Hesperidin, a Potential Antiviral Agent against SARS-CoV-2: The Influence of Citrus Consumption on COVID-19 Incidence and Severity in China
Abstract
1. Introduction
2. Hesperidin: Chemical Properties, Sources and Safety Profile
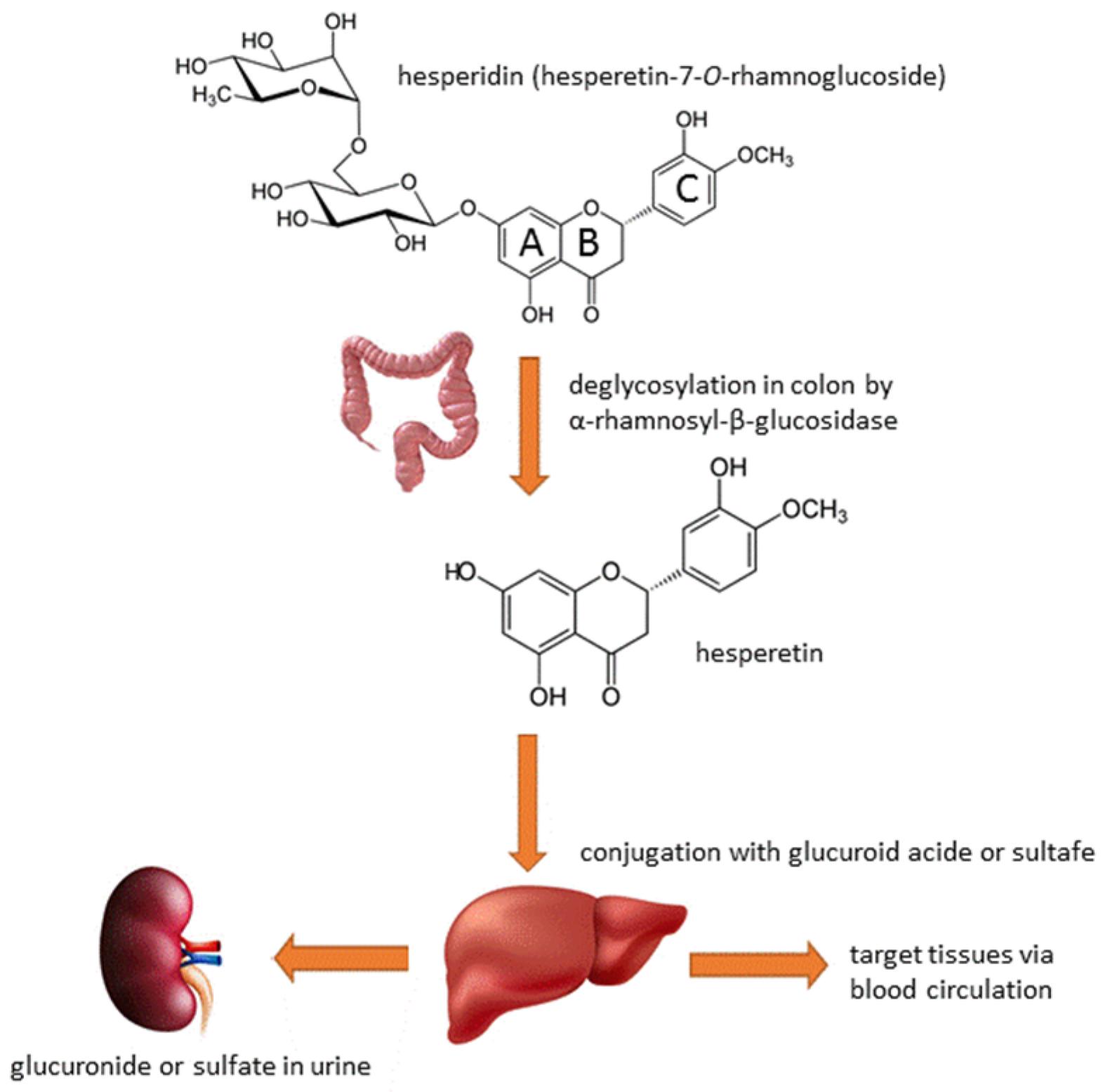
3. Hesperidin’s Metabolism and Bioavailability
4. Hesperidin Biological and Pharmacological Activity
4.1. Antioxidant Activity
4.2. Anti-Inflammatory Activity
4.3. Neuroprotective Activity
4.4. Impact on the Cardiovascular System
4.5. Hypolipidemic Activity
4.6. Anti-Carcinogenic Activity
4.7. Insulin-Sensitizing Activity
4.8. Antiviral Activity
5. Hesperidin versus SARS-CoV-2
5.1. Clinical Studies
5.2. In Vitro and In Silico Studies
6. Hesperidin Intake Levels and Some Potential Health Benefits
7. Hesperidin Contents in Some Citrus Juices
8. Citrus Fruits Consumption in China
9. Limitations and Challenges in Interpreting Findings from Clinical, In Vitro and In Silico Studies
10. Future Directions
11. Conclusions
Funding
Conflicts of Interest
References
- Pagani, I.; Ghezzi, S.; Alberti, S.; Poli, G.; Vicenzi, E. Origin and evolution of SARS-CoV-2. Eur. Phys. J. Plus 2023, 138, 157. [Google Scholar] [CrossRef] [PubMed]
- Brice, Y.; Morgan, L.; Kirmani, M.; Kirmani, M.; Udeh, M.C. COVID-19 vaccine evolution and beyond. Neurosci. Insights 2023, 18, 26331055231180543. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and efficacy of the BNT162b2 MRNA COVID-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Dagan, N.; Barda, N.; Kepten, E.; Miron, O.; Perchik, S.; Katz, M.A.; Hernán, M.A.; Lipsitch, M.; Reis, B.; Balicer, R.D. BNT162b2 MRNA COVID-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021, 384, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y. Potential interventions for novel coronavirus in China: A Systematic Review. J. Med. Virol. 2020, 92, 479–490. [Google Scholar] [CrossRef]
- Colunga Biancatelli, R.M.L.; Berrill, M.; Catravas, J.D.; Marik, P.E. Quercetin and Vitamin C: An experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19). Front. Immunol. 2020, 11, 1451. [Google Scholar] [CrossRef]
- Wang, J.; Qi, F. Traditional chinese medicine to treat COVID-19: The importance of evidence-based research. Drug Discov. Ther. 2020, 14, 149–150. [Google Scholar] [CrossRef]
- Ren, J.l.; Zhang, A.H.; Wang, X.J. Traditional chinese medicine for COVID-19 treatment. Pharmacol. Res. 2020, 155, 104743. [Google Scholar] [CrossRef]
- Kim, H.; Rebholz, C.M.; Hegde, S.; Lafiura, C.; Raghavan, M.; Lloyd, J.F.; Cheng, S.; Seidelmann, S.B. Plant-based diets, pescatarian diets and COVID-19 severity: A population-based case-control study in six countries. BMJ Nutr. Prev. Health 2021, 4, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Stanisic, D.; Liu, L.H.B.; Dos Santos, R.V.; Costa, A.F.; Durán, N.; Tasic, L. New sustainable process for hesperidin isolation and anti-ageing effects of hesperidin nanocrystals. Molecules 2020, 25, 4534. [Google Scholar] [CrossRef] [PubMed]
- Bisen, A.C.; Rawat, P.; Sharma, G.; Sanap, S.N.; Agrawal, S.; Kumar, S.; Kumar, A.; Choudhury, A.D.; Kamboj, S.; Narender, T.; et al. Hesperidin: Enrichment, forced degradation, and structural elucidation of potential degradation products using spectral techniques. Rapid Commun. Mass. Spectrom. 2023, 37, e9615. [Google Scholar] [CrossRef] [PubMed]
- Crescenti, A.; Caimari, A.; Alcaide-Hidalgo, J.M.; Mariné-Casadó, R.; Valls, R.M.; Companys, J.; Salamanca, P.; Calderón-Pérez, L.; Pla-Pagà, L.; Pedret, A.; et al. Hesperidin bioavailability is increased by the presence of 2S—Diastereoisomer and micronization-a randomized, crossover and double-blind clinical trial. Nutrients 2022, 14, 2481. [Google Scholar] [CrossRef] [PubMed]
- Ur Rehman, M.F.; Batool, A.I.; Qadir, R.; Aslam, M. A Centum of Valuable Plant Bioactives; Academic Press: Cambridge, MA, USA, 2021; pp. 403–444. [Google Scholar]
- Samota, M.K.; Kaur, M.; Sharma, M.; Sarita; Krishnan, V.; Thakur, J.; Rawat, M.; Phogat, B.; Guru, P.N. Hesperidin from citrus peel waste: Extraction and its health implications. Qual. Assur. Saf. Crop. Foods 2023, 15, 71–99. [Google Scholar] [CrossRef]
- Pyrzynska, K. Hesperidin: A review on extraction methods, stability and biological activities. Nutrients 2022, 14, 2387. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/?set=GRASNotices&sort=GRN_ (accessed on 16 April 2024).
- Available online: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/?set=GRASNotices&id=796&sort=GRN_No&order=DESC&startrow=1&type=basic&search=hesperidin (accessed on 10 May 2024).
- Available online: https://www.preprints.org/manuscript/202312.1353/v1 (accessed on 10 May 2024).
- Available online: https://www.webmd.com/vitamins/ai/ingredientmono-1033/hesperidin (accessed on 9 May 2024).
- Roy, J.; Azamthulla, M.; Mukkerjee, D. Hesperidin and diosmin—A novel drugs. Int. J. Pharm. Res. Technol. (IJPRT) 2020, 10, 25–33. [Google Scholar]
- Yamamoto, M.; Jokura, H.; Hashizume, K.; Ominami, H.; Shibuya, Y.; Suzuki, A.; Hase, T.; Shimotoyodome, A. Hesperidin metabolite hesperetin-7-O-Glucuronide, but not hesperetin-3-o-glucuronide, exerts hypotensive, vasodilatory, and anti-inflammatory activities. Food Funct. 2013, 4, 1346–1351. [Google Scholar] [CrossRef]
- Guo, X.; Li, K.; Guo, A.; Li, E. Intestinal absorption and distribution of naringin, hesperidin, and their metabolites in mice. J. Funct. Foods 2020, 74, 104158. [Google Scholar] [CrossRef]
- Ávila-gálvez, M.Á.; Giménez-bastida, J.A.; González-sarrías, A.; Espín, J.C. New insights into the metabolism of the flavanones eriocitrin and hesperidin: A comparative human pharmacokinetic study. Antioxidants 2021, 10, 435. [Google Scholar] [CrossRef]
- Mas-Capdevila, A.; Teichenne, J.; Domenech-Coca, C.; Caimari, A.; Bas, J.M.D.; Escoté, X.; Crescenti, A. Effect of hesperidin on cardiovascular disease risk factors: The role of intestinal microbiota on hesperidin bioavailability. Nutrients 2020, 12, 1488. [Google Scholar] [CrossRef] [PubMed]
- Van Rymenant, E.; Salden, B.; Voorspoels, S.; Jacobs, G.; Noten, B.; Pitart, J.; Possemiers, S.; Smagghe, G.; Grootaert, C.; Van Camp, J. A critical evaluation of in vitro hesperidin 2S bioavailability in a model combining luminal (Microbial) digestion and Caco-2 cell absorption in comparison to a randomized controlled human trial. Mol. Nutr. Food Res. 2018, 62, e1700881. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhong, X.; Yan, J.; Sun, C.; Zhao, X.; Wang, X. Potential roles of gut microbes in biotransformation of natural products: An overview. Front. Microbiol. 2022, 13, 956378. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Afzaal, M.; Saeed, F.; Ali, S.W.; Imran, A.; Zaidi, S.Y.R.; Saleem, M.A.; Hussain, M.; Al Jbawi, E. A comprehensive review of the therapeutic potential of citrus bioflavonoid hesperidin against lifestyle-related disorders. Cogent Food Agric. 2023, 9, 2226427. [Google Scholar] [CrossRef]
- Buzdağlı, Y.; Eyipınar, C.D.; Kacı, F.N.; Tekin, A. Effects of hesperidin on anti-inflammatory and antioxidant response in healthy people: A meta-analysis and meta-regression. Int. J. Environ. Health Res. 2023, 33, 1390–1405. [Google Scholar] [CrossRef] [PubMed]
- Lorzadeh, E.; Ramezani-Jolfaie, N.; Mohammadi, M.; Khoshbakht, Y.; Salehi-Abargouei, A. The effect of hesperidin supplementation on inflammatory markers in human adults: A systematic review and meta-analysis of randomized controlled clinical trials. Chem. Biol. Interact. 2019, 307, 5–15. [Google Scholar] [CrossRef]
- Li, X.; Huang, W.; Tan, R.; Xu, C.; Chen, X.; Li, S.; Liu, Y.; Qiu, H.; Cao, H.; Cheng, Q. The benefits of hesperidin in central nervous system disorders, based on the neuroprotective effect. Biomed. Pharmacother. 2023, 159, 114222. [Google Scholar] [CrossRef]
- Khorasanian, A.S.; Fateh, S.T.; Gholami, F.; Rasaei, N.; Gerami, H.; Khayyatzadeh, S.S.; Shiraseb, F.; Asbaghi, O. The effects of hesperidin supplementation on cardiovascular risk factors in adults: A systematic review and dose–response meta-analysis. Front. Nutr. 2023, 10, 1177708. [Google Scholar] [CrossRef] [PubMed]
- Bansal, K.; Bhati, H.; Vanshita; Bajpai, M. New Insights into therapeutic applications and nanoformulation approaches of hesperidin: An updated review. Pharmacol. Res.-Mod. Chin. Med. 2024, 10, 100363. [Google Scholar] [CrossRef]
- Ganeshpurkar, A.; Saluja, A. The pharmacological potential of hesperidin. Indian J. Biochem. Biophys. 2019, 56, 287–300. [Google Scholar]
- Dupuis, J.; Laurin, P.; Tardif, J.C.; Hausermann, L.; Rosa, C.; Guertin, M.C.; Thibaudeau, K.; Gagnon, L.; Cesari, F.; Robitaille, M.; et al. Fourteen-day evolution of COVID-19 symptoms during the third wave in nonvaccinated subjects and effects of hesperidin therapy: A randomized, double-blinded, placebo-controlled study. Evid.-Based Complement. Altern. Med. 2022, 2022, 3125662. [Google Scholar] [CrossRef] [PubMed]
- Tanta University. Hesperidin and Diosmin for Treatment of COVID-19. Available online: https://clinicaltrials.gov/study/NCT04452799?cond=Covid19&intr=hesperidin&rank=2 (accessed on 9 April 2024).
- Jahangirifard, A.; Mirtajani, S.B.; Karimzadeh, M.; Forouzmehr, M.; Kiani, A.; Moradi, M.; Soleimani, S.; Bezadmoghadam, M.; Abedini, A. The effect of hesperidin on laboratory parameters of patients with COVID 19: A preliminary report of a clinical trial study. J. Iran. Med. Counc. 2022, 5, 89–95. [Google Scholar] [CrossRef]
- Cheng, F.J.; Huynh, T.K.; Yang, C.S.; Hu, D.W.; Shen, Y.C.; Tu, C.Y.; Wu, Y.C.; Tang, C.H.; Huang, W.C.; Chen, Y.; et al. Hesperidin is a potential inhibitor against SARS-CoV-2 infection. Nutrients 2021, 13, 2800. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhao, Z.; Lan, Q.; Zhou, H.; Mai, Z.; Wang, Y.; Ding, X.; Zhang, W.; Pi, J.; Evans, C.E.; et al. Nasal delivery of hesperidin/chitosan nanoparticles suppresses cytokine storm syndrome in a mouse model of acute lung injury. Front. Pharmacol. 2021, 11, 592238. [Google Scholar] [CrossRef] [PubMed]
- Özgürbüz, U.; Vatansever, S.; Becer, E.; Kabadayı Ensarioğlu, H.; Akoğulları Çelik, D. Protective effects of citrus flavonoid hesperidin in enterocytes after induction with TNF-α and IFN-ɣ which mimic the COVID-19 disease. Cyprus J. Med. Sci. 2023, 8, 390–396. [Google Scholar] [CrossRef]
- Niazi, S.K.; Mariam, Z. Computer-aided drug design and drug discovery: A prospective analysis. Pharmaceuticals 2024, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Shaker, B.; Ahmad, S.; Lee, J.; Jung, C.; Na, D. In silico methods and tools for drug discovery. Comput. Biol. Med. 2021, 137, 104851. [Google Scholar] [CrossRef] [PubMed]
- Mushebenge, A.G.A.; Ugbaja, S.C.; Mbatha, N.A.; Khan, R.B.; Kumalo, H.M. Assessing the potential contribution of in silico studies in discovering drug candidates that interact with various SARS-CoV-2 receptors. Int. J. Mol. Sci. 2023, 24, 15518. [Google Scholar] [CrossRef]
- Chan, W.K.B.; Olson, K.M.; Wotring, J.W.; Sexton, J.Z.; Carlson, H.A.; Traynor, J.R. In silico analysis of SARS-CoV-2 proteins as targets for clinically available drugs. Sci. Rep. 2022, 12, 5320. [Google Scholar] [CrossRef]
- Yusuf, M. Insights into the in-silico research: Current scenario, advantages, limits, and future perspectives. Life Silico 2023, 1, 13–25. [Google Scholar]
- de Oliveira, I.S.; Silva, G.M.A.; Cordeiro, F.A.C.; Pinheiro Júnior, E.L.; Ferreira, I.G.; Cerni, F.A.; Zottic, U.; Pucca, M.B. Research models in biomedical sciences: Advantages and limitations. Open Access J. Biomed. Sci. 2020, 2, 464–476. [Google Scholar]
- Tallei, T.E.; Tumilaar, S.G.; Niode, N.J.; Fatimawali; Kepel, B.J.; Idroes, R.; Effendi, Y.; Sakib, S.A.; Emran, T.B. Potential of plant bioactive compounds as SARS-CoV-2 main protease (Mpro) and spike (S) glycoprotein inhibitors: A molecular docking study. Scientifica 2020, 2020, 6307457. [Google Scholar] [CrossRef] [PubMed]
- El Hawary, S.S.; Khattab, A.R.; Marzouk, H.S.; El Senousy, A.S.; Alex, M.G.A.; Aly, O.M.; Teleb, M.; Abdelmohsen, U.R. In silico identification of SARS-CoV-2 spike (S) protein-ACE2 complex inhibitors from eight tecoma species and cultivars analyzed by LC-MS. RSC Adv. 2020, 10, 43103–43108. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Sarkar, A.; Maulik, U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV-2 spike protein and human ACE2. Sci. Rep. 2020, 10, 17699. [Google Scholar] [CrossRef] [PubMed]
- Mahdian, S.; Ebrahim-Habibi, A.; Zarrabi, M. Drug repurposing using computational methods to identify therapeutic options for COVID-19. J. Diabetes Metab. Disord. 2020, 19, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Balmeh, N.; Mahmoudi, S.; Mohammadi, N.; Karabedianhajiabadi, A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Inform. Med. Unlocked 2020, 20, 100407. [Google Scholar] [CrossRef] [PubMed]
- Khater, S.; Kumar, P.; Dasgupta, N.; Das, G.; Ray, S.; Prakash, A. Combining SARS-CoV-2 proofreading exonuclease and RNA-dependent RNA polymerase inhibitors as a strategy to combat COVID-19: A high-throughput in silico screening. Front. Microbiol. 2021, 12, 647693. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A. Electron donor–acceptor capacity of selected pharmaceuticals against COVID-19. Antioxidants 2021, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; Mostafa, A.; Kutkat, O.; Moatasim, Y.; Al-karmalawy, A.A.; Rashad, A.A.; Kayed, A.E.; Kayed, A.E.; El-Shesheny, R.; Kayali, G.; et al. Bioactive polyphenolic compounds showing strong antiviral activities against severe acute respiratory syndrome coronavirus 2. Pathogens 2021, 10, 758. [Google Scholar] [CrossRef]
- Attia, G.H.; Moemen, Y.S.; Youns, M.; Ibrahim, A.M.; Abdou, R. Antiviral zinc oxide nanoparticles mediated by hesperidin and in silico comparison study between antiviral phenolics as anti-SARS-CoV-2. Colloids Surf. B Biointerfaces 2021, 203, 111724. [Google Scholar] [CrossRef]
- Bhowmik, D.; Nandi, R.; Prakash, A.; Kumar, D. Evaluation of flavonoids as 2019-NCoV cell entry inhibitor through molecular docking and pharmacological analysis. Heliyon 2021, 7, e06515. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Nandi, R.; Vishwakarma, P.; Prakash, A.; Kumar, D. Discovering potential RNA dependent RNA polymerase inhibitors as prospective drugs against COVID-19: An in silico approach. Front. Pharmacol. 2021, 12, 634047. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, C.; Sun, Y.; Sui, X.; Zhu, T.; Wang, Q.; Wang, S.; Yang, J.; Yang, W.; Liu, F.; et al. A novel screening strategy of anti-SARS-CoV-2 drugs via blocking interaction between spike RBD and ACE2. Environ. Int. 2021, 147, 106361. [Google Scholar] [CrossRef] [PubMed]
- Behloul, N.; Baha, S.; Guo, Y.; Yang, Z.; Shi, R. In silico identification of strong binders of the SARS-CoV-2 receptor-binding domain. Eur. J. Pharmacol. 2021, 890, 173701. [Google Scholar] [CrossRef]
- Satyam Singh, M.D.; Fulbabu, S.K.; Sonawane, A.; Kar, P.; Sadhukhan, S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: An in-silico analysis. J. Biomol. Struct. Dyn. 2021, 36, 6249–6264. [Google Scholar] [CrossRef]
- Das, S.; Sarmah, S.; Lyndem, S.; Roy, A.S. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2021, 39, 3347–3357. [Google Scholar] [CrossRef]
- Moovarkumudalvan, B.; Geethakumari, A.M.; Ramadoss, R.; Biswas, K.H.; Mifsud, B. Structure-based virtual screening and functional validation of potential hit molecules targeting the SARS-CoV-2 main protease. Biomolecules 2022, 12, 1754. [Google Scholar] [CrossRef] [PubMed]
- Ovchynnykova, O.; Kapusta, K.; Sizochenko, N.; Sukhyy, K.M.; Kolodziejczyk, W.; Hill, G.A.; Saloni, J. Homology modeling and molecular dynamics-driven search for natural inhibitors that universally target receptor-binding domain of spike glycoprotein in SARS-CoV-2 variants. Molecules 2022, 27, 7336. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, P.; Thakur, M.; Singh, N.; Shikha, D.; Kumar, S.; Baniwal, P.; Yadav, Y.S.; Sharma, M.; Sridhar, K.; Inbaraj, B.S. In silico evaluation of natural flavonoids as a potential inhibitor of coronavirus disease. Molecules 2022, 27, 6374. [Google Scholar] [CrossRef]
- Erlina, L.; Paramita, R.I.; Kusuma, W.A.; Fadilah, F.; Tedjo, A.; Pratomo, I.P.; Ramadhanti, N.S.; Nasution, A.K.; Surado, F.K.; Fitriawan, A.; et al. Virtual screening of indonesian herbal compounds as COVID-19 supportive therapy: Machine learning and pharmacophore modeling approaches. BMC Complement. Med. Ther. 2022, 22, 207. [Google Scholar] [CrossRef]
- Houchi, S.; Messasma, Z. Exploring the inhibitory potential of saussurea costus and saussurea involucrata phytoconstituents against the spike glycoprotein receptor binding domain of SARS-CoV-2 Delta (B.1.617.2) variant and the main protease (Mpro) as therapeutic candidates, using. J. Mol. Struct. 2022, 1263, 133032. [Google Scholar] [CrossRef] [PubMed]
- Adem, S.; Eyupoglu, V.; Ibrahim, I.M.; Sarfraz, I.; Rasul, A.; Ali, M.; Elfiky, A.A. Multidimensional in silico strategy for identification of natural polyphenols-based SARS-CoV-2 main protease (mpro) inhibitors to unveil a hope against COVID-19. Comput. Biol. Med. 2022, 145, 105452. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhou, W.; Sun, J.; Ou, G.; Zhong, N.S.; Liu, Z. Exploring the potential pharmacological mechanism of hesperidin and glucosyl hesperidin against COVID-19 based on bioinformatics analyses and antiviral assays. Am. J. Chin. Med. 2022, 50, 351–369. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zheng, W.; Cheng, L.; Li, M.; Huang, J.; Bao, S.; Xu, Q.; Ma, Z. Citrus fruits are rich in flavonoids for immunoregulation and potential targeting ACE2. Nat. Products Bioprospect. 2022, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Madanagopal, P.; Ramprabhu, N.; Jagadeesan, R. In silico prediction and structure-based multitargeted molecular docking analysis of selected bioactive compounds against mucormycosis. Bull. Natl. Res. Cent. 2022, 46, 24. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Paul, P.; Yadav, P.; Kaul, R.; Maitra, S.S.; Kumar Jha, S.; Chaari, A. A multi-targeted approach to identify potential flavonoids against three targets in the SARS-CoV-2 life cycle. Comput. Biol. Med. 2022, 141, 105231. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Singh, V.; Varadwaj, P.K.; Chakravartty, N.; Katta, A.V.S.K.M.; Lekkala, S.P.; Thomas, G.; Narasimhan, S.; Reddy, A.R.; Reddy Lachagari, V.B. Secondary metabolites from spice and herbs as potential multitarget inhibitors of SARS-CoV-2 proteins. J. Biomol. Struct. Dyn. 2022, 40, 2264–2283. [Google Scholar] [CrossRef] [PubMed]
- Meneguzzo, F.; Ciriminna, R.; Zabini, F.; Pagliaro, M. Review of evidence available on hesperidin-rich products as potential tools against COVID-19 and hydrodynamic cavitation-based extraction as a method of increasing their production. Processes 2020, 8, 549. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, L.; Manning, M.; Bonahoom, M.; Lotvola, A.; Yang, Z.; Yang, Z.Q. Structural analysis, virtual screening and molecular simulation to identify potential inhibitors targeting 2′-O-ribose methyltransferase of SARS-CoV-2 coronavirus. J. Biomol. Struct. Dyn. 2022, 40, 1331–1346. [Google Scholar] [CrossRef]
- Naidu, S.A.G.; Mustafa, G.; Clemens, R.A.; Naidu, A.S. Plant-derived natural non-nucleoside analog inhibitors (NNAIs) against RNA-dependent RNA polymerase complex (Nsp7/Nsp8/Nsp12) of SARS-CoV-2. J. Diet. Suppl. 2023, 20, 254–283. [Google Scholar] [CrossRef]
- Mohamed, E.A.R.; Abdel-Rahman, I.M.; Zaki, M.E.A.; Al-Khdhairawi, A.; Abdelhamid, M.M.; Alqaisi, A.M.; Rahim, L.B.A.; Abu-Hussein, B.; El-Sheikh, A.A.K.; Abdelwahab, S.F.; et al. In silico prediction of potential inhibitors for SARS-CoV-2 omicron variant using molecular docking and dynamics simulation-based drug repurposing. J. Mol. Model. 2023, 29, 70. [Google Scholar] [CrossRef] [PubMed]
- Kubra, B.; Badshah, S.L.; Faisal, S.; Sharaf, M.; Emwas, A.H.; Jaremko, M.; Abdalla, M. Inhibition of the Predicted allosteric site of the SARS-CoV-2 main protease through flavonoids. J. Biomol. Struct. Dyn. 2023, 41, 9103–9120. [Google Scholar] [CrossRef] [PubMed]
- Perche, O.; Vergnaud-Gauduchon, J.; Morand, C.; Dubray, C.; Mazur, A.; Vasson, M.P. Orange juice and its major polyphenol hesperidin consumption do not induce immunomodulation in healthy well-nourished humans. Clin. Nutr. 2014, 33, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Yari, Z.; Movahedian, M.; Imani, H.; Alavian, S.M.; Hedayati, M.; Hekmatdoost, A. The effect of hesperidin supplementation on metabolic profiles in patients with metabolic syndrome: A randomized, double-blind, placebo-controlled clinical trial. Eur. J. Nutr. 2020, 59, 2569–2577. [Google Scholar] [CrossRef] [PubMed]
- Valls, R.M.; Pedret, A.; Calderón-Pérez, L.; Llauradó, E.; Pla-Pagà, L.; Companys, J.; Moragas, A.; Martín-Luján, F.; Ortega, Y.; Giralt, M.; et al. Effects of hesperidin in orange juice on blood and pulse pressures in mildly hypertensive individuals: A randomized controlled trial (Citrus study). Eur. J. Nutr. 2021, 60, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Tadros, F.J.; Andrade, J.M. Impact of hesperidin in 100% orange juice on chronic disease biomarkers: A narrative systematic review and gap analysis. Crit. Rev. Food Sci. Nut. 2022, 62, 8335–8354. [Google Scholar] [CrossRef] [PubMed]
- Bellavite, P.; Donzelli, A. Hesperidin and SARS-CoV-2: New light on the healthy function of citrus fruits. Antioxidants 2020, 9, 742. [Google Scholar] [CrossRef] [PubMed]
- Richa, R.; Kohli, D.; Vishwakarma, D.; Mishra, A.; Kabdal, B.; Kothakota, A.; Richa, S.; Sirohi, R.; Kumar, R.; Naik, B. Citrus fruit: Classification, value addition, nutritional and medicinal values, and relation with pandemic and hidden hunger. J. Agric. Food Res. 2023, 14, 100718. [Google Scholar] [CrossRef]
- Visvanathan, R.; Williamson, G. Review of factors affecting citrus polyphenol bioavailability and their importance in designing in vitro, animal, and intervention studies. Compr. Rev. Food Sci. Food Saf. 2022, 21, 4509–4545. [Google Scholar] [CrossRef]
- Xu, B.; Gutierrez, B.; Mekaru, S.; Sewalk, K.; Goodwin, L.; Loskill, A.; Cohn, E.L.; Hswen, Y.; Hill, S.C.; Cobo, M.M.; et al. Epidemiological data from the COVID-19 outbreak, real-time case information. Sci. Data 2020, 7, 106. [Google Scholar] [CrossRef]
- Khan, I.; Haleem, A.; Javaid, M. Analysing COVID-19 pandemic through cases, deaths, and recoveries. J. Oral Biol. Craniofacial Res. 2020, 10, 450–469. [Google Scholar] [CrossRef] [PubMed]
- WHO COVID-19 Dashboard. Available online: https://data.who.int/dashboards/covid19/cases?m49=156&n=c (accessed on 15 April 2024).
- Citrus Fruit Fresh and Processed Statistical Bulletin 2020. Available online: https://www.fao.org/policy-support/tools-and-publications/resources-details/en/c/1439010 (accessed on 15 March 2024).
- Citrus: World Markets and Trade. Available online: https://fas.usda.gov/data/citrus-world-markets-and-trade (accessed on 20 March 2024).


| Reference | Trial ID | Year | Study Design | Study Population | Duration | Control | Outcome Measures | Results |
|---|---|---|---|---|---|---|---|---|
| [36] | NCT04715932 | 2022 | Randomized, double-blind, placebo-controlled | 216 | 14 days | Placebo | Yes | Yes |
| [37] | NCT04452799 | 2020 | Randomized double-blind | 100 | 14 days | No treatment | Yes | Yes |
| [38] | IRCT20150725023332N5 | 2022 | Randomized controlled | 20 | 5 days | No treatment | Yes | Yes |
| References | Computational Techniques | Biological Targets of SARS-CoV-2 | Effectiveness Outcomes |
|---|---|---|---|
| [15] | 1. Molecular docking 2. Molecular dynamic simulations | Mpro, S protein, RdRp, nsp13 | 1. Strong binding affinity with viral proteins |
| [48] | 1. Molecular docking 2. Quantum chemical density functional theory calculations | Mpro, S protein | 1. Binding affinity 2. Inhibitory effects on viral replication 3. Comparative analyses with standard antiviral drugs |
| [49] | 1. Molecular docking | S protein, hACE2 | 1. Interactions with key residues of the spike protein and ACE2 receptor |
| [50] | 1. Molecular docking 2. Molecular dynamics simulations | S protein, hACE2 | 1. Non-competitive modulator that destabilizes the interaction between the spike protein and the ACE2 receptor |
| [51] | 1. Molecular docking 2. Virtual screening | Mpro, hACE2, PLpro, HR1, RBD | 1. Affinity to bind 2. Interact with key viral proteins 3. Interfere with virus-host interactions, inhibit viral replication |
| [52] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro, S protein, RdRp, TMPRSS2, hACE2 | 1. Inhibitory effect on virus replication, entry and infectivity 2. Potential to modulate host immune response against SARS-CoV-2 |
| [53] | 1. Molecular docking 2. Molecular dynamics simulations | S protein, RdRp | 1. Binding affinity, stability and potential inhibitory effect on viral proteins |
| [54] | 1. Gaussian09 software for electronic calculations 2. Density functional theory 3. Conceptual density functional theory for antioxidant properties | Mpro, S protein, RdRp | 1. Evaluation of the ability to interact with viral components 2. Potential inhibition of viral replication |
| [55] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro | 1. Binding affinity to viral proteins, particularly Mpro 2. Potential inhibitor of viral replication and maturation |
| [56] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro | 1. Binding energy, binding sites, key interactions with viral proteins |
| [57] | 1. Molecular docking 2. Molecular dynamics simulations 3. Pharmacokinetic studies | S protein | 1. Inhibiting viral proteins or disrupting viral-host interactions, as evidenced by favorable binding affinities, pharmacokinetic properties 2. Potential inhibitory effects on viral entry or replication. |
| [58] | 1. Molecular docking 2. Molecular dynamics simulations 3. MM-GBSA analysis | Mpro, S protein, RdRp, N protein, E protein | 1. Binding energy values and key residue interactions 2. Drug-likeness assessments 3. ADMET properties |
| [59] | 1. Molecular docking 2. Molecular dynamics simulations 3. Virtual screening 4. Quantitative structure-activity relationship analysis | S protein, RBD, hACE2 | 1. Key interactions identification 2. Binding energies 3. Inhibition constants and mechanism of action |
| [60] | 1. Molecular docking 2. Structure-based virtual screening | RBD, hACE2 | 1. Inhibit the SARS-CoV-2-ACE2 interaction, suggesting a possible role in preventing viral cellular entry |
| [61] | 1. Molecular docking 2. Molecular dynamics simulations 3. Free energy calculations 4. Target prediction algorithms | Mpro, RdRp | 1. Binding affinity 2. Stability of protein-ligand complexes 3. Inhibitory activity against viral proteins |
| [62] | 1. Molecular docking 2. Blind docking analyses | Mpro | 1. Estimated free energy of binding for the main protease |
| [63] | 1. Molecular docking 2. Molecular dynamics simulations 3. Virtual screening 4. Deep learning tools for drug-target interaction predictions | Mpro | 1. Potential inhibitor of SARS-CoV-2. Targeting key viral proteins |
| [64] | 1. Molecular docking 2. Molecular dynamics simulations | S protein | 1. Binding affinity, stability, and specific interactions with viral |
| [65] | 1. Molecular docking 2. Molecular dynamics simulations 3. Binding free energy calculations | Mpro, S protein | 1. Inhibiting viral replication 2. Blocking viral entry into host cells 3. Modulating the host immune response |
| [66] | 1. Molecular docking 2. Machine learning approaches | Mpro, S protein | 1. Potential inhibitor of Mpro 2. Binding interactions and potential antiviral activity |
| [67] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro, S protein, hACE2 | 1. Binding affinity scores 2. Interaction energies 3. Key residues involved in hesperidin-protein interactions 4. Constant inhibition or IC50 values for quantifying the potency of hesperidin as an antiviral agent |
| [68] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro | 1. Promising binding energies 2. Interactions at Mpro active site |
| [69] | 1. Molecular docking 2. Binding affinity tests, including biolayer interferometry assay and isothermal titration calorimetry assay | Mpro, hACE2, S protein, RBD | 1. Binding affinity with ACE2, M, S, RBD proteins 2. Impact on immune, inflammation, virus infection, IC50 values (51.5 μM and 5.5 mM) |
| [70] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro, S protein, hACE2 | 1. Binding energies 2. Interaction patterns 3. Key amino acid residues 4. Evaluate stability and dynamics of complexes |
| [71] | 1. Molecular docking 2. Molecular dynamics simulations 3. Pharmacophore modeling | Mpro, S protein, RdRp, PLpro, nsp13 | 1. Evaluate binding affinity, stability, potential to inhibit viral replication |
| [72] | 1. Molecular docking 2. Molecular dynamics simulations 3. SwissADME and ProTox-II for drug-likeness and toxicity assessment | Mpro, TMPRSS2, PLpro | 1. Strong complex formation 2. Stable interactions with viral proteins |
| [73] | 1. Molecular docking 2. Molecular dynamics simulations 3. Molecular modeling techniques | nsp13, ExoN, Guanine-N7 methyltransferase | 1. Interactions with critical residues of target proteins |
| [74] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro, RdRp | 1. Binding affinity 2. Stability in forming complexes with viral enzymes 3. Potential multi-target inhibitory activity |
| [75] | 1. Molecular docking 2. Molecular dynamics simulations 3. Virtual screening | nsp16,2′-O-methyltransferase | 1. Promising interactions with key residues of the nsp16 protein |
| [76] | 1. Molecular docking 2. Molecular dynamics simulations 3. ADMET for drug properties | Mpro, RdRp | 1. Superior binding affinities with Mpro, RdRp compared to standard drugs 2. Strong interactions with catalytic residues |
| [77] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro, S protein, hACE2 | 1. Inhibitory effects on viral proteins 2. Disruption of viral entry mechanisms 3. High binding affinities to key viral targets |
| [78] | 1. Molecular docking 2. Molecular dynamics simulations | Mpro, allosteric site | 1. Identified potent allosteric inhibitors |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalczyk, A. Hesperidin, a Potential Antiviral Agent against SARS-CoV-2: The Influence of Citrus Consumption on COVID-19 Incidence and Severity in China. Medicina 2024, 60, 892. https://doi.org/10.3390/medicina60060892
Kowalczyk A. Hesperidin, a Potential Antiviral Agent against SARS-CoV-2: The Influence of Citrus Consumption on COVID-19 Incidence and Severity in China. Medicina. 2024; 60(6):892. https://doi.org/10.3390/medicina60060892
Chicago/Turabian StyleKowalczyk, Adam. 2024. "Hesperidin, a Potential Antiviral Agent against SARS-CoV-2: The Influence of Citrus Consumption on COVID-19 Incidence and Severity in China" Medicina 60, no. 6: 892. https://doi.org/10.3390/medicina60060892
APA StyleKowalczyk, A. (2024). Hesperidin, a Potential Antiviral Agent against SARS-CoV-2: The Influence of Citrus Consumption on COVID-19 Incidence and Severity in China. Medicina, 60(6), 892. https://doi.org/10.3390/medicina60060892





