Hepatitis E Virus: What More Do We Need to Know?
Abstract
:1. Introduction
2. HEV Genotypes
3. Virology
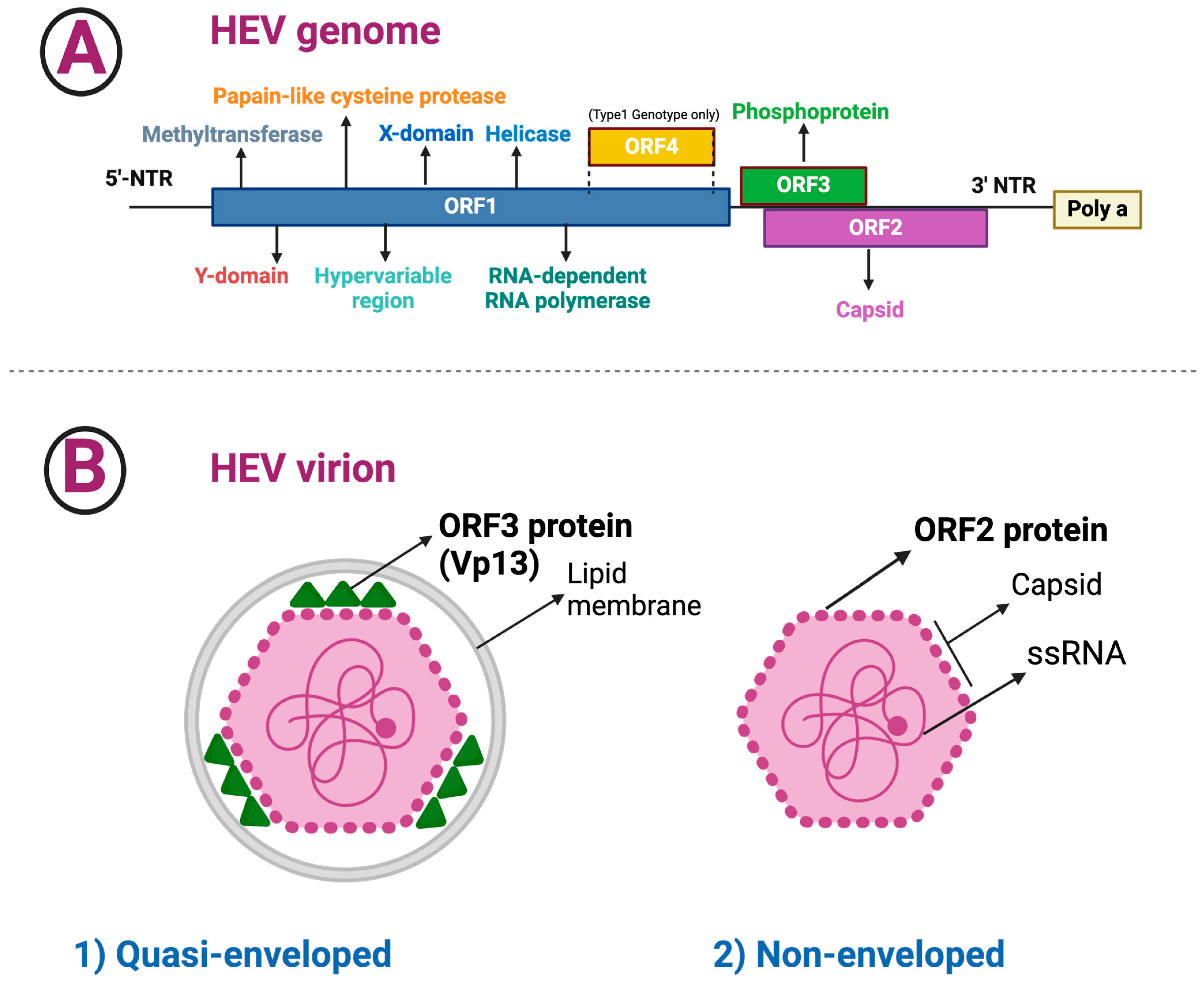
3.1. HEV Composition and Structure
- ▪
- The length of ORF1 varies among HEV genotypes; ORF1 codes for nonstructural proteins involved in virus replication (i.e., methyltransferase, Y domain, papain-like cysteine protease, hypervariable region, X or macro domain, helicase, and RNA-dependent RNA polymerase) [25].
- ▪
- The ORF2 has 112–114 codons that are divided into three structural domains [shell (S), middle (M), and protruding (P)] [20,26]. HEV encodes two isoforms of the ORF2 capsid protein, which are distributed in different subcellular compartments and serve distinct functions throughout the HEV lifecycle [27]. Recombinant ORF2 proteins expressed in bacterial and insect cells have been shown to self-assemble into virus-like particles, the crystal structures of which have been determined [20,21,28,29]. The ORF2 protein was found to bind specifically to the 5′ end of the HEV genome [23]. Several techniques for amplifying the ORF2 gene have been reported, as the ORF2 gene is frequently utilized to analyze and genotype HEV strains [30].
- ▪
- ORF3 has 123 codons and encodes a small phosphoprotein, a functional iron channel that appears to function as a viroporin, allowing infectious virions to be released from infected cells [31].
- ▪
- ORF4, a new ORF discovered within the coding region of ORF1, appears to be restricted to genotype 1 HEV and is translated into a protein that increases activity of the RNA-dependent RNA polymerase [1,18,32,33]; p-ORF4 consists of 124 aa, promotes replication of the virus, is indispensable for the life cycle of HEV genotype 1, and is possibly involved in the increased severity of infection in pregnancy [1,18].
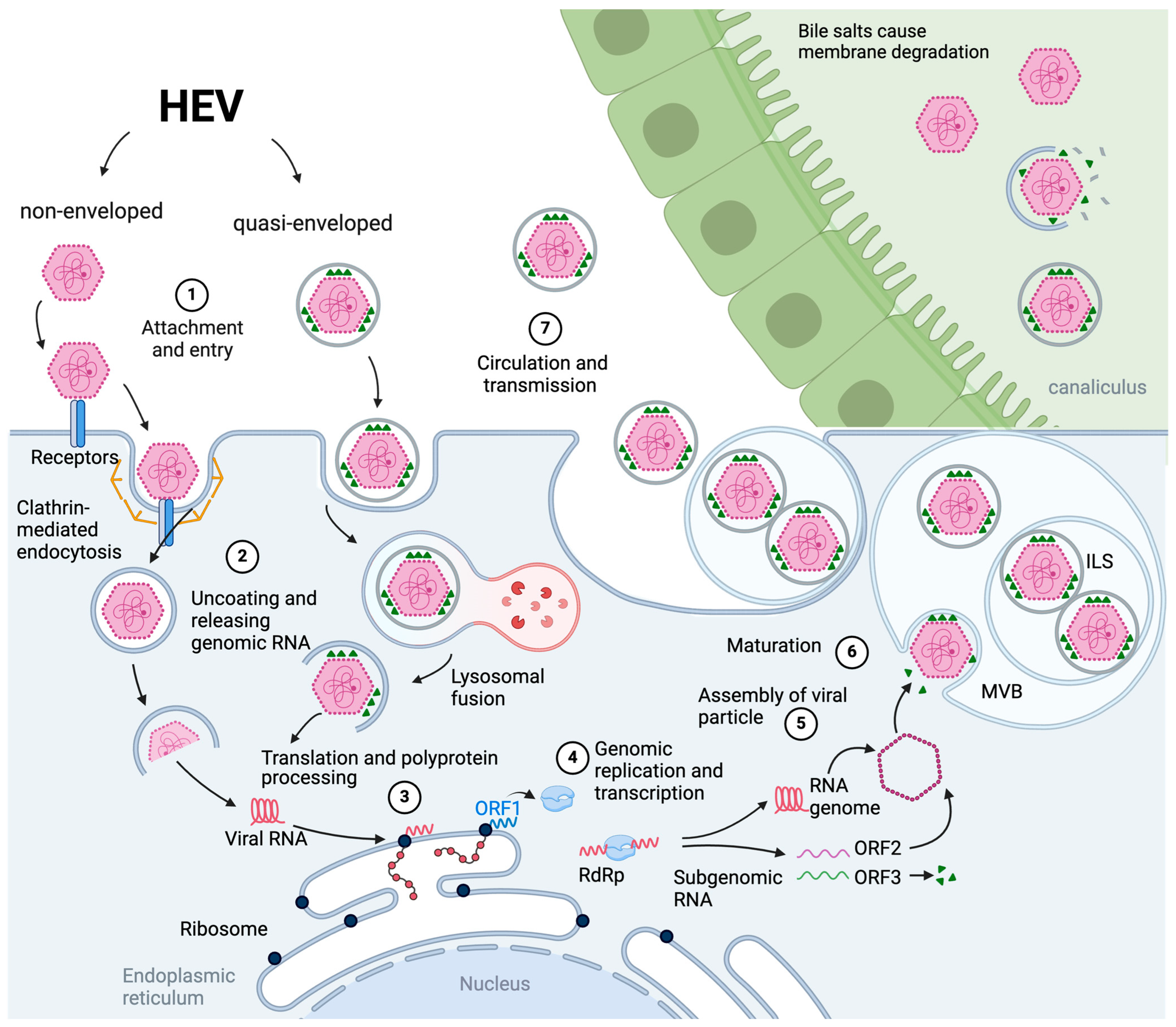
3.2. The Viral Cell Entry, Endosomal Transport and Uncoating
3.3. Virion Assembly and Infectious Particle Release
3.4. Therapeutic Options for HEV Infection
4. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nimgaonkar, I.; Ding, Q.; Schwartz, R.E.; Ploss, A. Hepatitis E Virus: Advances and Challenges. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 96–110. [Google Scholar] [CrossRef]
- Gupta, D.N.; Smetana, H.F. The Histopathology of Viral Hepatitis as Seen in the Delhi Epidemic (1955–56). Indian J. Med. Res. 1957, 45, 101–113. [Google Scholar]
- Tian, D.; Li, W.; Heffron, C.L.; Wang, B.; Mahsoub, H.M.; Sooryanarain, H.; Hassebroek, A.M.; Clark-Deener, S.; LeRoith, T.; Meng, X.-J. Hepatitis E Virus Infects Brain Microvascular Endothelial Cells, Crosses the Blood-Brain Barrier, and Invades the Central Nervous System. Proc. Natl. Acad. Sci. USA 2022, 119, e2201862119. [Google Scholar] [CrossRef]
- Rein, D.B.; Stevens, G.A.; Theaker, J.; Wittenborn, J.S.; Wiersma, S.T. The Global Burden of Hepatitis E Virus Genotypes 1 and 2 in 2005. Hepatology 2012, 55, 988–997. [Google Scholar] [CrossRef]
- Geng, Y.; Zhao, C.; Geng, K.; Wang, C.; Wang, X.; Liu, H.; Wang, Y. High Seroprevalence of Hepatitis E Virus in Rabbit Slaughterhouse Workers. Transbound. Emerg. Dis. 2019, 66, 1085–1089. [Google Scholar] [CrossRef]
- Halánová, M.; Veseliny, E.; Kalinová, Z.; Jarčuška, P.; Janičko, M.; Urbančíková, I.; Pella, D.; Dražilová, S.; Babinská, I.; HepaMeta Team. Seroprevalence of Hepatitis E Virus in Roma Settlements: A Comparison with the General Population in Slovakia. Int. J. Environ. Res. Public Health 2018, 15, 904. [Google Scholar] [CrossRef]
- Dalton, H.R.; Bendall, R.; Ijaz, S.; Banks, M. Hepatitis E: An Emerging Infection in Developed Countries. Lancet Infect. Dis. 2008, 8, 698–709. [Google Scholar] [CrossRef]
- Khuroo, M.S.; Khuroo, M.S.; Khuroo, N.S. Hepatitis E: Discovery, global impact, control and cure. World J. Gastroenterol. 2016, 22, 7030–7045. [Google Scholar] [CrossRef]
- Meng, X.J.; Purcell, R.H.; Halbur, P.G.; Lehman, J.R.; Webb, D.M.; Tsareva, T.S.; Haynes, J.S.; Thacker, B.J.; Emerson, S.U. A Novel Virus in Swine Is Closely Related to the Human Hepatitis E Virus. Proc. Natl. Acad. Sci. USA 1997, 94, 9860–9865. [Google Scholar] [CrossRef]
- Meng, X.-J. Expanding Host Range and Cross-Species Infection of Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005695. [Google Scholar] [CrossRef]
- Sridhar, S.; Yip, C.C.-Y.; Wu, S.; Chew, N.F.-S.; Leung, K.-H.; Chan, J.F.-W.; Zhao, P.S.; Chan, W.-M.; Poon, R.W.-S.; Tsoi, H.-W.; et al. Transmission of Rat Hepatitis E Virus Infection to Humans in Hong Kong: A Clinical and Epidemiological Analysis. Hepatology 2021, 73, 10–22. [Google Scholar] [CrossRef]
- Cangin, C.; Focht, B.; Harris, R.; Strunk, J.A. Hepatitis E Seroprevalence in the United States: Results for Immunoglobulins IGG and IGM. J. Med. Virol. 2019, 91, 124–131. [Google Scholar] [CrossRef]
- Sun, P.; Lin, S.; He, S.; Zhou, E.-M.; Zhao, Q. Avian Hepatitis E Virus: With the Trend of Genotypes and Host Expansion. Front. Microbiol. 2019, 10, 1696. [Google Scholar] [CrossRef]
- Ahmed, R.; Nasheri, N. Animal reservoirs for hepatitis E virus within the Paslahepevirus genus. Vet. Microbiol. 2023, 278, 109618. [Google Scholar] [CrossRef]
- Nicot, F.; Dimeglio, C.; Migueres, M.; Jeanne, N.; Latour, J.; Abravanel, F.; Ranger, N.; Harter, A.; Dubois, M.; Lameiras, S.; et al. Classification of the Zoonotic Hepatitis E Virus Genotype 3 Into Distinct Subgenotypes. Front. Microbiol. 2021, 11, 634430. [Google Scholar] [CrossRef] [PubMed]
- Andonov, A.; Robbins, M.; Borlang, J.; Cao, J.; Hatchette, T.; Stueck, A.; Deschambault, Y.; Murnaghan, K.; Varga, J.; Johnston, L. Rat Hepatitis E Virus Linked to Severe Acute Hepatitis in an Immunocompetent Patient. J. Infect. Dis. 2019, 220, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Balayan, M.S.; Andjaparidze, A.G.; Savinskaya, S.S.; Ketiladze, E.S.; Braginsky, D.M.; Savinov, A.P.; Poleschuk, V.F. Evidence for a Virus in Non-A, Non-B Hepatitis Transmitted via the Fecal-Oral Route. Intervirology 1983, 20, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Yadav, K.K.; Boley, P.A.; Fritts, Z.; Kenney, S.P. Ectopic Expression of Genotype 1 Hepatitis E Virus ORF4 Increases Genotype 3 HEV Viral Replication in Cell Culture. Viruses 2021, 13, 75. [Google Scholar] [CrossRef]
- Krawczynski, K.; Bradley, D.W. Enterically Transmitted Non-A, Non-B Hepatitis: Identification of Virus-Associated Antigen in Experimentally Infected Cynomolgus Macaques. J. Infect. Dis. 1989, 159, 1042–1049. [Google Scholar] [CrossRef]
- Yamashita, T.; Mori, Y.; Miyazaki, N.; Cheng, R.H.; Yoshimura, M.; Unno, H.; Shima, R.; Moriishi, K.; Tsukihara, T.; Li, T.C.; et al. Biological and Immunological Characteristics of Hepatitis E Virus-like Particles Based on the Crystal Structure. Proc. Natl. Acad. Sci. USA 2009, 106, 12986–12991. [Google Scholar] [CrossRef]
- Guu TS, Y.; Liu, Z.; Ye, Q.; Mata, D.A.; Li, K.; Yin, C.; Zhang, J.; Tao, Y.J. Structure of the Hepatitis E Virus-like Particle Suggests Mechanisms for Virus Assembly and Receptor Binding. Proc. Natl. Acad. Sci. USA 2009, 106, 12992–12997. [Google Scholar] [CrossRef] [PubMed]
- Primadharsini, P.P.; Nagashima, S.; Okamoto, H. Genetic Variability and Evolution of Hepatitis E Virus. Viruses 2019, 11, 456. [Google Scholar] [CrossRef]
- Purdy, M.A.; Drexler, J.F.; Meng, X.-J.; Norder, H.; Okamoto, H.; Van der Poel WH, M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Gorbalenya, A.E.; Purdy, M.A.; Rozanov, M.N.; Reyes, G.R.; Bradley, D.W. Computer-Assisted Assignment of Functional Domains in the Nonstructural Polyprotein of Hepatitis E Virus: Delineation of an Additional Group of Positive-Strand RNA Plant and Animal Viruses. Proc. Natl. Acad. Sci. USA 1992, 89, 8259–8263. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Meng, X.-J. Molecular Biology and Replication of Hepatitis E Virus. Emerg. Microbes Infect. 2012, 1, e17. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Ying, D.; Lhomme, S.; Tang, Z.; Walker, C.M.; Xia, N.; Zheng, Z.; Feng, Z. Origin, Antigenicity, and Function of a Secreted Form of ORF2 in Hepatitis E Virus Infection. Proc. Natl. Acad. Sci. USA 2018, 115, 4773–4778. [Google Scholar] [CrossRef] [PubMed]
- Hervouet, K.; Ferrié, M.; Ankavay, M.; Montpellier, C.; Camuzet, C.; Alexandre, V.; Dembélé, A.; Lecoeur, C.; Foe, A.T.; Bouquet, P.; et al. An Arginine-Rich Motif in the ORF2 capsid protein regulates the hepatitis E virus lifecycle and interactions with the host cell. PLoS Pathog. 2022, 18, e1010798. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Li, T.C.; Mayazaki, N.; Simon, M.N.; Wall, J.S.; Moore, M.; Wang, C.Y.; Takeda, N.; Wakita, T.; Miyamura, T.; et al. The structure of hepatitis E virion-sized particles reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 2010, 285, 33175–33183. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.J. Recent Advances in Hepatitis E Virus. J. Viral Hepat. 2010, 17, 153–161. [Google Scholar] [CrossRef]
- Ali, M.M.; Gul, M.; Imran, M.; Ijaz, M.; Azeem, S.; Ullah, A.; Yaqub, H.M.F. Molecular identification and genotyping of hepatitis E virus from Southern Punjab, Pakistan. Sci. Rep. 2024, 14, 223. [Google Scholar] [CrossRef]
- Ding, Q.; Heller, B.; Capuccino JM, V.; Song, B.; Nimgaonkar, I.; Hrebikova, G.; Contreras, J.E.; Ploss, A. Hepatitis E Virus ORF3 Is a Functional Ion Channel Required for Release of Infectious Particles. Proc. Natl. Acad. Sci. USA 2017, 114, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Shalimar; Nayak, B.; Ranjith Kumar, C.T.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005521. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Rivera-Serrano, E.E.; Yin, X.; Walker, C.M.; Feng, Z.; Lemon, S.M. Cell entry and release of quasi-enveloped human hepatitis viruses. Nat. Rev. Microbiol. 2023, 21, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Ambardekar, C.; Lu, Y.; Feng, Z. Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses. J. Virol. 2016, 90, 4232–4242. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Nishiyama, T.; Primadharsini, P.P.; Okamoto, H. Characterization of the Quasi-Enveloped Hepatitis E Virus Particles Released by the Cellular Exosomal Pathway. J. Virol. 2017, 91, e00822-17. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Jirintai, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Kouki, T.; Yashiro, T.; Okamoto, H. Hepatitis E Virus Egress Depends on the Exosomal Pathway, with Secretory Exosomes Derived from Multivesicular Bodies. J. Gen. Virol. 2014, 95 Pt 10, 2166–2175. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Tanaka, T.; Takahashi, H.; Hoshino, Y.; Nagashima, S.; Jirintai; Mizuo, H.; Yazaki, Y.; Takagi, T.; Azuma, M.; et al. Hepatitis E Virus (HEV) Strains in Serum Samples Can Replicate Efficiently in Cultured Cells despite the Coexistence of HEV Antibodies: Characterization of HEV Virions in Blood Circulation. J. Clin. Microbiol. 2010, 48, 1112–1125. [Google Scholar] [CrossRef]
- Okamoto, H. Culture Systems for Hepatitis E Virus. J. Gastroenterol. 2013, 48, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Yang, J.; Huang, W.; Harrison, T.J.; Zhou, Y.; Wen, Z.; Wang, Y. Virus Host Protein Interaction Network Analysis Reveals That the HEV ORF3 Protein May Interrupt the Blood Coagulation Process. PLoS ONE 2013, 8, e56320. [Google Scholar] [CrossRef]
- Subramani, C.; Nair, V.P.; Anang, S.; Mandal, S.D.; Pareek, M.; Kaushik, N.; Srivastava, A.; Saha, S.; Shalimar; Nayak, B.; et al. Host-Virus Protein Interaction Network Reveals the Involvement of Multiple Host Processes in the Life Cycle of Hepatitis E Virus. mSystems 2018, 3, e00135-17. [Google Scholar] [CrossRef]
- Fu, R.M.; Decker, C.C.; Dao Thi, V.L. Cell Culture Models for Hepatitis E Virus. Viruses 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed]
- Meister, T.L.; Bruening, J.; Todt, D.; Steinmann, E. Cell Culture Systems for the Study of Hepatitis E Virus. Antivir. Res. 2019, 163, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Marion, O.; Lhomme, S.; Nayrac, M.; Dubois, M.; Pucelle, M.; Requena, M.; Migueres, M.; Abravanel, F.; Peron, J.M.; Carrere, N.; et al. Hepatitis E Virus Replication in Human Intestinal Cells. Gut 2020, 69, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Drave, S.A.; Debing, Y.; Walter, S.; Todt, D.; Engelmann, M.; Friesland, M.; Wedemeyer, H.; Neyts, J.; Behrendt, P.; Steinmann, E. Extra-Hepatic Replication and Infection of Hepatitis E Virus in Neuronal-Derived Cells. J. Viral Hepat. 2016, 23, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Gouilly, J.; Chen, Q.; Siewiera, J.; Cartron, G.; Levy, C.; Dubois, M.; Al-Daccak, R.; Izopet, J.; Jabrane-Ferrat, N.; El Costa, H. Genotype Specific Pathogenicity of Hepatitis E Virus at the Human Maternal-Fetal Interface. Nat. Commun. 2018, 9, 4748. [Google Scholar] [CrossRef] [PubMed]
- Knegendorf, L.; Drave, S.A.; Dao Thi, V.L.; Debing, Y.; Brown RJ, P.; Vondran FW, R.; Resner, K.; Friesland, M.; Khera, T.; Engelmann, M.; et al. Hepatitis E Virus Replication and Interferon Responses in Human Placental Cells. Hepatol. Commun. 2018, 2, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.; Nguyen, H.T.; Torian, U.; Engle, R.E.; Faulk, K.; Dalton, H.R.; Bendall, R.P.; Keane, F.E.; Purcell, R.H.; Emerson, S.U. Cross-Species Infections of Cultured Cells by Hepatitis E Virus and Discovery of an Infectious Virus-Host Recombinant. Proc. Natl. Acad. Sci. USA 2011, 108, 2438–2443. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Huang, F.; Xu, L.; Lin, Z.; de Vrij FM, S.; Ayo-Martin, A.C.; van der Kroeg, M.; Zhao, M.; Yin, Y.; Wang, W.; et al. Hepatitis E Virus Infects Neurons and Brains. J. Infect. Dis. 2017, 215, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Bendall, R.; Legrand-Abravanel, F.; Xia, N.-S.; Ijaz, S.; Izopet, J.; Dalton, H.R. Hepatitis E. Lancet 2012, 379, 2477–2488. [Google Scholar] [CrossRef]
- Debing, Y.; Moradpour, D.; Neyts, J.; Gouttenoire, J. Update on Hepatitis E Virology: Implications for Clinical Practice. J. Hepatol. 2016, 65, 200–212. [Google Scholar] [CrossRef]
- Geng, Y.; Zhao, C.; Huang, W.; Harrison, T.J.; Zhang, H.; Geng, K.; Wang, Y. Detection and Assessment of Infectivity of Hepatitis E Virus in Urine. J. Hepatol. 2016, 64, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Abravanel, F.; Lhomme, S.; Rostaing, L.; Izopet, J. Hepatitis E Virus: Chronic Infection, Extra-Hepatic Manifestations, and Treatment. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 20–27. [Google Scholar] [CrossRef]
- Jemielity, S.; Wang, J.J.; Chan, Y.K.; Ahmed, A.A.; Li, W.; Monahan, S.; Bu, X.; Farzan, M.; Freeman, G.J.; Umetsu, D.T.; et al. TIM-Family Proteins Promote Infection of Multiple Enveloped Viruses through Virion-Associated Phosphatidylserine. PLoS Pathog. 2013, 9, e1003232. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Maury, W.; Lemon, S.M. TIM1 (HAVCR1): An Essential “Receptor” or an “Accessory Attachment Factor” for Hepatitis A Virus? J. Virol. 2019, 93, e01793-18. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, S.; Yang, C.; Wei, M.; Song, C.; Zheng, Z.; Gu, Y.; Du, H.; Zhang, J.; Xia, N. Homology Model and Potential Virus-Capsid Binding Site of a Putative HEV Receptor Grp78. J. Mol. Model. 2011, 17, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, Y.; Wen, Z.; Zhang, F.; Qi, Y.; Huang, W.; Zhang, H.; Wang, Y. Asialoglycoprotein Receptor Facilitates Infection of PLC/PRF/5 Cells by HEV through Interaction with ORF2. J. Med. Virol. 2016, 88, 2186–2195. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M.; Chandra, V.; Rahman, S.A.; Sehgal, D.; Jameel, S. Heparan Sulfate Proteoglycans Are Required for Cellular Binding of the Hepatitis E Virus ORF2 Capsid Protein and for Viral Infection. J. Virol. 2009, 83, 12714–12724. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Li, X.; Feng, Z. Role of Envelopment in the HEV Life Cycle. Viruses 2016, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Kapur, N.; Thakral, D.; Durgapal, H.; Panda, S.K. Hepatitis E Virus Enters Liver Cells through Receptor-Dependent Clathrin-Mediated Endocytosis. J. Viral Hepat. 2012, 19, 436–448. [Google Scholar] [CrossRef]
- Holla, P.; Ahmad, I.; Ahmed, Z.; Jameel, S. Hepatitis E Virus Enters Liver Cells through a Dynamin-2, Clathrin and Membrane Cholesterol-Dependent Pathway. Traffic 2015, 16, 398–416. [Google Scholar] [CrossRef]
- Chu, H.; Chan, C.-M.; Zhang, X.; Wang, Y.; Yuan, S.; Zhou, J.; Au-Yeung, R.K.-H.; Sze, K.-H.; Yang, D.; Shuai, H.; et al. Middle East Respiratory Syndrome Coronavirus and Bat Coronavirus HKU9 Both Can Utilize GRP78 for Attachment onto Host Cells. J. Biol. Chem. 2018, 293, 11709–11726. [Google Scholar] [CrossRef] [PubMed]
- Nain, M.; Mukherjee, S.; Karmakar, S.P.; Paton, A.W.; Paton, J.C.; Abdin, M.Z.; Basu, A.; Kalia, M.; Vrati, S. GRP78 Is an Important Host Factor for Japanese Encephalitis Virus Entry and Replication in Mammalian Cells. J. Virol. 2017, 91, e02274-16. [Google Scholar] [CrossRef] [PubMed]
- Triantafilou, K.; Fradelizi, D.; Wilson, K.; Triantafilou, M. GRP78, a Coreceptor for Coxsackievirus A9, Interacts with Major Histocompatibility Complex Class I Molecules Which Mediate Virus Internalization. J. Virol. 2002, 76, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Horie, M.; Daito, T.; Ikuta, K.; Tomonaga, K. Molecular Chaperone BiP Interacts with Borna Disease Virus Glycoprotein at the Cell Surface. J. Virol. 2009, 83, 12622–12625. [Google Scholar] [CrossRef] [PubMed]
- Fongsaran, C.; Jirakanwisal, K.; Kuadkitkan, A.; Wikan, N.; Wintachai, P.; Thepparit, C.; Ubol, S.; Phaonakrop, N.; Roytrakul, S.; Smith, D.R. Involvement of ATP Synthase β Subunit in Chikungunya Virus Entry into Insect Cells. Arch. Virol. 2014, 159, 3353–3364. [Google Scholar] [CrossRef]
- Ueda, K.; Suwanmanee, Y. ATP5B Is an Essential Factor for Hepatitis B Virus Entry. Int. J. Mol. Sci. 2022, 23, 9570. [Google Scholar] [CrossRef] [PubMed]
- Shiota, T.; Li, T.-C.; Nishimura, Y.; Yoshizaki, S.; Sugiyama, R.; Shimojima, M.; Saijo, M.; Shimizu, H.; Suzuki, R.; Wakita, T.; et al. Integrin A3 Is Involved in Non-Enveloped Hepatitis E Virus Infection. Virology 2019, 536, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Barth, H.; Schafer, C.; Adah, M.I.; Zhang, F.; Linhardt, R.J.; Toyoda, H.; Kinoshita-Toyoda, A.; Toida, T.; Van Kuppevelt, T.H.; Depla, E.; et al. Cellular Binding of Hepatitis C Virus Envelope Glycoprotein E2 Requires Cell Surface Heparan Sulfate. J. Biol. Chem. 2003, 278, 41003–41012. [Google Scholar] [CrossRef] [PubMed]
- Choy, M.Y.; Richman, P.I.; Horton, M.A.; MacDonald, T.T. Expression of the VLA Family of Integrins in Human Intestine. J. Pathol. 1990, 160, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Ye, X.; Simon, S. The Integrins. Genome Biol. 2007, 8, 215. [Google Scholar] [CrossRef]
- Oechslin, N.; Moradpour, D.; Gouttenoire, J. Hepatitis E Virus Finds Its Path through the Gut. Gut 2020, 69, 796–798. [Google Scholar] [CrossRef]
- de Melker, A.A.; Sterk, L.M.; Delwel, G.O.; Fles, D.L.; Daams, H.; Weening, J.J.; Sonnenberg, A. The A and B Variants of the Alpha 3 Integrin Subunit: Tissue Distribution and Functional Characterization. Lab. Investig. 1997, 76, 547–563. [Google Scholar]
- Volpes, R.; van den Oord, J.J.; Desmet, V.J. Distribution of the VLA Family of Integrins in Normal and Pathological Human Liver Tissue. Gastroenterology 1991, 101, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Volpes, R.; van den Oord, J.J.; Desmet, V.J. Integrins as Differential Cell Lineage Markers of Primary Liver Tumors. Am. J. Pathol. 1993, 142, 1483–1492. [Google Scholar]
- Dubland, J.A.; Francis, G.A. Lysosomal Acid Lipase: At the Crossroads of Normal and Atherogenic Cholesterol Metabolism. Front. Cell Dev. Biol. 2015, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Bubeck, D.; Filman, D.J.; Cheng, N.; Steven, A.C.; Hogle, J.M.; Belnap, D.M. The Structure of the Poliovirus 135S Cell Entry Intermediate at 10-Angstrom Resolution Reveals the Location of an Externalized Polypeptide That Binds to Membranes. J. Virol. 2005, 79, 7745–7755. [Google Scholar] [CrossRef] [PubMed]
- Cavaldesi, M.; Caruso, M.; Sthandier, O.; Amati, P.; Garcia, M.I. Conformational Changes of Murine Polyomavirus Capsid Proteins Induced by Sialic Acid Binding. J. Biol. Chem. 2004, 279, 41573–41579. [Google Scholar] [CrossRef]
- Sapp, M.; Bienkowska-Haba, M. Viral Entry Mechanisms: Human Papillomavirus and a Long Journey from Extracellular Matrix to the Nucleus. FEBS J. 2009, 276, 7206–7216. [Google Scholar] [CrossRef]
- Rivera-Serrano, E.E.; González-López, O.; Das, A.; Lemon, S.M. Cellular Entry and Uncoating of Naked and Quasi-Enveloped Human Hepatoviruses. Elife 2019, 8, e43983. [Google Scholar] [CrossRef]
- Tyagi, S.; Korkaya, H.; Zafrullah, M.; Jameel, S.; Lal, S.K. The Phosphorylated Form of the ORF3 Protein of Hepatitis E Virus Interacts with Its Non-Glycosylated Form of the Major Capsid Protein, ORF2. J. Biol. Chem. 2002, 277, 22759–22767. [Google Scholar] [CrossRef]
- Menzo, S.; Clementi, M.; Alfani, E.; Bagnarelli, P.; Iacovacci, S.; Manzin, A.; Dandri, M.; Natoli, G.; Levrero, M.; Carloni, G. Trans-Activation of Epidermal Growth Factor Receptor Gene by the Hepatitis B Virus X-Gene Product. Virology 1993, 196, 878–882. [Google Scholar] [CrossRef]
- Lupberger, J.; Zeisel, M.B.; Xiao, F.; Thumann, C.; Fofana, I.; Zona, L.; Davis, C.; Mee, C.J.; Turek, M.; Gorke, S.; et al. EGFR and EphA2 Are Host Factors for Hepatitis C Virus Entry and Possible Targets for Antiviral Therapy. Nat. Med. 2011, 17, 589–595. [Google Scholar] [CrossRef]
- Suppiah, S.; Zhou, Y.; Frey, T.K. Lack of processing of the expressed ORF1 gene product of hepatitis E virus. Virol. J. 2011, 8, 245. [Google Scholar] [CrossRef] [PubMed]
- Cancela, F.; Noceti, O.; Arbiza, J.; Mirazo, S. Structural aspects of hepatitis E virus. Arch. Virol. 2022, 167, 2457–2481. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, D.; Thomas, S.; Chakraborty, M.; Jameel, S. Expression and processing of the Hepatitis E virus ORF1 nonstructural polyprotein. Virol. J. 2006, 3, 38. [Google Scholar] [CrossRef]
- Graff, J.; Zhou, Y.-H.; Torian, U.; Nguyen, H.; St Claire, M.; Yu, C.; Purcell, R.H.; Emerson, S.U. Mutations within Potential Glycosylation Sites in the Capsid Protein of Hepatitis E Virus Prevent the Formation of Infectious Virus Particles. J. Virol. 2008, 82, 1185–1194. [Google Scholar] [CrossRef]
- Montpellier, C.; Wychowski, C.; Sayed, I.M.; Meunier, J.-C.; Saliou, J.-M.; Ankavay, M.; Bull, A.; Pillez, A.; Abravanel, F.; Helle, F.; et al. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology 2018, 154, 211–223.e8. [Google Scholar] [CrossRef] [PubMed]
- Ankavay, M.; Montpellier, C.; Sayed, I.M.; Saliou, J.-M.; Wychowski, C.; Saas, L.; Duvet, S.; Aliouat-Denis, C.-M.; Farhat, R.; de Masson d’Autume, V.; et al. New Insights into the ORF2 Capsid Protein, a Key Player of the Hepatitis E Virus Lifecycle. Sci. Rep. 2019, 9, 6243. [Google Scholar] [CrossRef]
- Lenggenhager, D.; Gouttenoire, J.; Malehmir, M.; Bawohl, M.; Honcharova-Biletska, H.; Kreutzer, S.; Semela, D.; Neuweiler, J.; Hürlimann, S.; Aepli, P.; et al. Visualization of Hepatitis E Virus RNA and Proteins in the Human Liver. J. Hepatol. 2017, 67, 471–479. [Google Scholar] [CrossRef]
- Jameel, S.; Zafrullah, M.; Ozdener, M.H.; Panda, S.K. Expression in Animal Cells and Characterization of the Hepatitis E Virus Structural Proteins. J. Virol. 1996, 70, 207–216. [Google Scholar] [CrossRef]
- Zafrullah, M.; Ozdener, M.H.; Kumar, R.; Panda, S.K.; Jameel, S. Mutational Analysis of Glycosylation, Membrane Translocation, and Cell Surface Expression of the Hepatitis E Virus ORF2 Protein. J. Virol. 1999, 73, 4074–4082. [Google Scholar] [CrossRef] [PubMed]
- Emerson, S.U.; Nguyen, H.; Torian, U.; Purcell, R.H. ORF3 Protein of Hepatitis E Virus Is Not Required for Replication, Virion Assembly, or Infection of Hepatoma Cells In Vitro. J. Virol. 2006, 80, 10457–10464. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Takahashi, M.; Hoshino, Y.; Takahashi, H.; Ichiyama, K.; Nagashima, S.; Tanaka, T.; Okamoto, H. ORF3 Protein of Hepatitis E Virus Is Essential for Virion Release from Infected Cells. J. Gen. Virol. 2009, 90 Pt 8, 1880–1891. [Google Scholar] [CrossRef] [PubMed]
- Capelli, N.; Marion, O.; Dubois, M.; Allart, S.; Bertrand-Michel, J.; Lhomme, S.; Abravanel, F.; Izopet, J.; Chapuy-Regaud, S. Vectorial Release of Hepatitis E Virus in Polarized Human Hepatocytes. J. Virol. 2019, 93, e01207-18. [Google Scholar] [CrossRef] [PubMed]
- Dao Thi, V.L.; Wu, X.; Belote, R.L.; Andreo, U.; Takacs, C.N.; Fernandez, J.P.; Vale-Silva, L.A.; Prallet, S.; Decker, C.C.; Fu, R.M.; et al. Stem Cell-Derived Polarized Hepatocytes. Nat. Commun. 2020, 11, 1677. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Lemon, S.M. Peek-a-Boo: Membrane Hijacking and the Pathogenesis of Viral Hepatitis. Trends Microbiol. 2014, 22, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Hurley, J.H. Proline-Rich Regions and Motifs in Trafficking: From ESCRT Interaction to Viral Exploitation. Traffic 2011, 12, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Radulovic, M.; Stenmark, H. The Many Functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Jirintai, S.; Tanaka, T.; Nishizawa, T.; Yasuda, J.; Okamoto, H. Tumour Susceptibility Gene 101 and the Vacuolar Protein Sorting Pathway Are Required for the Release of Hepatitis E Virions. J. Gen. Virol. 2011, 92 Pt 12, 2838–2848. [Google Scholar] [CrossRef]
- Nagashima, S.; Takahashi, M.; Jirintai, N.; Tanaka, T.; Yamada, K.; Nishizawa, T.; Okamoto, H. A PSAP Motif in the ORF3 Protein of Hepatitis E Virus Is Necessary for Virion Release from Infected Cells. J. Gen. Virol. 2011, 92 Pt 2, 269–278. [Google Scholar] [CrossRef]
- Feng, Z.; Hirai-Yuki, A.; McKnight, K.L.; Lemon, S.M. Naked Viruses That Aren’t Always Naked: Quasi-Enveloped Agents of Acute Hepatitis. Annu. Rev. Virol. 2014, 1, 539–560. [Google Scholar] [CrossRef] [PubMed]
- Zafrullah, M.; Ozdener, M.H.; Panda, S.K.; Jameel, S. The ORF3 Protein of Hepatitis E Virus Is a Phosphoprotein That Associates with the Cytoskeleton. J. Virol. 1997, 71, 9045–9053. [Google Scholar] [CrossRef] [PubMed]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Côté, M.; Rich, R.L.; et al. Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. HIV-1 and Ebola Virus Encode Small Peptide Motifs That Recruit Tsg101 to Sites of Particle Assembly to Facilitate Egress. Nat. Med. 2001, 7, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Jirintai, S.; Tanggis; Kobayashi, T.; Nishizawa, T.; Okamoto, H. The Membrane on the Surface of Hepatitis E Virus Particles Is Derived from the Intracellular Membrane and Contains Trans-Golgi Network Protein 2. Arch. Virol. 2014, 159, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, F.; Zhang, L.; Harrison, T.J.; Huang, W.; Zhao, C.; Kong, W.; Jiang, C.; Wang, Y. Hepatitis E Virus Produced from Cell Culture Has a Lipid Envelope. PLoS ONE 2015, 10, e0132503. [Google Scholar] [CrossRef] [PubMed]
- Gouttenoire, J.; Pollán, A.; Abrami, L.; Oechslin, N.; Mauron, J.; Matter, M.; Oppliger, J.; Szkolnicka, D.; Dao Thi, V.L.; van der Goot, F.G.; et al. Palmitoylation Mediates Membrane Association of Hepatitis E Virus ORF3 Protein and Is Required for Infectious Particle Secretion. PLoS Pathog. 2018, 14, e1007471. [Google Scholar] [CrossRef] [PubMed]
- Blaskovic, S.; Blanc, M.; van der Goot, F.G. What Does S-Palmitoylation Do to Membrane Proteins? FEBS J. 2013, 280, 2766–2774. [Google Scholar] [CrossRef] [PubMed]
- Chapuy-Regaud, S.; Dubois, M.; Plisson-Chastang, C.; Bonnefois, T.; Lhomme, S.; Bertrand-Michel, J.; You, B.; Simoneau, S.; Gleizes, P.-E.; Flan, B.; et al. Characterization of the Lipid Envelope of Exosome Encapsulated HEV Particles Protected from the Immune Response. Biochimie 2017, 141, 70–79. [Google Scholar] [CrossRef]
- Chahar, H.S.; Bao, X.; Casola, A. Exosomes and Their Role in the Life Cycle and Pathogenesis of RNA Viruses. Viruses 2015, 7, 3204–3225. [Google Scholar] [CrossRef]
- Kamar, N.; Dalton, H.R.; Abravanel, F.; Izopet, J. Hepatitis E Virus Infection. Clin. Microbiol. Rev. 2014, 27, 116–138. [Google Scholar] [CrossRef] [PubMed]
- Dao Thi, V.L.; Debing, Y.; Wu, X.; Rice, C.M.; Neyts, J.; Moradpour, D.; Gouttenoire, J. Sofosbuvir Inhibits Hepatitis E Virus Replication In Vitro and Results in an Additive Effect When Combined with Ribavirin. Gastroenterology 2016, 150, 82–85.e4. [Google Scholar] [CrossRef] [PubMed]
- Cornberg, M.; Koch, A.; Lohse, A.; Hardtke, S.; Manns, M.P.; Wedemeyer, H. Sofosbuvir Monotherapy Fails to Achieve HEV RNA Elimination in Patients with Chronic Hepatitis E—The HepNet SofE Pilot Study. J. Hepatol. 2020, 73, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Rostaing, L.; Abravanel, F.; Garrouste, C.; Lhomme, S.; Esposito, L.; Basse, G.; Cointault, O.; Ribes, D.; Nogier, M.B.; et al. Ribavirin Therapy Inhibits Viral Replication on Patients with Chronic Hepatitis E Virus Infection. Gastroenterology 2010, 139, 1612–1618. [Google Scholar] [CrossRef] [PubMed]
- Mallet, V.; Nicand, E.; Sultanik, P.; Chakvetadze, C.; Tessé, S.; Thervet, E.; Mouthon, L.; Sogni, P.; Pol, S. Brief Communication: Case Reports of Ribavirin Treatment for Chronic Hepatitis E. Ann. Intern. Med. 2010, 153, 85–89. [Google Scholar] [CrossRef]
- Alric, L.; Bonnet, D.; Beynes-Rauzy, O.; Izopet, J.; Kamar, N. Definitive Clearance of a Chronic Hepatitis E Virus Infection with Ribavirin Treatment. Am. J. Gastroenterol. 2011, 106, 1562–1563. [Google Scholar] [CrossRef]
- Chaillon, A.; Sirinelli, A.; De Muret, A.; Nicand, E.; d’Alteroche, L.; Goudeau, A. Sustained Virologic Response with Ribavirin in Chronic Hepatitis E Virus Infection in Heart Transplantation. J. Heart Lung Transplant. 2011, 30, 841–843. [Google Scholar] [CrossRef]
- de Niet, A.; Zaaijer, H.L.; ten Berge, I.; Weegink, C.J.; Reesink, H.W.; Beuers, U. Chronic Hepatitis E after Solid Organ Transplantation. Neth. J. Med. 2012, 70, 261–266. [Google Scholar] [PubMed]
- Del Bello, A.; Arné-Bes, M.C.; Lavayssière, L.; Kamar, N. Hepatitis E Virus-Induced Severe Myositis. J. Hepatol. 2012, 57, 1152–1153. [Google Scholar] [CrossRef]
- Pischke, S.; Stiefel, P.; Franz, B.; Bremer, B.; Suneetha, P.V.; Heim, A.; Ganzenmueller, T.; Schlue, J.; Horn-Wichmann, R.; Raupach, R.; et al. Chronic Hepatitis E in Heart Transplant Recipients. Am. J. Transplant. 2012, 12, 3128–3133. [Google Scholar] [CrossRef]
- Hajji, H.; Gérolami, R.; Solas, C.; Moreau, J.; Colson, P. Chronic Hepatitis E Resolution in a Human Immunodeficiency Virus (HIV)-Infected Patient Treated with Ribavirin. Int. J. Antimicrob. Agents 2013, 41, 595–597. [Google Scholar] [CrossRef]
- Junge, N.; Pischke, S.; Baumann, U.; Goldschmidt, I.; Manns, M.; Wedemeyer, H.; Pfister, E.-D. Results of Single-Center Screening for Chronic Hepatitis E in Children after Liver Transplantation and Report on Successful Treatment with Ribavirin. Pediatr. Transplant. 2013, 17, 343–347. [Google Scholar] [CrossRef]
- Koning, L.; Pas, S.D.; de Man, R.A.; Balk, A.H.M.M.; de Knegt, R.J.; ten Kate, F.J.; Osterhaus, A.D.M.E.; van der Eijk, A.A. Clinical Implications of Chronic Hepatitis E Virus Infection in Heart Transplant Recipients. J. Heart Lung Transplant. 2013, 32, 78–85. [Google Scholar] [CrossRef]
- Neukam, K.; Barreiro, P.; Macías, J.; Avellón, A.; Cifuentes, C.; Martín-Carbonero, L.; Echevarría, J.M.; Vargas, J.; Soriano, V.; Pineda, J.A. Chronic Hepatitis E in HIV Patients: Rapid Progression to Cirrhosis and Response to Oral Ribavirin. Clin. Infect. Dis. 2013, 57, 465–468. [Google Scholar] [CrossRef]
- Pischke, S.; Hardtke, S.; Bode, U.; Birkner, S.; Chatzikyrkou, C.; Kauffmann, W.; Bara, C.L.; Gottlieb, J.; Wenzel, J.; Manns, M.P.; et al. Ribavirin Treatment of Acute and Chronic Hepatitis E: A Single-Centre Experience. Liver Int. 2013, 33, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Riezebos-Brilman, A.; Puchhammer-Stöckl, E.; van der Weide, H.Y.; Haagsma, E.B.; Jaksch, P.; Bejvl, I.; Niesters, H.G.; Verschuuren, E.A.M. Chronic Hepatitis E Infection in Lung Transplant Recipients. J. Heart Lung Transplant. 2013, 32, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Schlevogt, B.; Kinast, V.; Reusch, J.; Kerkhoff, A.; Praditya, D.; Todt, D.; Schmidt, H.H.; Steinmann, E.; Behrendt, P. Chronic Hepatitis E Virus Infection during Lymphoplasmacytic Lymphoma and Ibrutinib Treatment. Pathogens 2019, 8, 129. [Google Scholar] [CrossRef]
- Hooda, P.; Chaudhary, M.; Parvez, M.K.; Sinha, N.; Sehgal, D. Inhibition of Hepatitis E Virus Replication by Novel Inhibitor Targeting Methyltransferase. Viruses 2022, 14, 1778. [Google Scholar] [CrossRef] [PubMed]
- Bhise, N.; Agarwal, M.; Thakur, N.; Akshay, P.S.; Cherian, S.; Lole, K. Repurposing of Artesunate, an Antimalarial Drug, as a Potential Inhibitor of Hepatitis E Virus. Arch. Virol. 2023, 168, 147. [Google Scholar] [CrossRef] [PubMed]
- Feld, J.J.; Hoofnagle, J.H. Mechanism of Action of Interferon and Ribavirin in Treatment of Hepatitis C. Nature 2005, 436, 967–972. [Google Scholar] [CrossRef]
- Nasir, M.; Wu, G.Y. HEV and HBV Dual Infection: A Review. J. Clin. Transl. Hepatol. 2020, 8, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, M.P.; Scott, R.M.; Joshi, D.M.; Mammen, M.P., Jr.; Thapa, G.B.; Thapa, N.; Myint, K.S.; Fourneau, M.; Kuschner, R.A.; Shrestha, S.K.; et al. Safety and efficacy of a recombinant hepatitis E vaccine. New Engl. J. Med. 2007, 356, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, J.; Xia, N. Lessons from hepatitis E vaccine design. Curr. Opin. Virol. 2015, 11, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, F.; Alberti, F.; Russo, C.; Cursaro, C.; Seferi, H.; Margotti, M.; Andreone, P. Treatment Options for Hepatitis A and E: A Non-Systematic Review. Viruses 2023, 15, 1080. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.F.; Huang, S.J.; Wu, T.; Hu, Y.M.; Wang, Z.Z.; Wang, H.; Jiang, H.M.; Wang, Y.J.; Yan, Q.; et al. Long-term efficacy of a hepatitis E vaccine. New Engl. J. Med. 2015, 372, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.Y.; Huang, S.J.; Guo, M.; Zhao, J.; Yu, H.; He, W.G.; Jiang, H.M.; Wang, Y.J.; Zhang, X.F.; Cai, J.P.; et al. Persistence of antibodies acquired by natural hepatitis E virus infection and effects of vaccination. Clin. Microbiol.Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2017, 23, 336.e1–336.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.C.; Zhang, J.; Zhang, X.F.; Zhou, C.; Wang, Z.Z.; Huang, S.J.; Wang, H.; Yang, C.L.; Jiang, H.M.; Cai, J.P.; et al. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: A large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet 2010, 376, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, X.; Zhang, Z.; Li, S.; Zhang, J.; Xia, N.; Zhao, Q. Prophylactic Hepatitis E Vaccines: Antigenic Analysis and Serological Evaluation. Viruses 2020, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Dao Thi, V.L.; Liu, P.; Takacs, C.N.; Xiang, K.; Andrus, L.; Gouttenoire, J.; Moradpour, D.; Rice, C.M. Pan-Genotype Hepatitis E Virus Replication in Stem Cell-Derived Hepatocellular Systems. Gastroenterology 2018, 154, 663–674.e7. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.; Hafirassou, M.L.; Couderc, T.; Bonnet-Madin, L.; Kril, V.; Kümmerer, B.M.; Labeau, A.; Brugier, A.; Simon-Loriere, E.; Burlaud-Gaillard, J.; et al. FHL1 Is a Major Host Factor for Chikungunya Virus Infection. Nature 2019, 574, 259–263. [Google Scholar] [CrossRef]
- Zhang, R.; Kim, A.S.; Fox, J.M.; Nair, S.; Basore, K.; Klimstra, W.B.; Rimkunas, R.; Fong, R.H.; Lin, H.; Poddar, S.; et al. Mxra8 Is a Receptor for Multiple Arthritogenic Alphaviruses. Nature 2018, 557, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; tenOever, B.; Manicassamy, B. Genome-Wide CRISPR/Cas9 Screen Identifies Host Factors Essential for Influenza Virus Replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.B.; Ohlson, M.B.; Eitson, J.L.; Kumar, A.; McDougal, M.B.; Boys, I.N.; Mar, K.B.; De La Cruz-Rivera, P.C.; Douglas, C.; Konopka, G.; et al. A CRISPR Screen Identifies IFI6 as an ER-Resident Interferon Effector That Blocks Flavivirus Replication. Nat. Microbiol. 2018, 3, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Flint, M.; Chatterjee, P.; Lin, D.L.; McMullan, L.K.; Shrivastava-Ranjan, P.; Bergeron, É.; Lo, M.K.; Welch, S.R.; Nichol, S.T.; Tai, A.W.; et al. A Genome-Wide CRISPR Screen Identifies N-Acetylglucosamine-1-Phosphate Transferase as a Potential Antiviral Target for Ebola Virus. Nat. Commun. 2019, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- V’kovski, P.; Gerber, M.; Kelly, J.; Pfaender, S.; Ebert, N.; Braga Lagache, S.; Simillion, C.; Portmann, J.; Stalder, H.; Gaschen, V.; et al. Determination of Host Proteins Composing the Microenvironment of Coronavirus Replicase Complexes by Proximity-Labeling. Elife 2019, 8, e42037. [Google Scholar] [CrossRef]
| Author | Year | Study Type | Immunosuppressed Patient Group | N. of pts | HEV Genotype (N. pts) | Drug | Mechanism of Action | Dosage/Daily | Treatment Duration (mo) | Virological Response, % | Adverse Events |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kamar [114] | 2010 | Pilot study | Kidney transplantation/simultaneous kidney-pancreas transplantation | 6 | 3f (4), 3 c (2) | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3 | 100 | Anemia |
| Mallet [115] | 2010 | Case report | Simultaneous kidney-pancreas transplantation, hematological disease | 2 | 3f (1), 3c (1) | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3 | 100 | Not relevant |
| Alric [116] | 2011 | Case report | Hematological disease | 1 | 3c | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3 | 100 | Not relevant |
| Chaillon [117] | 2011 | Case report | Heart transplantation | 1 | 3c | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3 | 100 | Well-tolerated anemia |
| de Niet [118] | 2012 | Case report | Kidney transplantation | 1 | 3 | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3 | 100 | Not relevant |
| Del Bello [119] | 2012 | Case report | Liver transplantation | 1 | 3f | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3 | 100 | Not relevant |
| Pischke [120] | 2012 | Prospective | Heart transplantation | 4 | n.a. | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 5 | 75 | Not relevant |
| Hajji [121] | 2013 | Case report | HIV | 1 | 3f | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3 | 100 | Not relevant |
| Junge [122] | 2013 | Case report | Liver transplantation | 1 | n.a. | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 6 | 100 | Not relevant |
| Koning [123] | 2013 | Retrospective | Heart transplantation | 4 | 3 | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 3–12 | 75 | Not relevant |
| Neukam [124] | 2013 | Case report | HIV | 1 | 3 | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 6 | 100 | Not relevant |
| Pischke [125] | 2013 | Prospective case series | kidney transplantation, heart transplantation, lung transplantation | 11 | n.a. | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 5 | 82 | Not relevant |
| Riezebos-Brilman [126] | 2013 | Case report | Lung transplantation | 2 | 3 | RBV monotherapy | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 4 | 100 | Not relevant |
| Schlevogt B [127] | 2019 | Case report + in vitro studies (hepatoma cell line HepG2) | Lymphoplasmacytic lymphoma | 1 | 3c | RBV | Guanosine nucleoside analog (viral polymerase inhibitor) | 600–1000 mg | 2 | 100 | Severe exanthema |
| 1–3 | Ibrutinib | BTK inhibitor | 3.33 microM | 48 h | Moderate antiviral effect against HEV replicating isolates | No cytotoxicity | |||||
| Hooda P [128] | 2022 | In silico (Docking analysis) | n.a. | n.a. | N.A. | HPPA | HEV MTase inhibitor | 800 M | n.a. | HEV-RNA copies decreased significantly from ~3.2 × 106 in untreated cells to ~4.3 × 102.8 copies | Because the homology between human and HEV-MTase is so low, HPPA is unlikely to affect the host Mtase |
| Bhise N [129] | 2023 | Drug repurposing strategy: experimental + in silico (Docking analysis) | n.a. | n.a. | 1–3 | ART | (A) binding to the HEV helicase active site, potentially affecting ATP hydrolysis activity; (B) inhibition of the HEV RdRp | 19.5 μM (EC50) and 78 μM | n.a. | (A) ART showed a 24% and 55% inhibition of the helicase; (B) ART showed 26% and 40% inhibition of the RdRp activity | In silico toxicity studies revealed no risk of reproductive or developmental toxicity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahini, E.; Argentiero, A.; Andriano, A.; Losito, F.; Maida, M.; Facciorusso, A.; Cozzolongo, R.; Villa, E. Hepatitis E Virus: What More Do We Need to Know? Medicina 2024, 60, 998. https://doi.org/10.3390/medicina60060998
Shahini E, Argentiero A, Andriano A, Losito F, Maida M, Facciorusso A, Cozzolongo R, Villa E. Hepatitis E Virus: What More Do We Need to Know? Medicina. 2024; 60(6):998. https://doi.org/10.3390/medicina60060998
Chicago/Turabian StyleShahini, Endrit, Antonella Argentiero, Alessandro Andriano, Francesco Losito, Marcello Maida, Antonio Facciorusso, Raffaele Cozzolongo, and Erica Villa. 2024. "Hepatitis E Virus: What More Do We Need to Know?" Medicina 60, no. 6: 998. https://doi.org/10.3390/medicina60060998






