Investigating the Role of the Zinc Finger Protein ZC2HC1C on Autism Spectrum Disorder Susceptibility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Next-Generation Sequencing
2.2. Data Analysis
3. Results
3.1. Clinical Report
3.2. Diagnostic Evaluations
- Mullen Scales of Early Learning (MSEL): At 3 years of age, scores were significantly below average in expressive and receptive language domains (expressive language age equivalent of 18 months, receptive language age equivalent of 21 months). Fine and gross motor skills were slightly delayed but within the lower range of normal (age equivalent 30–33 months).
- Autism Diagnostic Observation Schedule, Second Edition (ADOS-2): The patient had a score within the autism spectrum range with deficits in communication, social interaction, and play behaviors. At the ADOS examination, the patient was placed at Level 2 (phrase speech).
- Vineland Adaptive Behavior Scales, Third Edition (VABS-3): The patient showed delays in adaptive functioning, particularly in the communication and socialization domains. The adaptive behavior composite was significantly below average, with an age equivalent of 2 years in communication and personal–social skills.
3.3. Ongoing Management
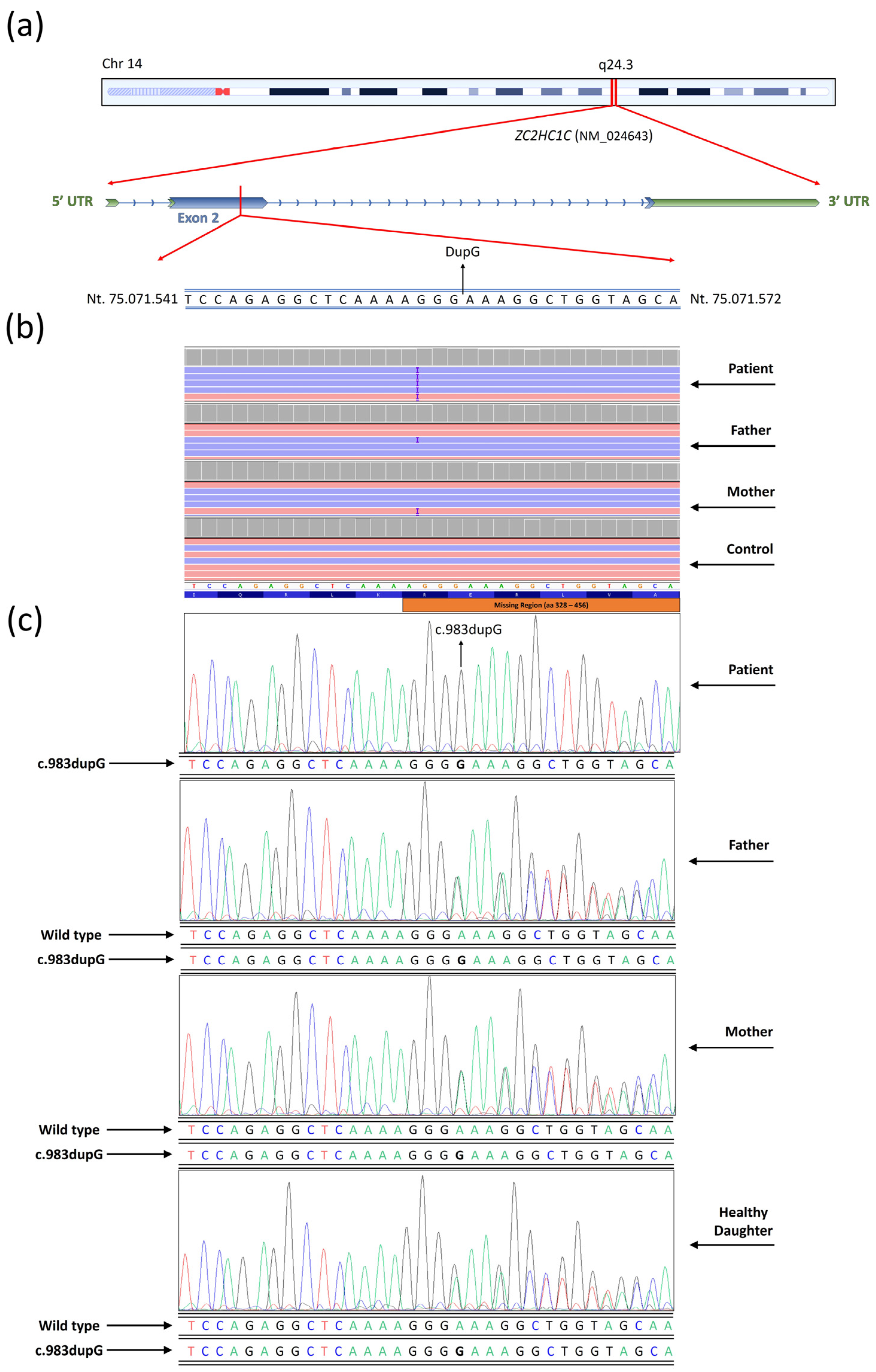
3.4. Next Generation Sequencing
3.5. Allele Frequency Prediction and Bayes Statistical Probability Model
4. Discussion
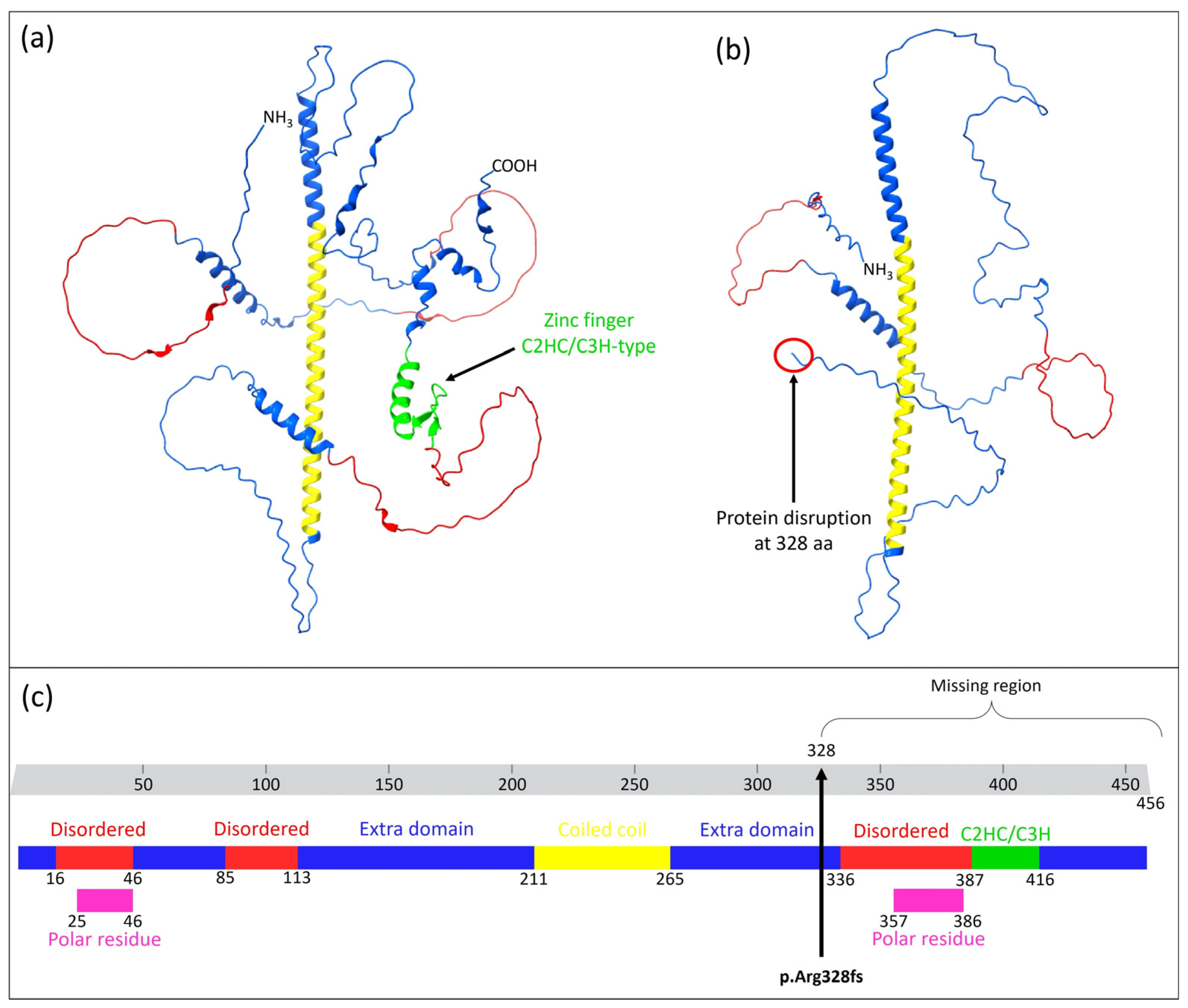
4.1. Patient’s Phenotype and Variant Identification
4.2. Allele Frequency and Bayes Statistical Model
4.3. Protein–Protein Interaction Prediction
4.4. Further Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD | Autism spectrum disorder |
| AR | Autosomal recessive |
| CNVs | Copy number variants |
| EEG | Electroencephalogram |
| ESDM | Early Start Denver Model |
| ITF | Intraflagellar transport |
| NGS | Next-generation sequencing |
| WES | Whole-exome sequencing |
References
- Bu, S.; Lv, Y.; Liu, Y.; Qiao, S.; Wang, H. Zinc Finger Proteins in Neuro-Related Diseases Progression. Front. Neurosci. 2021, 15, 760567. [Google Scholar] [CrossRef] [PubMed]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-Finger Proteins in Health and Disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.A.; Yates, B.; Seal, R.L.; Wright, M.W.; Bruford, E.A. Genenames.Org: The HGNC Resources in 2015. Nucleic Acids Res. 2015, 43, D1079–D1085. [Google Scholar] [CrossRef] [PubMed]
- Anitha, A.; Thanseem, I.; Nakamura, K.; Vasu, M.M.; Yamada, K.; Ueki, T.; Iwayama, Y.; Toyota, T.; Tsuchiya, K.J.; Iwata, Y.; et al. Zinc Finger Protein 804A (ZNF804A) and Verbal Deficits in Individuals with Autism. J. Psychiatry Neurosci. 2014, 39, 294–303. [Google Scholar] [CrossRef]
- Jones, K.A.; Luo, Y.; Dukes-Rimsky, L.; Srivastava, D.P.; Koul-Tewari, R.; Russell, T.A.; Shapiro, L.P.; Srivastava, A.K.; Penzes, P. Neurodevelopmental Disorder-Associated ZBTB20 Gene Variants Affect Dendritic and Synaptic Structure. PLoS ONE 2018, 13, e0203760. [Google Scholar] [CrossRef]
- Al-Naama, N.; Mackeh, R.; Kino, T. C2H2-Type Zinc Finger Proteins in Brain Development, Neurodevelopmental, and Other Neuropsychiatric Disorders: Systematic Literature-Based Analysis. Front. Neurol. 2020, 11, 32. [Google Scholar] [CrossRef]
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive Zinc-Binding Domains in the Protein Transcription Factor IIIA from Xenopus Oocytes. EMBO J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef]
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M. The Dof Domain, a Zinc Finger DNA-binding Domain Conserved Only in Higher Plants, Truly Functions as a Cys2/Cys2 Zn Finger Domain. Plant J. 2004, 37, 741–749. [Google Scholar] [CrossRef]
- Ghosh, S.; Chatterji, D. Two Zinc Finger Proteins from Mycobacterium Smegmatis: DNA Binding and Activation of Transcription. Genes Cells 2017, 22, 699–714. [Google Scholar] [CrossRef]
- Zhou, C.; Li, L.-Y.; Lu, G.-X. Molecular Cloning and Character Analysis of the Mouse Zinc Finger Protein Gene Zfp474 Exclusively Expressed in Testis and Ovary. Yi Chuan Xue Bao 2005, 32, 155–162. [Google Scholar]
- Redon, E.; Bosseboeuf, A.; Rocancourt, C.; Da Silva, C.; Wincker, P.; Mazan, S.; Sourdaine, P. Stage-Specific Gene Expression during Spermatogenesis in the Dogfish (Scyliorhinus canicula). Reproduction 2010, 140, 57–71. [Google Scholar] [CrossRef]
- McClintock, T.S.; Glasser, C.E.; Bose, S.C.; Bergman, D.A. Tissue Expression Patterns Identify Mouse Cilia Genes. Physiol. Genom. 2008, 32, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Shao, T.; Chen, Y.; Chen, J.; Sun, X.; Chen, X. Screening of Candidate Genes at GLC3B and GLC3C Loci in Chinese Primary Congenital Glaucoma Patients with Targeted next Generation Sequencing. Ophthalmic Genet. 2023, 44, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Wang, D.; Shen, X.; Qiu, J.; Wu, L.; Yan, J.; Ji, H. Identification of LncRNA Expression Profiles and Analysis of CeRNA in the Hippocampus of Perinatal Glyphosate-exposed Mice. Int. J. Dev. Neurosci. 2021, 81, 312–323. [Google Scholar] [CrossRef]
- Liu, S.; Trupiano, M.X.; Simon, J.; Guo, J.; Anton, E.S. The Essential Role of Primary Cilia in Cerebral Cortical Development and Disorders. Curr. Top. Dev. Biol. 2021, 142, 99–146. [Google Scholar]
- Yin, W.; Yang, T.; Wan, G.; Zhou, X. Identification of Image Genetic Biomarkers of Alzheimer’s Disease by Orthogonal Structured Sparse Canonical Correlation Analysis Based on a Diagnostic Information Fusion. Math. Biosci. Eng. 2023, 20, 16648–16662. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual Proteome-Scale Networks Reveal Cell-Specific Remodeling of the Human Interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef]
- Shi, L.; Shi, X.; Shen, Y. Intraflagellar Transport 46 (IFT46) Is Essential for Trafficking IFT Proteins between Cilia and Cytoplasm in Paramecium. Sci. Rep. 2018, 8, 9259. [Google Scholar] [CrossRef]
- Baker, K.; Beales, P.L. Abnormalities of the Central Nervous System Across the Ciliopathy Spectrum. In Cilia and Nervous System Development and Function; Springer: Dordrecht, The Netherlands, 2013; pp. 229–273. [Google Scholar]
- Trulioff, A.; Ermakov, A.; Malashichev, Y. Primary Cilia as a Possible Link between Left-Right Asymmetry and Neurodevelopmental Diseases. Genes 2017, 8, 48. [Google Scholar] [CrossRef]
- Alhassen, W.; Chen, S.; Vawter, M.; Robbins, B.K.; Nguyen, H.; Myint, T.N.; Saito, Y.; Schulmann, A.; Nauli, S.M.; Civelli, O.; et al. Patterns of Cilia Gene Dysregulations in Major Psychiatric Disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 109, 110255. [Google Scholar] [CrossRef]
- Karalis, V.; Donovan, K.E.; Sahin, M. Primary Cilia Dysfunction in Neurodevelopmental Disorders beyond Ciliopathies. J. Dev. Biol. 2022, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Calì, F.; Vinci, M.; Treccarichi, S.; Papa, C.; Gloria, A.; Musumeci, A.; Federico, C.; Vitello, G.A.; Nicotera, A.G.; Di Rosa, G.; et al. PLEKHG1: New Potential Candidate Gene for Periventricular White Matter Abnormalities. Genes 2024, 15, 1096. [Google Scholar] [CrossRef] [PubMed]
- Treccarichi, S.; Failla, P.; Vinci, M.; Musumeci, A.; Gloria, A.; Vasta, A.; Calabrese, G.; Papa, C.; Federico, C.; Saccone, S.; et al. UNC5C: Novel Gene Associated with Psychiatric Disorders Impacts Dysregulation of Axon Guidance Pathways. Genes 2024, 15, 306. [Google Scholar] [CrossRef]
- Desvignes, J.-P.; Bartoli, M.; Delague, V.; Krahn, M.; Miltgen, M.; Béroud, C.; Salgado, D. VarAFT: A Variant Annotation and Filtration System for Human next Generation Sequencing Data. Nucleic Acids Res. 2018, 46, W545–W553. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The Human Genomic Variant Search Engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Mayo, O. A Century of Hardy–Weinberg Equilibrium. Twin Res. Hum. Genet. 2008, 11, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.; Gaunt, T.R.; Day, I.N.M. Hardy-Weinberg Equilibrium Testing of Biological Ascertainment for Mendelian Randomization Studies. Am. J. Epidemiol. 2009, 169, 505–514. [Google Scholar] [CrossRef]
- Stephens, M.; Balding, D.J. Bayesian Statistical Methods for Genetic Association Studies. Nat. Rev. Genet. 2009, 10, 681–690. [Google Scholar] [CrossRef]
- Scattoni, M.L.; Fatta, L.M.; Micai, M.; Sali, M.E.; Bellomo, M.; Salvitti, T.; Fulceri, F.; Castellano, A.; Molteni, M.; Gambino, G.; et al. Autism Spectrum Disorder Prevalence in Italy: A Nationwide Study Promoted by the Ministry of Health. Child Adolesc. Psychiatry Ment. Health 2023, 17, 125. [Google Scholar] [CrossRef]
- Hubbard, R.E.; Kamran Haider, M. Hydrogen Bonds in Proteins: Role and Strength. In Encyclopedia of Life Sciences; Wiley: Hoboken, NJ, USA, 2010. [Google Scholar]
- Scheiner, S. Forty Years of Progress in the Study of the Hydrogen Bond. Struct. Chem. 2019, 30, 1119–1128. [Google Scholar] [CrossRef]
- Kemp, M.T.; Lewandowski, E.M.; Chen, Y. Low Barrier Hydrogen Bonds in Protein Structure and Function. Biochim. Biophys. Acta BBA Proteins Proteom. 2021, 1869, 140557. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, M.; Saito, Y.; Takasaki, A.; Nakano, K.; Suzuki, C.; Kawamura, N.; Hattori, A.; Nagashima, S.; Oikawa, M.; Yanagi, S.; et al. Role of Immature Choroid Plexus in the Pathology of Autism Spectrum Disorder. bioRxiv 2023. [Google Scholar] [CrossRef]
- Pavăl, D.; Micluția, I.V. The Dopamine Hypothesis of Autism Spectrum Disorder Revisited: Current Status and Future Prospects. Dev. Neurosci. 2021, 43, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, S.; Dianzani, I.; Lattanzio, P.; Spada, M.; Romano, V.; Calì, F.; Andria, G.; Ponzone, A.; Marra, E.; Piazza, A. Genetic Heterogeneity in Five Italian Regions: Analysis of PAH Mutations and Minihaplotypes. Hum. Hered. 2001, 52, 154–159. [Google Scholar] [CrossRef]
- Mirisola, M.G.; Cali, F.; Gloria, A.; Schinocca, P.; D’Amato, M.; Cassara, G.; De Leo, G.; Palillo, L.; Meli, C.; Romano, V. PAH Gene Mutations in the Sicilian Population: Association with Minihaplotypes and Expression Analysis. Mol. Genet. Metab. 2001, 74, 353–361. [Google Scholar] [CrossRef]
- Tsunematsu, R.; Nishiyama, M.; Kotoshiba, S.; Saiga, T.; Kamura, T.; Nakayama, K.I. Fbxw8 Is Essential for Cul1-Cul7 Complex Formation and for Placental Development. Mol. Cell Biol. 2006, 26, 6157–6169. [Google Scholar] [CrossRef]
- Tsutsumi, T.; Kuwabara, H.; Arai, T.; Xiao, Y.; DeCaprio, J.A. Disruption of the Fbxw8 Gene Results in Pre- and Postnatal Growth Retardation in Mice. Mol. Cell Biol. 2008, 28, 743–751. [Google Scholar] [CrossRef]
- Livesay, S.B.; Collier, S.E.; Bitton, D.A.; Bähler, J.; Ohi, M.D. Structural and Functional Characterization of the N Terminus of Schizosaccharomyces Pombe Cwf10. Eukaryot. Cell. 2013, 12, 1472–1489. [Google Scholar] [CrossRef]
- Mishra, S.K.; Ammon, T.; Popowicz, G.M.; Krajewski, M.; Nagel, R.J.; Ares, M.; Holak, T.A.; Jentsch, S. Role of the Ubiquitin-like Protein Hub1 in Splice-Site Usage and Alternative Splicing. Nature 2011, 474, 173–178. [Google Scholar] [CrossRef]
- Krogan, N.J.; Cagney, G.; Yu, H.; Zhong, G.; Guo, X.; Ignatchenko, A.; Li, J.; Pu, S.; Datta, N.; Tikuisis, A.P.; et al. Global Landscape of Protein Complexes in the Yeast Saccharomyces Cerevisiae. Nature 2006, 440, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D.; et al. A Global Genetic Interaction Network Maps a Wiring Diagram of Cellular Function. Science 2016, 353, aaf1420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Rigo, N.; Dybkov, O.; Fourmann, J.-B.; Will, C.L.; Kumar, V.; Urlaub, H.; Stark, H.; Lührmann, R. Structural Insights into How Prp5 Proofreads the Pre-MRNA Branch Site. Nature 2021, 596, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Lucker, B.F.; Miller, M.S.; Dziedzic, S.A.; Blackmarr, P.T.; Cole, D.G. Direct Interactions of Intraflagellar Transport Complex B Proteins IFT88, IFT52, and IFT46. J. Biol. Chem. 2010, 285, 21508–21518. [Google Scholar] [CrossRef]
- Baron Gaillard, C.L.; Pallesi-Pocachard, E.; Massey-Harroche, D.; Richard, F.; Arsanto, J.-P.; Chauvin, J.-P.; Lecine, P.; Krämer, H.; Borg, J.-P.; Le Bivic, A. Hook2 Is Involved in the Morphogenesis of the Primary Cilium. Mol. Biol. Cell 2011, 22, 4549–4562. [Google Scholar] [CrossRef]
- Lee, M.-S.; Hwang, K.-S.; Oh, H.-W.; Ji-Ae, K.; Kim, H.-T.; Cho, H.-S.; Lee, J.-J.; Yeong Ko, J.; Choi, J.-H.; Jeong, Y.-M.; et al. IFT46 Plays an Essential Role in Cilia Development. Dev. Biol. 2015, 400, 248–257. [Google Scholar] [CrossRef]
- Schwarz, T.; Prieler, B.; Schmid, J.A.; Grzmil, P.; Neesen, J. Ccdc181 Is a Microtubule-Binding Protein That Interacts with Hook1 in Haploid Male Germ Cells and Localizes to the Sperm Tail and Motile Cilia. Eur. J. Cell Biol. 2017, 96, 276–288. [Google Scholar] [CrossRef]
- Yang, W.-T.; Hong, S.-R.; He, K.; Ling, K.; Shaiv, K.; Hu, J.; Lin, Y.-C. The Emerging Roles of Axonemal Glutamylation in Regulation of Cilia Architecture and Functions. Front. Cell Dev. Biol. 2021, 9, 622302. [Google Scholar] [CrossRef]
- Hoffmann, F.; Bolz, S.; Junger, K.; Klose, F.; Stehle, I.F.; Ueffing, M.; Boldt, K.; Beyer, T. Paralog-Specific TTC30 Regulation of Sonic Hedgehog Signaling. Front. Mol. Biosci. 2023, 10, 1268722. [Google Scholar] [CrossRef]
- Hoffmann, F.; Bolz, S.; Junger, K.; Klose, F.; Schubert, T.; Woerz, F.; Boldt, K.; Ueffing, M.; Beyer, T. TTC30A and TTC30B Redundancy Protects IFT Complex B Integrity and Its Pivotal Role in Ciliogenesis. Genes 2022, 13, 1191. [Google Scholar] [CrossRef]
- Dwivedi, D.; Chawla, P.; Sharma, M. Incorporating Motility in the Motor: Role of the Hook Protein Family in Regulating Dynein Motility. Biochemistry 2019, 58, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Gorilák, P. Identification and Characterization of Ciliary Tip Proteins. Ph.D. Thesis, Univerzita Karlova, Prague, Czech Republic, 2022. [Google Scholar]
- Kondziella, D.; Lycke, J. Autism Spectrum Disorders: Does Cilia Dysfunction in Embryogenesis Play. a Role? Acta Neuropsychiatr. 2008, 20, 227–228. [Google Scholar] [CrossRef]
- Youn, Y.H.; Han, Y.-G. Primary Cilia in Brain Development and Diseases. Am. J. Pathol. 2018, 188, 11–22. [Google Scholar] [CrossRef]
- Love, S.L.; Emerson, J.D.; Koide, K.; Hoskins, A.A. Pre-MRNA Splicing-Associated Diseases and Therapies. RNA Biol. 2023, 20, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Guemez-Gamboa, A.; Coufal, N.G.; Gleeson, J.G. Primary Cilia in the Developing and Mature Brain. Neuron 2014, 82, 511–521. [Google Scholar] [CrossRef]
- Suciu, S.K.; Caspary, T. Cilia, Neural Development and Disease. Semin. Cell Dev. Biol. 2021, 110, 34–42. [Google Scholar] [CrossRef]
- Chaki, M.; Airik, R.; Ghosh, A.K.; Giles, R.H.; Chen, R.; Slaats, G.G.; Wang, H.; Hurd, T.W.; Zhou, W.; Cluckey, A.; et al. Exome Capture Reveals ZNF423 and CEP164 Mutations, Linking Renal Ciliopathies to DNA Damage Response Signaling. Cell 2012, 150, 533–548. [Google Scholar] [CrossRef]
- Hildebrandt, F. Exome Resequencing Identifies Novel NPHP Genes, Implicating DNA Damage Response Signaling in the Pathogenesis of Ciliopathies. Cilia 2012, 1, O2. [Google Scholar] [CrossRef]
- Castro-Sánchez, S.; Álvarez-Satta, M.; Tohamy, M.A.; Beltran, S.; Derdak, S.; Valverde, D. Whole Exome Sequencing as a Diagnostic Tool for Patients with Ciliopathy-like Phenotypes. PLoS ONE 2017, 12, e0183081. [Google Scholar] [CrossRef]

| Clinical Features | |
|---|---|
| Family | Non-consanguineous healthy parents |
| Sex | Female |
| Pregnancy history | Uneventful |
| Delivery | Vaginal at term |
| Growth (at birth—40 weeks) | |
| Weight | 3630 gr |
| Lenght | 50 cm |
| Occipitofrontal circumference | 33.5 cm |
| Growth | |
| Height | 108 cm (25° centile) |
| Weight | 19 kg (50° centile) |
| Occipitofrontal circumference | 50 cm (25° centile) |
| Craniofacial anomalies | Brachicephaly, short neck, epicanthus, wide nasal bridge, flat philtrum, prognatism, large anteverted ears, brachidactily, brachycephaly |
| Other anomalies | Tireoglossus duct cist, supranumerary nipple |
| Development | Global developmental delay, iq (scale) 82, normal gross motor skills, normal fine motor skills, language delay, autism spectrum disorder, aggressive behavior/self-injurious behavior, stereotypic behavior, hyperactivity |
| Neurological features | Normal muscle tone, limb hypertonia, tetraplegia, no seizures, no EEG abnormalities |
| Node 2 | Annotation | Score | Organism | Reference |
|---|---|---|---|---|
| CDC5L | Cell division cycle 5-like protein | 0.786 | S. pombe | [41] |
| SART1 | U4/U6.U5 tri-snRNP-associated protein 1 | 0.638 | S. cerevisiae | [42] |
| SNRPE | Small nuclear ribonucleoprotein E | 0.62 | S. cerevisiae | [43] |
| IFT46 | Intraflagellar transport protein 46 homolog | 0.606 | H. sapiens | [17] |
| TTC30A | Intraflagellar transport protein 88 homolog | 0.606 | H. sapiens | [17] |
| IFT88 | Intraflagellar transport protein 88 homolog | 0.594 | H. sapiens | [17] |
| DDX46 | Probable ATP-dependent RNA helicase DDX46 | 0.584 | S. cerevisiae | [44] |
| RBM25 | RNA-binding protein 25 | 0.564 | S. cerevisiae | [45] |
| IFT52 | Intraflagellar transport protein 52 homolog | 0.551 | H. sapiens | [17] |
| SF3B3 | Splicing factor 3B subunit 3 | 0.526 | S. cerevisiae | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Treccarichi, S.; Vinci, M.; Musumeci, A.; Rando, R.G.; Papa, C.; Saccone, S.; Federico, C.; Failla, P.; Ruggieri, M.; Calì, F.; et al. Investigating the Role of the Zinc Finger Protein ZC2HC1C on Autism Spectrum Disorder Susceptibility. Medicina 2025, 61, 574. https://doi.org/10.3390/medicina61040574
Treccarichi S, Vinci M, Musumeci A, Rando RG, Papa C, Saccone S, Federico C, Failla P, Ruggieri M, Calì F, et al. Investigating the Role of the Zinc Finger Protein ZC2HC1C on Autism Spectrum Disorder Susceptibility. Medicina. 2025; 61(4):574. https://doi.org/10.3390/medicina61040574
Chicago/Turabian StyleTreccarichi, Simone, Mirella Vinci, Antonino Musumeci, Rosanna Galati Rando, Carla Papa, Salvatore Saccone, Concetta Federico, Pinella Failla, Martino Ruggieri, Francesco Calì, and et al. 2025. "Investigating the Role of the Zinc Finger Protein ZC2HC1C on Autism Spectrum Disorder Susceptibility" Medicina 61, no. 4: 574. https://doi.org/10.3390/medicina61040574
APA StyleTreccarichi, S., Vinci, M., Musumeci, A., Rando, R. G., Papa, C., Saccone, S., Federico, C., Failla, P., Ruggieri, M., Calì, F., Polizzi, A., & Praticò, A. (2025). Investigating the Role of the Zinc Finger Protein ZC2HC1C on Autism Spectrum Disorder Susceptibility. Medicina, 61(4), 574. https://doi.org/10.3390/medicina61040574









