In Silico Analysis of the Effect of Hydrastis canadensis on Controlling Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Target Genes
2.2. Identification of Control Drugs
2.3. Optimization of the Docking Algorithm
2.4. Identification of Potential Phytochemicals
2.5. Identification of Potential Phytochemicals
2.6. Evaluation of Ligand–Target Complex
2.6.1. Molecular Dynamic Simulation
2.6.2. MM-GBSA and MM-PBSA Analysis
3. Results
3.1. Identification of Targets
3.2. Identification of Control Drugs
3.3. Optimization of the Docking Algorithm
3.4. Identification of Phytochemicals from Hydrastis canadensis
3.5. Molecular Docking
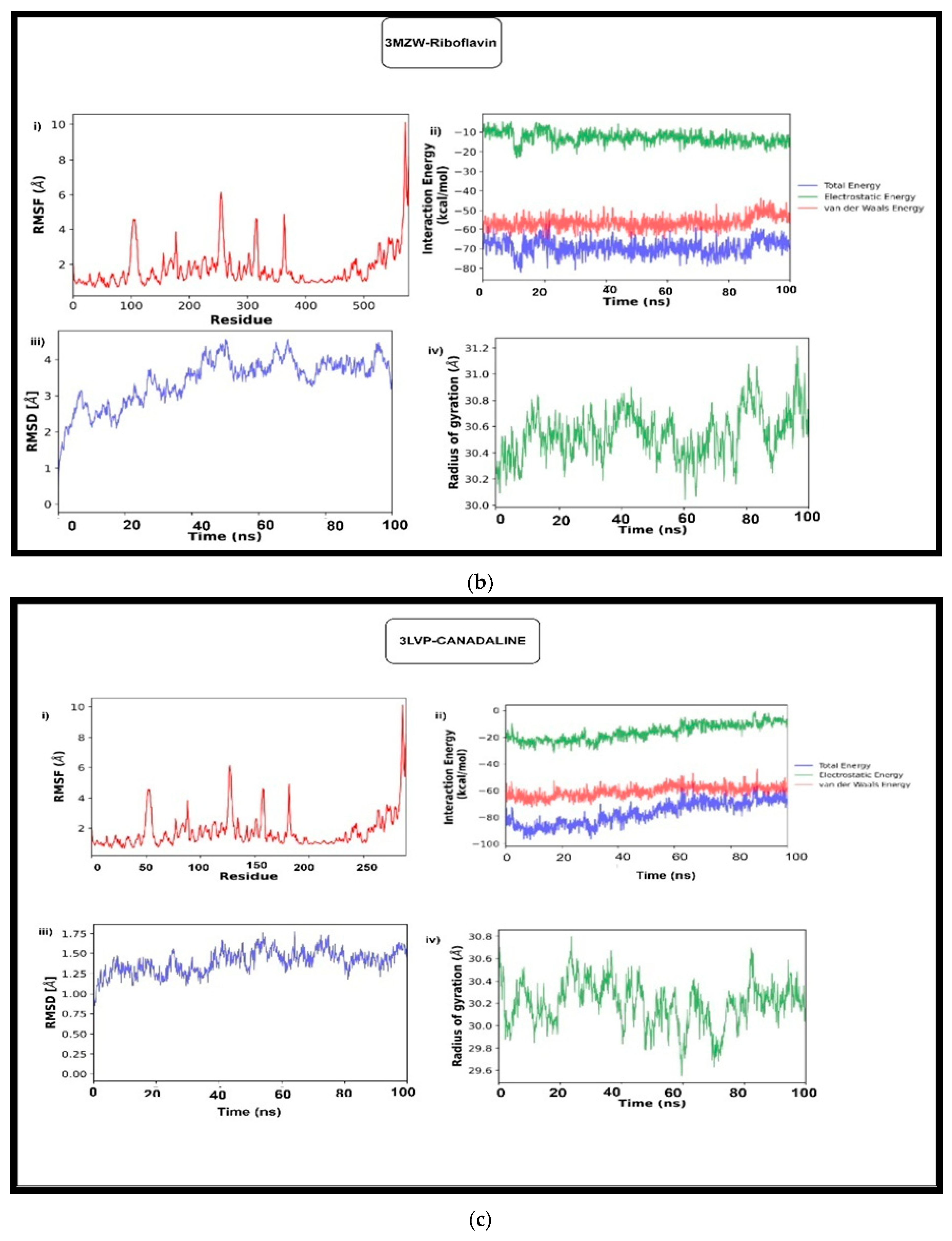
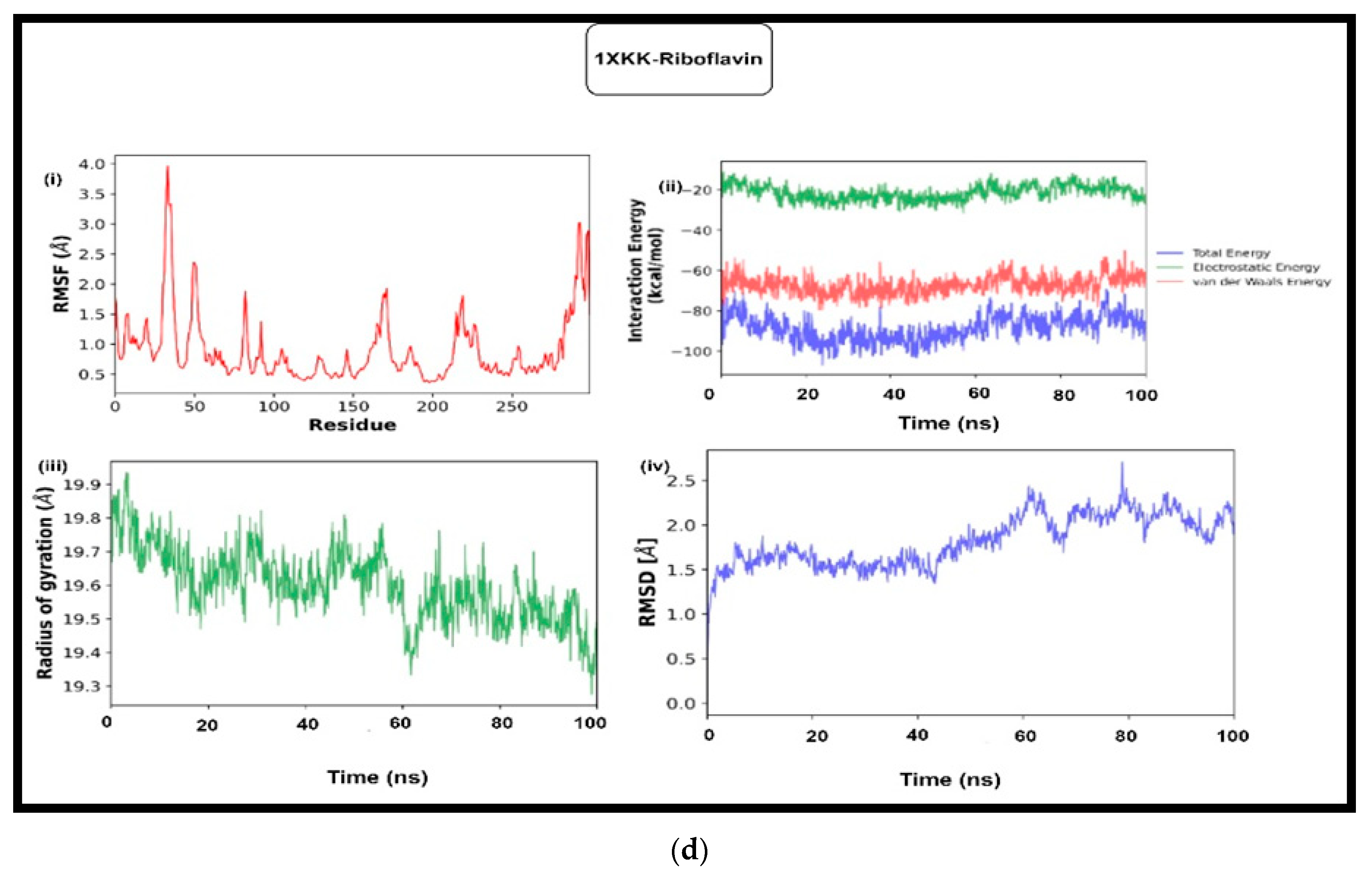
3.5.1. Molecular Dynamic Simulation
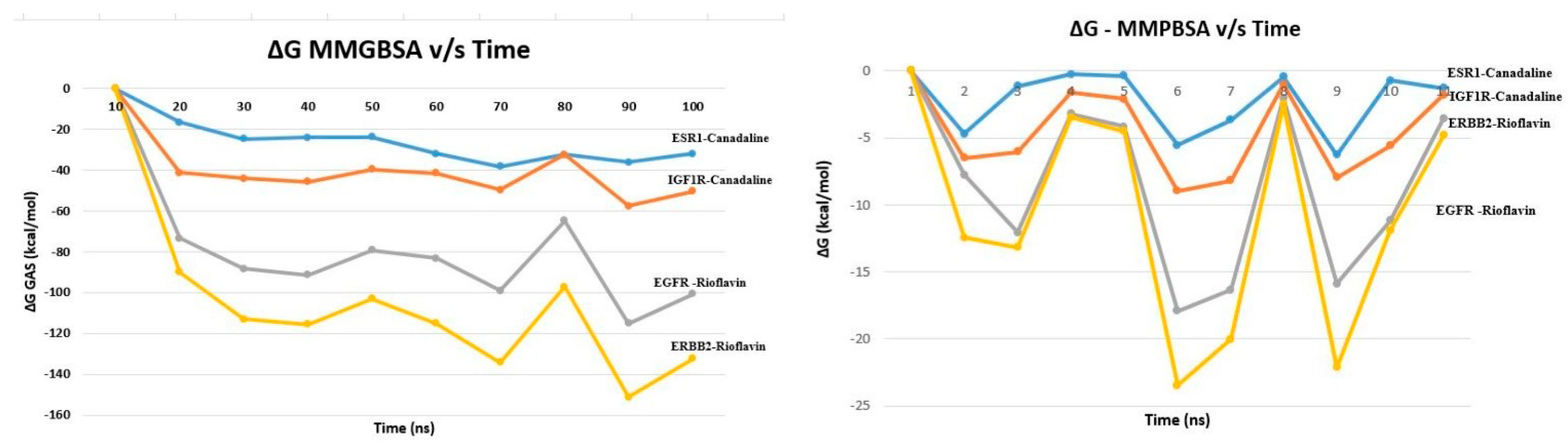
3.5.2. MM-GBSA and MM-PBSA Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alkabban, F.M.; Ferguson, T. Breast Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Beckmann, M.W.; Niederacher, D.; Schnürch, H.G.; Gusterson, B.A.; Bender, H.G. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J. Mol. Med. 1997, 75, 429–439. [Google Scholar]
- Mathur, P.; Sathishkumar, K.; Chaturvedi, M.; Das, P.; Sudarshan, K.L.; Santhappan, S.; Nallasamy, V.; John, A.; Narasimhan, S.; Roselind, F.S.; et al. Cancer Statistics, 2020: Report from National Cancer Registry Programme, India. JCO Glob. Oncol. 2020, 6, 1063–1075. [Google Scholar] [PubMed]
- Pal, S.K.; Childs, B.H.; Pegram, M. Triple negative breast cancer: Unmet medical needs. Breast Cancer Res. Treat. 2011, 125, 627–636. [Google Scholar] [PubMed] [Green Version]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar]
- He, J.; McLaughlin, R.P.; van der Noord, V.; Foekens, J.A.; Martens, J.W.M.; van Westen, G.; Zhang, Y.; van de Water, B. Multi-targeted kinase inhibition alleviates mTOR inhibitor resistance in triple-negative breast cancer. Breast Cancer Res. Treat. 2019, 178, 263–274. [Google Scholar]
- McLaughlin, R.P.; He, J.; van der Noord, V.E.; Redel, J.; Foekens, J.A.; Martens, J.W.M.; Smid, M.; Zhang, Y.; van de Water, B. A kinase inhibitor screen identifies a dual cdc7/CDK9 inhibitor to sensitise triple-negative breast cancer to EGFR-targeted therapy. Breast Cancer Res. 2019, 21, 77. [Google Scholar]
- Panicker, P.S.; Melge, A.R.; Biswas, L.; Keechilat, P.; Keechilat, P.; Mohan, C.G. Epidermal growth factor receptor (EGFR) structure-based bioactive pharmacophore models for identifying next-generation inhibitors against clinically relevant EGFR mutations. Chem. Biol. Drug Des. 2017, 90, 629–636. [Google Scholar]
- Koshland, D.E. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA 1958, 44, 98–104. [Google Scholar] [PubMed]
- Trott, O.; Olson, A.J. AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [PubMed] [Green Version]
- Gogoi, B.; Chowdhury, P.; Goswami, N.; Gogoi, N.; Naiya, T.; Chetia, P.; Mahanta, S.; Chetia, D.; Tanti, B.; Borah, P.; et al. Identification of potential plant-based inhibitor against viral proteases of SARS-CoV-2 through molecular docking, MM-PBSA binding energy calculations and molecular dynamics simulation. Mol. Divers. 2021, 25, 963–1977. [Google Scholar]
- Raj, M.R.; Sreeja, A. Analysis of Computational Gene Prioritization Approaches. Procedia Comput. Sci. 2018, 143, 395–410. [Google Scholar] [CrossRef]
- Sun, L.; Zhu, Y.; Mahmood, A.S.M.A.; Tudor, C.O.; Ren, J.; Vijay-Shanker, K.; Chen, J.; Schmidt, C.J. WebGIVI: A web-based gene enrichment analysis and visualization tool. BMC Bioinform. 2017, 18, 237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar]
- Masoudi-Nejad, A.; Meshkin, A.; Haji-Eghrari, B.; Gholamreza Bidkhori. Retracted Article: Candidate gene prioritization. Mol. Genet. Genom. 2012, 287, 679–698. [Google Scholar]
- Janwa, H.; Massey, S.E.; Velev, J.; Mishra, B. On the Origin of Biomolecular Networks. Front. Genet. 2019, 10, 240. [Google Scholar] [CrossRef] [Green Version]
- Jensen, L.; Kuhn, M.; Stark, M.; Chaffron, S.; von Mering, C.; Bork, P. STRING and STITCH: Known and predicted interactions between proteins and chemicals. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Roccatano, D.; Barthel, A.; Zacharias, M. Structural flexibility of the nucleosome core particle at atomic resolution studied by molecular dynamics simulation. Biopolymers 2007, 85, 407–421. [Google Scholar]
- Sharma, S.; Ding, F.; Dokholyan, N.V. Multiscale modeling of nucleosome dynamics. Biophys. J. 2007, 92, 1457–1470. [Google Scholar]
- Swegat, W.; Schlitter, J.; Krüger, P.; Wollmer, A. MD simulation of protein-ligand interaction: Formation and dissociation of an insulin-phenol complex. Biophys. J. 2003, 84, 1493–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hospital, A.; Goñi, J.R.; Orozco, M.; Gelpí, J.L.l. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar] [PubMed] [Green Version]
- Khan, S.; Nayak, D.; Khurana, A.; Manchanda, R.K.; Tandon, C.; Tandon, S. In Vitro Assessment of Homeopathic Potencies of Hydrastiscanadensis on Hormone-Dependent and Independent Breast Cancer. Homeopathy 2020, 109, 198–206. [Google Scholar] [PubMed]
- Li, Y.; Li, S.; Meng, X.; Gan, R.Y.; Zhang, J.J.; Li, H.B. Dietary Natural Products for Prevention and Treatment of Breast Cancer. Nutrients 2017, 9, 728. [Google Scholar]
- ShahiThakuri, P.; Gupta, M.; Singh, S.; Joshi, R.; Glasgow, E.; Lekan, A.; Agarwal, S.; Luker, G.D.; Tavana, H. Phytochemicals inhibit migration of triple negative breast cancer cells by targeting kinase signaling. BMC Cancer 2020, 20, 4. [Google Scholar]
- Shareef, M.; Ashraf, M.A.; Sarfraz, M. Natural cures for breast cancer treatment. Saudi Pharm. J. 2016, 24, 233–240. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353. [Google Scholar]
- Chen, J.; Xu, H.; Aronow, B.J.; Jegga, A.G. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinform. 2007, 8, 392. [Google Scholar] [CrossRef] [Green Version]
- wwPDB Consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef] [Green Version]
- Hulo, N.; Bairoch, A.; Bulliard, V.; Cerutti, L.; De Castro, E.; Langendijk-Genevaux, P.S.; Pagni, M.; Sigrist, C.J. The PROSITE database. Nucleic Acids Res. 2006, 34, D227. [Google Scholar] [CrossRef] [Green Version]
- Gradishar, W.J.; Anderson, B.O.; Abraham, J.; Aft, R.; Agnese, D.; Allison, K.H.; Blair, S.L.; Burstein, H.J.; Dang, C.; Elias, A.D.; et al. Breast Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 452–478. [Google Scholar] [CrossRef] [Green Version]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202. [Google Scholar] [CrossRef] [PubMed]
- López-López, E.; Jesús Naveja, J.; Medina-Franco, J.L. DataWarrior: An evaluation of the open-source drug discovery tool. Expert Opin. Drug Discov. 2009, 14, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Emon, N.U.; Alam, S.; Rudra, S.; Riya, S.R.; Paul, A.; Hossen, S.M.M.; Kulsum, U.; Ganguly, A. Antidepressant, anxiolytic, antipyretic, and thrombolytic profiling of methanol extract of the aerial part of Piper nigrum: In vivo, in vitro, and in silico approaches. Food Sci. Nutr. 2021, 9, 833–846. [Google Scholar] [PubMed]
- HimaVyshnavi, A.M.; Lakshmi Anand, C.; Deepak, O.M.; Krishnan Namboori, P.K. Evaluation of colorectal cancer (CRC) epidemiology a pharmacogenomic approach. J. Young Pharm. 2017, 9, 36–39. [Google Scholar]
- Sangeetha, M.; Saranya, T.S.; Sathianarayanan, S.; Vyshnavi, A.M.H.; Namboori, P.K.K. Design and development of potential flavonoid moiety for PBP2a inhibition for MRSA therapy-a computational technique. Biomed. Pharmacol. J. 2020, 13, 687–692. [Google Scholar] [CrossRef]
- Kumar, S.A.; Bhaskar, B. Structural elucidation of antihemorrhage drug molecule Diethylammonium 2,5-dihydroxybenzene sulfonate—An insilico approach. IOP Conf. Ser. Mater. Sci. Eng. 2018, 310, 012124. [Google Scholar] [CrossRef]
- Carneiro, T.; Da Nóbrega, R.V.M.; Nepomuceno, T.; Bian, G.; de Albuquerque, V.H.C.; Filho, P. Performance Analysis of Google Colaboratory as a Tool for Accelerating Deep Learning Applications. IEEE Access 2018, 6, 61677–61685. [Google Scholar] [CrossRef]
- Available online: https://github.com/pablo-arantes/making-it-rain/blob/main/Protein_ligand.ipynb (accessed on 23 August 2022).
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 4, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Dittrich, A.; Gautrey, H.; Browell, D.; Tyson-Capper, A. The HER2 Signaling Network in Breast Cancer—Like a Spider in its Web. J. Mammary Gland. Biol. Neoplasia 2014, 19, 253–270. [Google Scholar] [PubMed]
- Al-Hussaini, H.; Subramanyam, D.; Reedijk, M.; Sridhar, S.S. Notch signaling pathway as a therapeutic target in breast cancer. Mol. Cancer Ther. 2011, 10, 9–15. [Google Scholar]
- Ignatiadis, M.; Sotiriou, C. Luminal breast cancer: From biology to treatment. Nat. Rev. Clin. Oncol. 2013, 10, 494–506. [Google Scholar] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wang, H.; Zheng, H. A Mini Review of Node Centrality Metrics in Biological Networks. IJNDI 2022, 1, 99–110. [Google Scholar]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar]
- Garza, A.Z.; Park, S.B.; Kocz, R. Drug Elimination. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK547662/ (accessed on 23 August 2022).
- Martínez, L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar]
- Al-Karmalawy, A.A.; Dahab, M.A.; Metwaly, A.M.; Elhady, S.S.; Elkaeed, E.B.; Eissa, I.H.; Darwish, K.M. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of ACEIs Against SARS-CoV-2 Targeting the hACE2 Receptor. Front. Chem. 2021, 9, 661230. [Google Scholar]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, H.U.; Ahmad, N.; Abdalla, M.; Khan, K.; Martines, M.A.U.; Shabana, S. Molecular docking and dynamic simulations of Cefixime, Etoposide and Nebrodenside A against the pathogenic proteins of SARS-CoV-2. J. Mol. Struct. 2022, 1247, 131296. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 16–9478. [Google Scholar] [CrossRef]
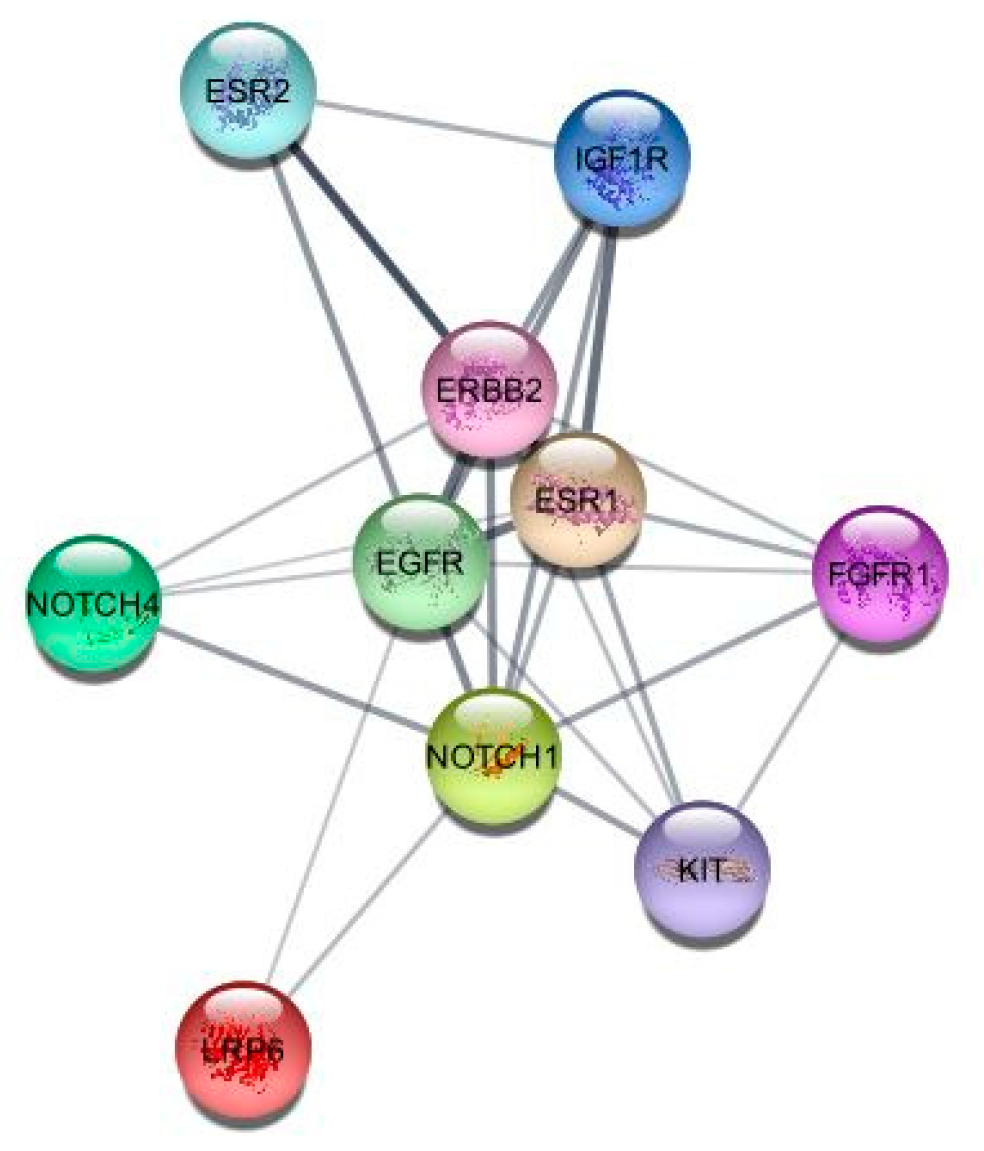
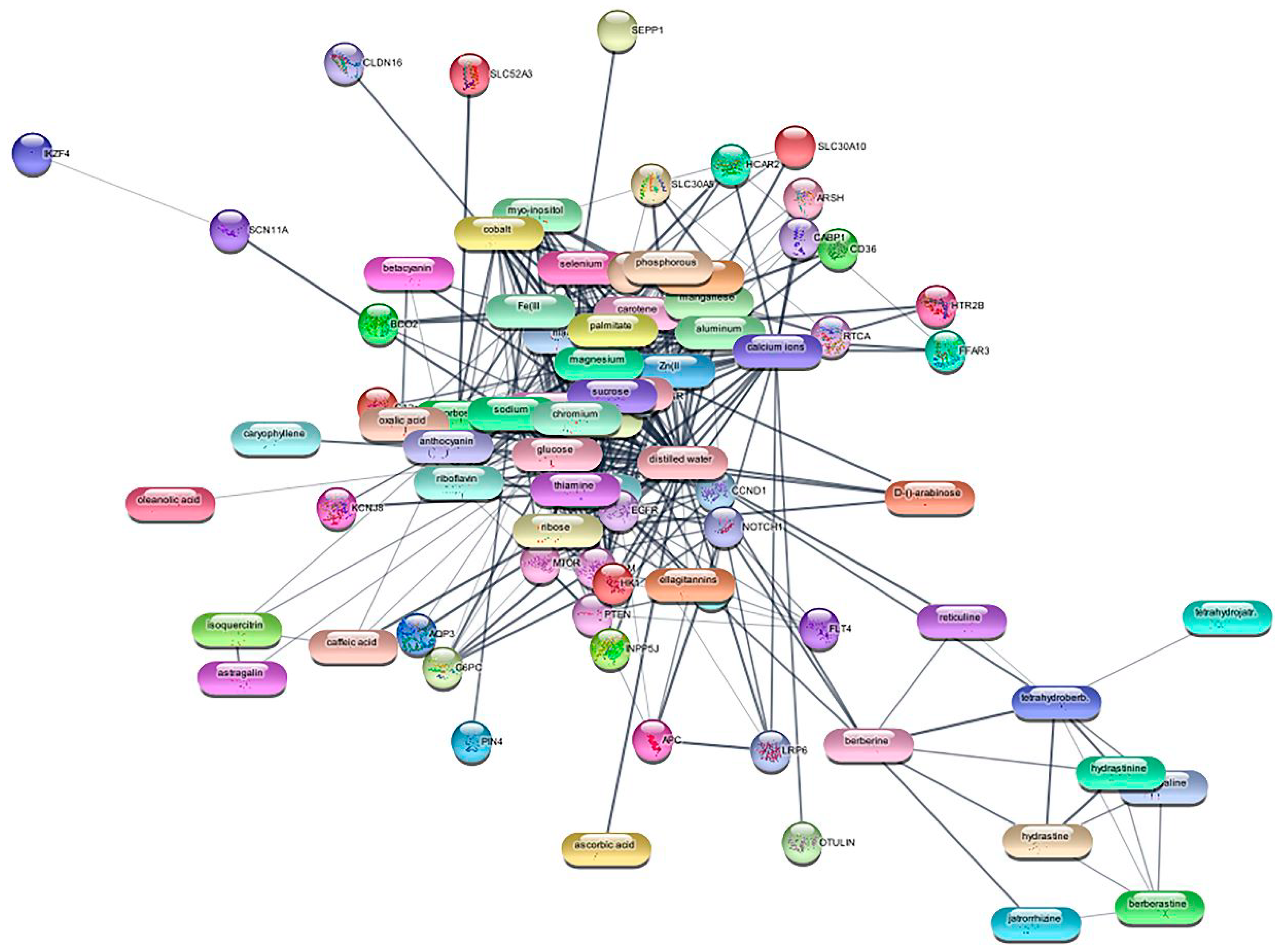



| Genes | Betweenness | Closeness | Degree |
|---|---|---|---|
| EGFR | 13 | 9 | 9 |
| IGF1R | 5 | 8.5 | 8 |
| ERBB2 | 8.5 | 8.5 | 8 |
| ESR1 | 5 | 8.5 | 8 |
| Target | Parameters | D1 | D2 | D3 | D4 | D5 | D6 | D7 | D8 | D9 |
|---|---|---|---|---|---|---|---|---|---|---|
| Hormone-Independent Breast Cancer Targets | ||||||||||
| 1xkk (EGFR) | Glide | −6.99 | −7.45 | −7.45 | −8.68 | −7.99 | −8.39 | −6.35 | −7.87 | −7.57 |
| Autodock | −8.39 | −9.65 | −7.88 | −8.59 | −9.57 | −8.98 | −8.94 | −9.26 | −7.96 | |
| RMSD | 2.36 | 1.54 | 1.51 | 2.37 | 2.06 | 1.98 | 2.37 | 1.70 | 3.09 | |
| MMGBSA_ΔG | −57.18 | −60.28 | −51.47 | −61.66 | −57.51 | −67.27 | −47.17 | −50.15 | −40.43 | |
| MMGBSA_ΔG(NS) | −63.00 | −66.92 | −59.53 | −70.56 | −63.55 | −73.10 | −52.79 | −59.60 | −51.12 | |
| 3LVP (IGF1R) | Glide | −6.64 | −7.31 | −6.41 | −4.62 | −7.14 | −6.24 | −6.50 | −7.91 | −7.40 |
| Autodock | −7.51 | −6.96 | −7.07 | −7.16 | −7.69 | −8.26 | −7.27 | −7.50 | −6.81 | |
| RMSD | 0.949 | 1.59 | 1.27 | 2.30 | 1.18 | 3.47 | 2.86 | 2.58 | 2.03 | |
| MMGBSA_ΔG | −31.48 | −44.22 | −48.01 | −34.77 | −45.00 | −42.16 | −52.71 | −61.92 | −53.57 | |
| MMGBSA_ΔG(NS) | −39.72 | −50.81 | −58.40 | −39.90 | −56.14 | −55.52 | −55.9 | −64.70 | −64.36 | |
| Hormone-Dependent Breast Cancer Targets | ||||||||||
| 3MZW (ERBB2) | Glide | −5.04 | −5.46 | −2.93 | −3.56 | −5.69 | −4.89 | −3.49 | −3.16 | −3.99 |
| Autodock | −5.96 | −6.08 | −6.79 | −6.25 | −6.39 | −7.00 | −5.95 | −6.08 | −5.96 | |
| RMSD | 2.02 | 1.83 | 2.39 | 1.38 | 1.65 | 2.69 | 2.75 | 2.39 | 4.62 | |
| MMGBSA_ΔG | −41.41 | −41.38 | −36.34 | −27.72 | −49.83 | −44.73 | −29.31 | −24.42 | −35.30 | |
| MMGBSA_ΔG(NS) | −48.45 | −48.43 | −38.31 | −31.82 | −52.95 | −47.80 | −34.15 | −30.26 | −39.34 | |
| 1EER (ESR1) | Glide | −7.16 | −7.25 | −7.84 | −6.48 | −6.94 | −5.15 | −5.28 | −6.05 | −5.25 |
| Autodock | −8.38 | −8.03 | −8.39 | −7.35 | −8.57 | −7.78 | −7.40 | −7.63 | −6.46 | |
| RMSD | 1.17 | 1.98 | 1.09 | 1.68 | 2.76 | 1.12 | 2.40 | 2.04 | 3.30 | |
| MMGBSA_ΔG | −52.38 | −51.17 | −29.77 | −56.45 | −66.48 | −45.21 | −50.13 | −39.92 | −36.70 | |
| MMGBSA_ΔG(NS) | −54.39 | −56.67 | −34.57 | −58.99 | −68.72 | −51.86 | −52.21 | −49.03 | −42.32 | |
| Target | D1 | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 |
|---|---|---|---|---|---|---|---|---|---|---|
| Hormone-Independent Breast Cancer Targets | ||||||||||
| 1XKK | −9.570 | −6.060 | −4.660 | −4.710 | −3.896 | −5.980 | −5.330 | −5.950 | −5.450 | −8.280 |
| 3lvp | −7.690 | −6.830 | −6.240 | −6.680 | −7.060 | −6.560 | −6.170 | −6.150 | −6.060 | −6.770 |
| Hormone-Dependent Breast Cancer Targets | ||||||||||
| 1ERR | −5.890 | −6.070 | −6.600 | −5.10 | −6.820 | −5.840 | −6.140 | −5.970 | −5.290 | −6.750 |
| 3MZW | −6.330 | −6.520 | −6.820 | −4.510 | −6.000 | −6.350 | −5.960 | −6.120 | −6.500 | −7.380 |
| Target | 1XKK | 3LVP | 3MZW | IERR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | ΔG_D | ΔG_G | ΔG_P | ΔG_D | ΔG_G | ΔG_P | ΔG_D | ΔG_G | ΔG_P | ΔG_D | ΔG_G | ΔG_P |
| D1 | −9.57 | −23.74 | −0.36 | −7.69 | −32.11 | −1.27 | −5.890 | −24.70 | −1.34 | −6.330 | −24.08 | −0.26 |
| C4 | −3.90 | −24.82 | −25.32 | −7.06 | −38.24 | −5.55 | −6.820 | −15.89 | −1.72 | −6.000 | −35.24 | −3.67 |
| C9 | −8.28 | −25.86 | −24.95 | −6.77 | −31.94 | −3.67 | −6.750 | −32.90 | −0.49 | −7.380 | −31.94 | −5.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vyshnavi AM, H.; Sankaran, S.; Namboori PK, K.; Venkidasamy, B.; Hirad, A.H.; Alarfaj, A.A.; Vinayagam, R. In Silico Analysis of the Effect of Hydrastis canadensis on Controlling Breast Cancer. Medicina 2023, 59, 1412. https://doi.org/10.3390/medicina59081412
Vyshnavi AM H, Sankaran S, Namboori PK K, Venkidasamy B, Hirad AH, Alarfaj AA, Vinayagam R. In Silico Analysis of the Effect of Hydrastis canadensis on Controlling Breast Cancer. Medicina. 2023; 59(8):1412. https://doi.org/10.3390/medicina59081412
Chicago/Turabian StyleVyshnavi AM, Hima, Sathianarayanan Sankaran, Krishnan Namboori PK, Baskar Venkidasamy, Abdurahman Hajinur Hirad, Abdullah A. Alarfaj, and Ramachandran Vinayagam. 2023. "In Silico Analysis of the Effect of Hydrastis canadensis on Controlling Breast Cancer" Medicina 59, no. 8: 1412. https://doi.org/10.3390/medicina59081412
APA StyleVyshnavi AM, H., Sankaran, S., Namboori PK, K., Venkidasamy, B., Hirad, A. H., Alarfaj, A. A., & Vinayagam, R. (2023). In Silico Analysis of the Effect of Hydrastis canadensis on Controlling Breast Cancer. Medicina, 59(8), 1412. https://doi.org/10.3390/medicina59081412








