a Inhibition Attenuates Hypoxia/Reoxygenation-Induced Myocardiocyte Apoptosis by Targeting Smad7. PLoS ONE 2014, 9, e100298. [Google Scholar] [CrossRef] [PubMed]
- Sayed, D.; He, M.; Hong, C.; Gao, S.; Rane, S.; Yang, Z.; Abdellatif, M. MicroRNA-21 Is a Downstream Effector of AKT That Mediates Its Antiapoptotic Effects via Suppression of Fas Ligand. J. Biol. Chem. 2010, 285, 20281. [Google Scholar] [CrossRef]
- Tijsen, A.; Creemers, E.; Moerland, P.; Windt, L.; Wal, A.; Kok, W.; Pinto, Y. MiR423-5p as a circulating biomarker for heart failure. Circ. Res. 2010, 106, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Goren, Y.; Kushnir, M.; Zafrir, B.; Tabak, S.; Lewis, B.; Amir, O. Serum levels of microRNAs in patients with heart failure. Eur. J. Heart Fail. 2012, 14, 147–154. [Google Scholar] [CrossRef]
- Tutarel, O.; Dangwal, S.; Bretthauer, J.; Westhoff-Bleck, M.; Roentgen, P.; Anker, S.; Bauersachs, J.; Thum, T. Circulating miR-423-5p fails as a biomarker for systemic ventricular function in adults after atrial repair for transposition of the great arteries. Int. J. Cardiol. 2013, 167, 63–66. [Google Scholar] [CrossRef]
- Scrutinio, D.; Conserva, F.; Passantino, A.; Iacoviello, M.; Lagioia, R.; Gesualdo, L. Circulating microRNA-150-5p as a novel biomarker for advanced heart failure: A genome-wide prospective study: A genome-wide prospective study. J. Heart Lung Transplant. 2017, 36, 616–624. [Google Scholar] [CrossRef]
- Wong, L.; Zou, R.; Zhou, L.; Lim, J.; Phua, D.; Liu, C.; Chong, J.; Ng, J.; Liew, O.; Chan, S.; et al. Combining Circulating MicroRNA and NT-proBNP to Detect and Categorize Heart Failure Subtypes. J. Am. Coll. Cardiol. 2019, 73, 1300–1313. [Google Scholar] [CrossRef]
- Zhang, J.; Xing, Q.; Zhou, X.; Li, J.; Li, Y.; Zhang, L.; Zhou, Q.; Tang, B. Circulating miRNA-21 is a promising biomarker for heart failure. Mol. Med. Rep. 2017, 16, 7766–7774. [Google Scholar] [CrossRef]
- Masson, S.; Batkai, S.; Beermann, J.; Bär, C.; Pfanne, A.; Thum, S.; Magnoli, M.; Balconi, G.; Nicolosi, G.; Tavazzi, L.; et al. Circulating microRNA-132 levels improve risk prediction for heart failure hospitalization in patients with chronic heart failure. Eur. J. Heart Fail. 2018, 20, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Seronde, M.; Vausort, M.; Gayat, E.; Goretti, E.; Ng, L.; Squire, I.; Vodovar, N.; Sadoune, M.; Samuel, J.; Thum, T.; et al. Circulating microRNAs and outcome in patients with acute heart failure. PLoS ONE 2015, 10, e0142237. [Google Scholar] [CrossRef] [PubMed]
- Van Boven, N.; Akkerhuis, K.; Anroedh, S.; Rizopoulos, D.; Pinto, Y.; Battes, L.; Hillege, H.; Caliskan, K.; Germans, T.; Manintveld, O.; et al. Serially measured circulating miR-22-3p is a biomarker for adverse clinical outcome in patients with chronic heart failure: The Bio-SHiFT study: The Bio-SHiFT study. Int. J. Cardiol. 2017, 235, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, H.; Coskunpinar, E.; Ikitimur, B.; Barman, H.; Karadag, B.; Tiryakioglu, N.; Kahraman, K.; Vural, V. The prognostic value of circulating microRNAs in heart failure. J. Cardiovasc. Med. 2015, 16, 431–437. [Google Scholar] [CrossRef]
- Duong, J.; Huyen, V.; Tible, M.; Gay, A.; Guillemain, R.; Aubert, O.; Varnous, S.; Iserin, F.; Rouvier, P.; François, A.; et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur. Heart J. 2014, 35, 3194–3202. [Google Scholar] [CrossRef] [PubMed]
- Sukma Dewi, I.; Torngren, K.; Gidlöf, O.; Kornhall, B.; Öhman, J. Altered serum miRNA profiles during acute rejection after heart transplantation: Potential for non-invasive allograft surveillance: Potential for non-invasive allograft surveillance. J. Heart Lung Transplant. 2013, 32, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, A.; Počivavšek, K.; Petrovič, D.; Peterlin, B. The Role of microRNAs in Heart Failure: A Systematic Review: A Systematic Review. Front. Cardiovasc. Med. 2020, 7, 161. [Google Scholar] [CrossRef]
| MiRNA | Diseases | Pathophysiological Significance | Ref. |
|---|---|---|---|
| miR-33 | Atherosclerosis | Uptake of cholesterol by HDL particles Transport of cholesterol through hepatocytes into the bile ducts | [29,30,34] |
| miR-27b | Atherosclerosis | Formation of lipoproteins | [31] |
| miR-148 | Atherosclerosis | LDL lipoprotein uptake | [32] |
| miR-223 | Atherosclerosis | LDL lipoprotein uptake by the scavenger receptor | [33] |
| miR-126, miR-126-5p | IHD, atherosclerosis | Regulation of endothelial cell function and enhancement of endothelial regeneration capacity Lower plasma miR-126-5p levels are associated with atherosclerotic plaque formation | [35] |
| MiRNA | Diagnostic Potential | MiRNA Alteration | Ref. |
|---|---|---|---|
| miR-17 | IHD biomarker | ↓ | [26] |
| miR-92a | IHD biomarker | ↓ | [26] |
| miR-126 | IHD biomarker | ↓ | [26] |
| miR-145 | IHD biomarker | ↓ | [26] |
| miR-155 | IHD biomarker | ↓ | [26] |
| miR-133, miR-208a | IHD biomarker | ↑ | [26] |
| panel miR-155, miR-145 and let-7c | IHD biomarker | ↓ | [26] |
| miR-31, miR-720a | Biomarker of the early phase of IHD | ↓ | [36] |
| miR-206, miR-574-5p | Biomarker of the early phase of IHD | ↑ | [37] |
| miR-149 miR-765, miR-424 | IHD biomarker-differentiating patients with stable and unstable AP from those without the disease | ↑ ↓ | [38] |
| miR-21, miR-23a, miR-34a | Biomarker of IHD and disease progression | ↑ | [39] |
| miR-145-3p, miR-190a-5p miR-196-5p, miR-3163-3p | Biomarker of the early phase of IHD | ↓ | [40] |
| miR-133a | Prediction of the presence and severity of coronary lesions in patients with IHD Biomarker AMI | ↑ | [41] |
| MiRNA | Diagnostic Potential | MiRNA Alteration | Ref. |
|---|---|---|---|
| miR-499 | Diagnostic or prognostic significance for all CVD Biomarker AMI Distinguishing STEMI and non-STEMI AMI | ↑ ↑ STEMI | [47,48,49,54,55,58,59] |
| miR-208a/b | Diagnostic or prognostic significance for all CVD Biomarker AMI Differentiation between STEMI and non-STEMI AMI Biomarker AP and differentiation between AMI and AP Prognostic marker of long-term complications after AMI | ↑ ↑ STEMI | [47,48,49,54,55,56,59] |
| miR-1 | Biomarker AMI Biomarker AP-differentiation of AMI and AP | ↑ | [47,48,49,54,55,56] |
| miR-133a/b | Diagnostic or prognostic significance for all CVD Biomarker of AMI and AP Distinguishing AMI and AP Distinguishing STEMI and non-STEMI AMI | ↑ ↑ STEMI | [47,48,49,55,56] |
| miR-145 | Biomarker of STEMI and worse AMI Distinguishing STEMI and non-STEMI AMI | ↓ ↓ STEMI | [49,60] |
| miR-451 | Recognition of plaque rupture Recognition of STEMI and non-STEMI AMI | ↓ ↑ STEMI | [50,55] |
| miR-155, miR-155-5p | Prognostic impact-risk of cardiac death after AMI Recognition of plaque rupture | ↑ | [50,61] |
| miR-483-5p | Recognition of plaque rupture | ↑ | [50] |
| miR-19a | Biomarker AMI | ↑ | [51] |
| Panel: miR-132, miR-150, miR-186 | Early diagnosis of unstable angina | ↑ | [52] |
| miR-21 | Biomarker AMI | ↑ | [54] |
| miR-320a | Biomarker AMI | ↑ | [55] |
| miR-134 | Distinguishing STEMI and non-STEMI AMI | ↑ STEMI | [59] |
| miR-380 | Prognostic impact-risk of cardiac death after AMI | ↑ in patients at risk of cardiac death | [61] |
| Panel: miR-16, miR-27a miR-101, miR-150 | Prognostic impact-risk of contractility failure after AMI | ↑ ↓ | [62] |
| miR-106a-5p, miR-424-5p, let-7g-5p, miR-144-3p, miR-660-5p | Prediction of future IM | ↑ | [63] |
| miR-132, miR-140-3p, miR-210 | Predictors of cardiovascular death | ↑ | [64] |
| miR-126, miRNA-197, miRNA-223 | Prediction of future MI | ↑ | [65,66] |
| miRNA | Diagnostic Potential | MiRNA Alteration | Ref. |
|---|---|---|---|
| miR-423-5p | Diagnosis of HF Prognostic biomarker of acute HF | ↑ ↓ | [69,78,79,85] |
| miR-622 | Biomarker HF | ↑ | [69,78] |
| miR-92a/b | Diagnosis of HF and correlation with important clinical prognostic parameters Non-invasive biomarker for risk of heart transplant rejection | ↑ | [69,79,88] |
| miR-150, miR-150-5p | Biomarker of advanced HF, association with maladaptive remodelling, disease severity | ↓ | [69,81] |
| miR-21 | HF biomarker and prediction of cardiovascular mortality and risk of rehospitalization | ↑ | [69,83] |
| miR-1 | Biomarker HF | ↑ | [69] |
| miR-30a | Biomarker HF | ↑ | [69] |
| miR-126 | Biomarker HF | ↑↓ * | [69] |
| miR-195 | Biomarker HF | ↓ | [69] |
| miR-210 | Biomarker HF | ↑ | [69] |
| miR-342-3p | Biomarker HF | ↓ | [69] |
| miR-499-5p | Acute heart failure | ↑ | [69] |
| miR-18b | Biomarker HF | ↑ | [78] |
| miR-129-5p | Biomarker HF | ↑ | [78] |
| miR-675 | Biomarker HF | ↑ | [78] |
| miR-1254 | Biomarker HF | ↑ | [78] |
| miR-22, miR-22-3p | Diagnosis of HF and correlation with important clinical prognostic parameters Prediction of prognosis in chronic HF | ↑ | [79,86] |
| miR-320a | Diagnosis of HF and correlation with important clinical prognostic parameters | ↑ | [79] |
| miR-132 | Biomarker HF Predication readmission and risk of rehospitalization | ↑ ↓ | [84] |
| miR-182 | Prediction of cardiovascular mortality | ↑ | [87] |
| miR-10a | A non-invasive biomarker for the risk of heart transplant rejection | ↓ | [88] |
| miR-31 | A non-invasive biomarker for the risk of heart transplant rejection | ↑ | [88] |
| miR-155 | A non-invasive biomarker for the risk of heart transplant rejection | ↑ | [88] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kramna, D.; Riedlova, P.; Jirik, V. MicroRNAs as a Potential Biomarker in the Diagnosis of Cardiovascular Diseases. Medicina 2023, 59, 1329. https://doi.org/10.3390/medicina59071329
Kramna D, Riedlova P, Jirik V. MicroRNAs as a Potential Biomarker in the Diagnosis of Cardiovascular Diseases. Medicina. 2023; 59(7):1329. https://doi.org/10.3390/medicina59071329
Chicago/Turabian StyleKramna, Dagmar, Petra Riedlova, and Vitezslav Jirik. 2023. "MicroRNAs as a Potential Biomarker in the Diagnosis of Cardiovascular Diseases" Medicina 59, no. 7: 1329. https://doi.org/10.3390/medicina59071329
APA StyleKramna, D., Riedlova, P., & Jirik, V. (2023). MicroRNAs as a Potential Biomarker in the Diagnosis of Cardiovascular Diseases. Medicina, 59(7), 1329. https://doi.org/10.3390/medicina59071329






