Machine Learning Tools for Image-Based Glioma Grading and the Quality of Their Reporting: Challenges and Opportunities
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Artificial Intelligence, Machine Learning, and Radiomics
1.2. Machine Learning Applications in Neuro-Oncology
1.3. Image-Based Machine Learning Models for Glioma Grading
2. Workflow for Developing Prediction Models
3. Algorithms for Glioma Grade Classification
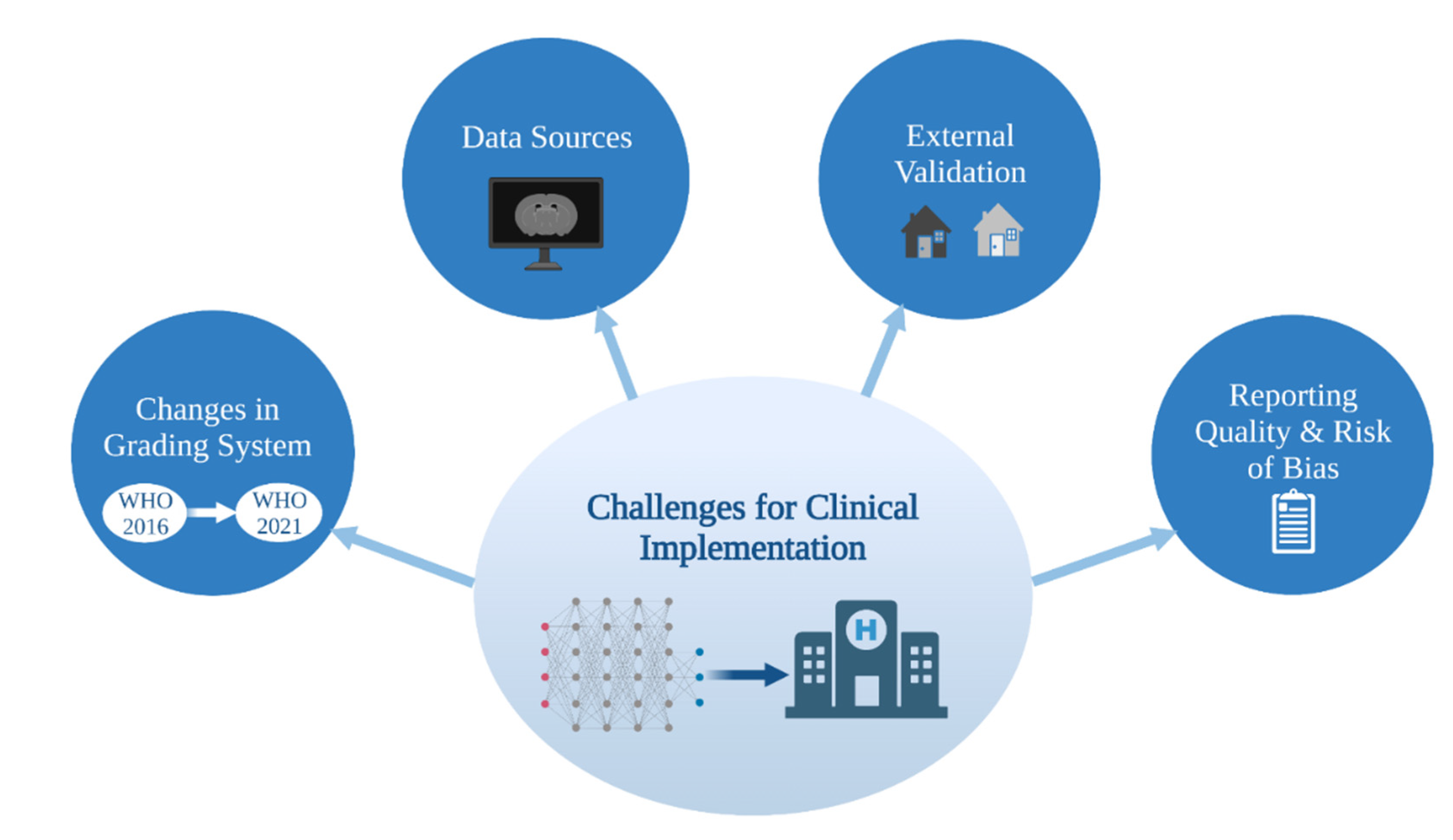
4. Challenges in Image-Based ML Glioma Grading
4.1. Data Sources
4.2. External Validation
4.3. Glioma Grade Classification Systems
4.4. Reporting Quality and Risk of Bias
4.4.1. Overview of Current Guidelines and Tools for Assessment
4.4.2. Reporting Quality and Risk of Bias in Image-Based Glioma Grade Prediction
4.4.3. Future of Reporting Guidelines and Risk of Bias Tools for ML Studies
4.4.4. Recommendations
- Clearly signifying the development of a prediction model in their titles;
- Increasing the number of participants included in training/testing/validation sets;
- Justifying their choice of sample/sample size (whether that be on practical or logistical grounds) and approach to handling missing data (e.g., imputation);
- Specifying all components of model development (including data pre-processing and model calibration) and a full slate of performance metrics (accuracy, area under the receiver operating characteristic curve (AUC), sensitivity, specificity, positive predictive value, negative predictive value, and F1 score as well as associated confidence intervals) for training/testing/validation. While accuracy is the most comprehensive measure of model performance, AUC is more sensitive to performance differences between classes (e.g., within imbalanced datasets) and should always be reported [69];
- Providing open access to the source code of their algorithms.
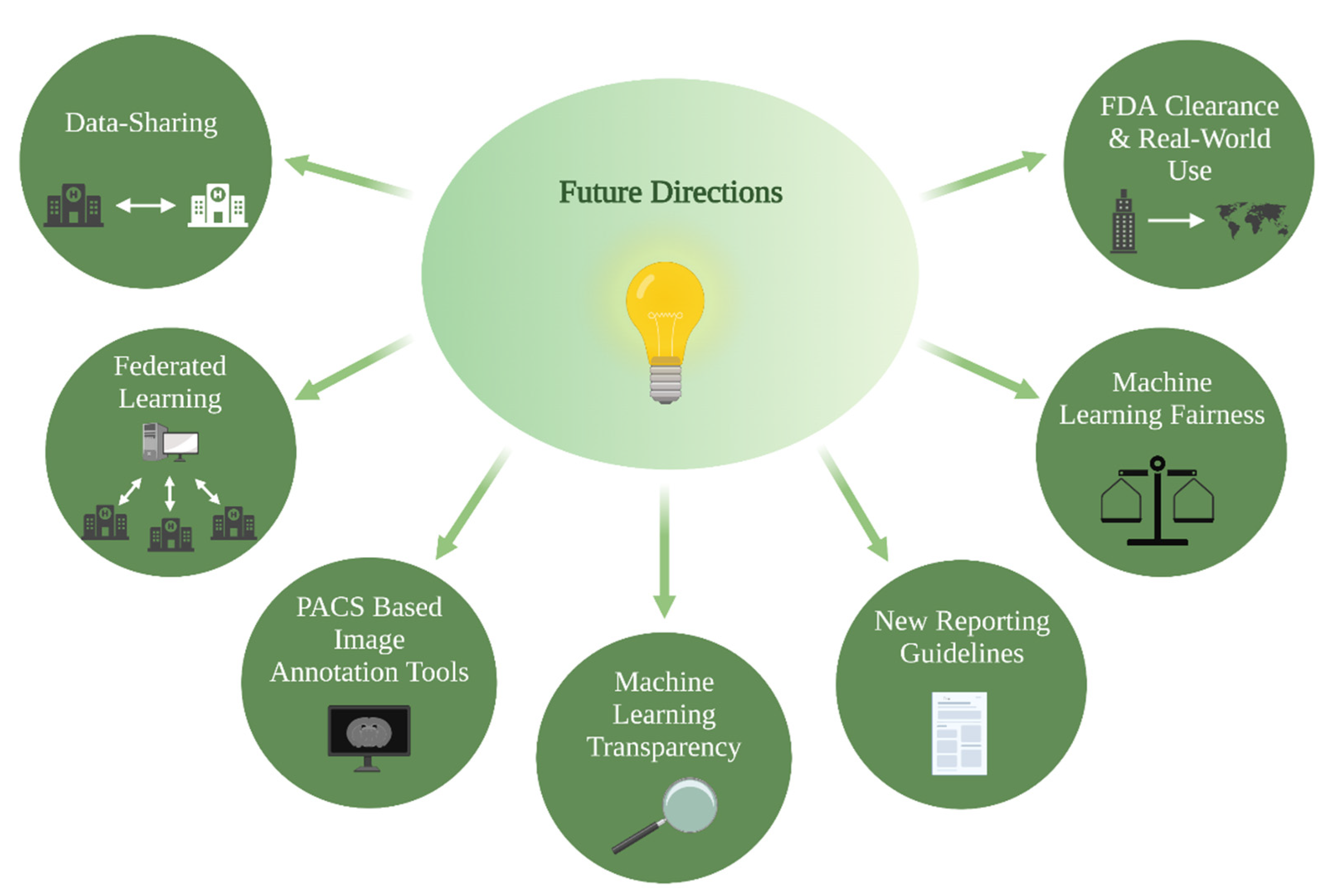
5. Future Directions
5.1. PACS-Based Image Annotation Tools
5.2. Data-Sharing and Federated Learning
5.3. ML Fairness
5.4. ML Transparency
5.5. FDA Clearance and Real-World Use
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeo, M.; Kok, H.K.; Kutaiba, N.; Maingard, J.; Thijs, V.; Tahayori, B.; Russell, J.; Jhamb, A.; Chandra, R.V.; Brooks, M.; et al. Artificial intelligence in clinical decision support and outcome prediction–Applications in stroke. J. Med. Imaging Radiat. Oncol. 2021, 65, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Kaka, H.; Zhang, E.; Khan, N. Artificial Intelligence and Deep Learning in Neuroradiology: Exploring the New Frontier. Can. Assoc. Radiol. J. 2021, 72, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Giger, M.L. Machine Learning in Medical Imaging. J. Am. Coll. Radiol. 2018, 15, 512–520. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-Oncology 2019, 21 Suppl. S5, v1–v100. [Google Scholar] [CrossRef]
- Abdel Razek, A.A.K.; Alksas, A.; Shehata, M.; AbdelKhalek, A.; Abdel Baky, K.; El-Baz, A.; Helmy, E. Clinical applications of artificial intelligence and radiomics in neuro-oncology imaging. Insights Imaging 2021, 12, 152. [Google Scholar] [CrossRef]
- Thon, N.; Tonn, J.-C.; Kreth, F.-W. The surgical perspective in precision treatment of diffuse gliomas. OncoTargets Ther. 2019, 12, 1497–1508. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.S.; Hawkins-Daarud, A.; Wang, L.; Li, J.; Swanson, K.R. Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett. 2020, 477, 97–106. [Google Scholar] [CrossRef]
- Seow, P.; Wong, J.H.D.; Annuar, A.A.; Mahajan, A.; Abdullah, N.A.; Ramli, N. Quantitative magnetic resonance imaging and radiogenomic biomarkers for glioma characterisation: A systematic review. Br. J. Radiol. 2018, 91, 20170930. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchlak, Q.D.; Esmaili, N.; Leveque, J.-C.; Bennett, C.; Farrokhi, F.; Piccardi, M. Machine learning applications to neuroimaging for glioma detection and classification: An artificial intelligence augmented systematic review. J. Clin. Neurosci. 2021, 89, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Chow, D.; Chang, P.; Weinberg, B.D.; Bota, D.A.; Grinband, J.; Filippi, C.G. Imaging Genetic Heterogeneity in Glioblastoma and Other Glial Tumors: Review of Current Methods and Future Directions. Am. J. Roentgenol. 2018, 210, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Codari, M.; Sardanelli, F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur. Radiol. Exp. 2018, 2, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pemberton, H.G.; Zaki, L.A.M.; Goodkin, O.; Das, R.K.; Steketee, R.M.E.; Barkhof, F.; Vernooij, M.W. Technical and clinical validation of commercial automated volumetric MRI tools for dementia diagnosis-a systematic review. Neuroradiology 2021, 63, 1773–1789. [Google Scholar] [CrossRef] [PubMed]
- Richens, J.G.; Lee, C.M.; Johri, S. Improving the accuracy of medical diagnosis with causal machine learning. Nat. Commun. 2020, 11, 3923. [Google Scholar] [CrossRef]
- Rubin, D.L. Artificial Intelligence in Imaging: The Radiologist’s Role. J. Am. Coll. Radiol. 2019, 16, 1309–1317. [Google Scholar] [CrossRef]
- Wu, J.T.; Wong, K.C.L.; Gur, Y.; Ansari, N.; Karargyris, A.; Sharma, A.; Morris, M.; Saboury, B.; Ahmad, H.; Boyko, O.; et al. Comparison of Chest Radiograph Interpretations by Artificial Intelligence Algorithm vs Radiology Residents. JAMA Netw. Open 2020, 3, e2022779. [Google Scholar] [CrossRef]
- Cassinelli Petersen, G.I.; Shatalov, J.; Verma, T.; Brim, W.R.; Subramanian, H.; Brackett, A.; Bahar, R.C.; Merkaj, S.; Zeevi, T.; Staib, L.H.; et al. Machine Learning in Differentiating Gliomas from Primary CNS Lymphomas: A Systematic Review, Reporting Quality, and Risk of Bias Assessment. AJNR Am. J. Neuroradiol. 2022, 43, 526–533. [Google Scholar] [CrossRef]
- Rauschecker, A.M.; Rudie, J.D.; Xie, L.; Wang, J.; Duong, M.T.; Botzolakis, E.J.; Kovalovich, A.M.; Egan, J.; Cook, T.C.; Bryan, R.N.; et al. Artificial Intelligence System Approaching Neuroradiologist-level Differential Diagnosis Accuracy at Brain MRI. Radiology 2020, 295, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Decuyper, M.; Bonte, S.; Van Holen, R. Binary Glioma Grading: Radiomics versus Pre-trained CNN Features. Med. Image Comput. Comput. Assist. Interv. 2018, 11072, 498–505. [Google Scholar] [CrossRef]
- Gao, M.; Huang, S.; Pan, X.; Liao, X.; Yang, R.; Liu, J. Machine Learning-Based Radiomics Predicting Tumor Grades and Expression of Multiple Pathologic Biomarkers in Gliomas. Front. Oncol. 2020, 10, 1676. [Google Scholar] [CrossRef] [PubMed]
- Haubold, J.; Demircioglu, A.; Gratz, M.; Glas, M.; Wrede, K.; Sure, U.; Antoch, G.; Keyvani, K.; Nittka, M.; Kannengiesser, S.; et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric 18F-FET PET-MRI and MR Fingerprinting. Eur. J. Pediatr. 2020, 47, 1435–1445. [Google Scholar] [CrossRef]
- Sengupta, A.; Ramaniharan, A.K.; Gupta, R.K.; Agarwal, S.; Singh, A. Glioma grading using a machine-learning framework based on optimized features obtained from T 1 perfusion MRI and volumes of tumor components. J. Magn. Reson. Imaging 2019, 50, 1295–1306. [Google Scholar] [CrossRef]
- Tian, Q.; Yan, L.-F.; Zhang, X.; Hu, Y.-C.; Han, Y.; Liu, Z.-C.; Nan, H.-Y.; Sun, Q.; Sun, Y.-Z.; Yang, Y.; et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J. Magn. Reson. Imaging 2018, 48, 1518–1528. [Google Scholar] [CrossRef]
- Abdolmaleki, P.; Mihara, F.; Masuda, K.; Buadu, L.D. Neural networks analysis of astrocytic gliomas from MRI appearances. Cancer Lett. 1997, 118, 69–78. [Google Scholar] [CrossRef]
- Christy, P.S.; Tervonen, O.; Scheithauer, B.W.; Forbes, G.S. Use of a Neural-Network and a Multiple-Regression Model to Predict Histologic Grade of Astrocytoma from Mri Appearances. Neuroradiology 1995, 37, 89–93. [Google Scholar] [CrossRef]
- Dandil, E.; Biçer, A. Automatic grading of brain tumours using LSTM neural networks on magnetic resonance spectroscopy signals. IET Image Process 2020, 14, 1967–1979. [Google Scholar] [CrossRef]
- Ji, B.; Wang, S.; Liu, Z.; Weinberg, B.D.; Yang, X.; Liu, T.; Wang, L.; Mao, H. Revealing hemodynamic heterogeneity of gliomas based on signal profile features of dynamic susceptibility contrast-enhanced MRI. NeuroImage Clin. 2019, 23, 101864. [Google Scholar] [CrossRef]
- Tabatabaei, M.; Razaei, A.; Sarrami, A.H.; Saadatpour, Z.; Singhal, A.; Sotoudeh, H. Current Status and Quality of Machine Learning-Based Radiomics Studies for Glioma Grading: A Systematic Review. Oncology 2021, 99, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Kotsiantis, S.B.; Kanellopoulos, D.; Pintelas, P.E. Data preprocessing for supervised leaning. Int. J. Comput. Sci. 2006, 1, 111–117. [Google Scholar]
- Tillmanns, N.; Lum, A.; Brim, W.R.; Subramanian, H.; Lin, M.; Bousabarah, K.; Malhotra, A.; Cui, J.; Brackett, A.; Payabvash, S.; et al. NIMG-71. Identifying clinically applicable machine learning algorithms for glioma segmentation using a systematic literature review. Neuro-Oncology 2021, 23, vi145. [Google Scholar] [CrossRef]
- Shaver, M.M.; Kohanteb, P.A.; Chiou, C.; Bardis, M.D.; Chantaduly, C.; Bota, D.; Filippi, C.G.; Weinberg, B.; Grinband, J.; Chow, D.S.; et al. Optimizing Neuro-Oncology Imaging: A Review of Deep Learning Approaches for Glioma Imaging. Cancers 2019, 11, 829. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef]
- Rios Velazquez, E.; Meier, R.; Dunn, W.D., Jr.; Alexander, B.; Wiest, R.; Bauer, S.; Aerts, H.J. Fully automatic GBM segmentation in the TCGA-GBM dataset: Prognosis and correlation with VASARI features. Sci. Rep. 2015, 5, 16822. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Brereton, R.G.; Lloyd, G.R. Support Vector Machines for classification and regression. Analyst 2010, 135, 230–267. [Google Scholar] [CrossRef]
- Sohn, C.K.; Bisdas, S. Diagnostic Accuracy of Machine Learning-Based Radiomics in Grading Gliomas: Systematic Review and Meta-Analysis. Contrast Media Mol. Imaging 2020, 2020, 2127062. [Google Scholar] [CrossRef] [PubMed]
- Gordillo, N.; Montseny, E.; Sobrevilla, P. State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 2013, 31, 1426–1438. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. RadioGraphics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [Green Version]
- Ge, C.; Gu, I.Y.-H.; Jakola, A.S.; Yang, J. Deep Learning and Multi-Sensor Fusion for Glioma Classification Using Multistream 2D Convolutional Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–22 July 2018; pp. 5894–5897. [Google Scholar] [CrossRef]
- Kabir-Anaraki, A.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2019, 39, 63–74. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.-H.; Jakola, A.S.; Yang, J. Deep semi-supervised learning for brain tumor classification. BMC Med. Imaging 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Sharif, M.I.; Li, J.P.; Khan, M.A.; Saleem, M.A. Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recognit. Lett. 2020, 129, 181–189. [Google Scholar] [CrossRef]
- Hayashi, Y. Toward the transparency of deep learning in radiological imaging: Beyond quantitative to qualitative artificial intelligence. J. Med. Artif. Intell. 2019, 2, 19. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Bahar, R.C.; Merkaj, S.; Cassinelli Petersen, G.I.; Tillmanns, N.; Subramanian, H.; Brim, W.R.; Zeevi, T.; Staib, L.; Kazarian, E.; Lin, M.; et al. NIMG-35. Machine Learning Models for Classifying High- and Low-Grade Gliomas: A Systematic Review and Quality of Reporting Analysis. Front. Oncol. 2022, 12, 856231. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Prior, F. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhavan, S.; Zenklusen, J.-C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping Personalized Medicine Become a Reality through Integrative Translational Research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajan, P.V.; Karnuta, J.M.; Haeberle, H.S.; Spitzer, A.I.; Schaffer, J.L.; Ramkumar, P.N. Response to letter to the editor on “Significance of external validation in clinical machine learning: Let loose too early?”. Spine J. 2020, 20, 1161–1162. [Google Scholar] [CrossRef]
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef]
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Weller, M. cIMPACT-NOW update 5: Recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020, 139, 603–608. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Br. J. Surg. 2015, 102, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, H.S.; Kim, D.; Park, S.Y.; Kim, J.Y.; Cho, S.J.; Kim, J.H. A systematic review reporting quality of radiomics research in neuro-oncology: Toward clinical utility and quality improvement using high-dimensional imaging features. BMC Cancer 2020, 20, 29. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef] [Green Version]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Wolff, R.F.; Moons, K.G.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andaur Navarro, C.L.; Damen, J.A.A.; Takada, T.; Nijman, S.W.J.; Dhiman, P.; Ma, J.; Collins, G.S.; Bajpai, R.; Riley, R.D.; Moons, K.G.M.; et al. Risk of bias in studies on prediction models developed using supervised machine learning techniques: Systematic review. BMJ 2021, 20, n2281. [Google Scholar]
- Yao, A.D.; Cheng, D.L.; Pan, I.; Kitamura, F. Deep Learning in Neuroradiology: A Systematic Review of Current Algorithms and Approaches for the New Wave of Imaging Technology. Radiol. Artif. Intell. 2020, 2, e190026. [Google Scholar] [CrossRef]
- Collins, G.S.; Dhiman, P.; Navarro, C.L.A.; Ma, J.; Hooft, L.; Reitsma, J.B.; Logullo, P.; Beam, A.L.; Peng, L.; Van Calster, B.; et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 2021, 11, e048008. [Google Scholar] [CrossRef]
- Collins, G.S.; Moons, K.G.M. Reporting of artificial intelligence prediction models. Lancet 2019, 393, 1577–1579. [Google Scholar] [CrossRef]
- Sounderajah, V.; Ashrafian, H.; Rose, S.; Shah, N.H.; Ghassemi, M.; Golub, R.; Kahn, C.E.; Esteva, A.; Karthikesalingam, A.; Mateen, B.; et al. A quality assessment tool for artificial intelligence-centered diagnostic test accuracy studies: QUADAS-AI. Nat. Med. 2021, 27, 1663–1665. [Google Scholar] [CrossRef]
- Ling, C.X.; Huang, J.; Zhang, H. AUC: A Better Measure than Accuracy in Comparing Learning Algorithms. Lect. Notes Artif. Int. 2003, 2671, 329–341. [Google Scholar] [CrossRef]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [Green Version]
- Zaharchuk, G.; Gong, E.; Wintermark, M.; Rubin, D.; Langlotz, C. Deep Learning in Neuroradiology. Am. J. Neuroradiol. 2018, 39, 1776–1784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, E. Strengthening Research through Data Sharing. N. Engl. J. Med. 2016, 375, 401–403. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The future of digital health with federated learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Sheller, M.J.; Reina, G.A.; Edwards, B.; Martin, J.; Bakas, S. Multi-Institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation. Brainlesion 2019, 11383, 92–104. [Google Scholar]
- Mehrabi, N.; Morstatter, F.; Saxena, N.; Lerman, K.; Galstyan, A. A Survey on Bias and Fairness in Machine Learning. ACM Comput. Surv. 2021, 54, 115. [Google Scholar] [CrossRef]
- Fry, A.; Littlejohns, T.J.; Sudlow, C.; Doherty, N.; Adamska, L.; Sprosen, T.; Collins, R.; Allen, N.E. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants with Those of the General Population. Am. J. Epidemiol. 2017, 186, 1026–1034. [Google Scholar] [CrossRef] [Green Version]
- Vickers, S.M.; Fouad, M.N. An overview of EMPaCT and fundamental issues affecting minority participation in cancer clinical trials: Enhancing minority participation in clinical trials (EMPaCT): Laying the groundwork for improving minority clinical trial accrual. Cancer 2014, 120 Suppl. S7, 1087–1090. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.Y.; Szolovits, P.; Ghassemi, M. Can AI Help Reduce Disparities in General Medical and Mental Health Care? AMA J. Ethics 2019, 21, E167–E179. [Google Scholar]
- Bird, S.; Dudík, M.; Edgar, R.; Horn, B.; Lutz, R.; Milan, V.; Sameki, M.; Wallach, H.; Walker, K. Fairlearn: A toolkit for assessing and improving fairness in ai. Tech. Rep. 2020, pp. 1–7. Available online: https://www.microsoft.com/en-us/research/publication/fairlearn-a-toolkit-for-assessing-and-improving-fairness-in-ai/ (accessed on 24 May 2022).
- Bellamy, R.K.E.; Dey, K.; Hind, M.; Hoffman, S.C.; Houde, S.; Kannan, K.; Lohia, P.; Martino, J.; Mehta, S.; Mojsilovic, A.; et al. AI Fairness 360: An extensible toolkit for detecting and mitigating algorithmic bias. IBM J. Res. Dev. 2019, 63, 4:1–4:15. [Google Scholar] [CrossRef]
- Gashi, M.; Vuković, M.; Jekic, N.; Thalmann, S.; Holzinger, A.; Jean-Quartier, C.; Jeanquartier, F. State-of-the-Art Explainability Methods with Focus on Visual Analytics Showcased by Glioma Classification. BioMedInformatics 2022, 2, 139–158. [Google Scholar] [CrossRef]
- Königstorfer, F.; Thalmann, S. Software documentation is not enough! Requirements for the documentation of AI. Digit. Policy Regul. Gov. 2021, 23, 475–488. [Google Scholar] [CrossRef]
- Castelvecchi, D. Can we open the black box of AI? Nat. News 2016, 538, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Holzinger, A. Usability engineering methods for software developers. Commun. ACM 2005, 48, 71–74. [Google Scholar] [CrossRef]
- Chou, J.R.; Hsiao, S.W. A usability study of human-computer interface for middle-aged learners. Comput. Hum. Behav. 2007, 23, 2040–2063. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. WIREs Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [Green Version]
- Shin, D. The effects of explainability and causability on perception, trust, and acceptance: Implications for explainable AI. Int. J. Hum. Comput. Stud. 2021, 146, 102551. [Google Scholar] [CrossRef]
- Institute ACoRDS. FDA Cleared AI Algorithms. Available online: https://models.acrdsi.org/ (accessed on 3 December 2021).
- Ebrahimian, S.; Kalra, M.K.; Agarwal, S.; Bizzo, B.C.; Elkholy, M.; Wald, C.; Allen, B.; Dreyer, K.J. FDA-regulated AI Algorithms: Trends, Strengths, and Gaps of Validation Studies. Acad. Radiol. 2021, 29, 559–566. [Google Scholar] [CrossRef]
- Lin, M. What’s Needed to Bridge the Gap between US FDA Clearance and Real-world Use of AI Algorithms. Acad. Radiol. 2021, 29, 567–568. [Google Scholar] [CrossRef]

| Feature Type | Explanation |
|---|---|
| Clinical | Describe patient demographics, e.g., gender and age. |
| Deep learning extracted | Derived from pre-trained deep neural networks. |
| First-order | Create a three-dimensional (3D) histogram out of tumor volume characteristics, from which mean, median, range, skewness, kurtosis, etc., can be calculated [35]. |
| Higher-order | Identify repetitiveness in image patterns, suppress noise, or highlight details [35]. |
| Qualitative | Describe visible tumor characteristics on imaging using controlled vocabulary, e.g., VASARI features (tumor location, side of lesion center, enhancement quality, etc.). |
| Second-order | Classify texture characteristics, e.g., contrast, correlation, dissimilarity, maximum probability, grey level run length features, etc. [35] |
| Shape and size | Describe the statistical inter-relationships between neighboring voxels, e.g., total volume or surface area, surface-to-volume ratio, tumor compactness, sphericity, etc. [35] |
| Guideline/Tool | Full Name | Year Published | Articles Targeted | Purpose | Specific to ML? |
|---|---|---|---|---|---|
| QUADAS-2 4 | Quality Assessment of Diagnostic Accuracy Studies | 2011 (original QUADAS 4: 2003) | Diagnostic accuracy studies | Evaluates study risk of bias and applicability | No; QUADAS-AI 4 is in development |
| TRIPOD 6 | Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis | 2015 | Studies developing, validating, or updating a diagnostic or prognostic prediction model | Provides a set of recommendations for study reporting | No; TRIPOD-AI 6 is in development |
| RQS 5 | Radiomics quality score | 2017 | Radiomic studies | Assesses study quality (emulating TRIPOD 6) | No |
| PROBAST 3 | Prediction model Risk Of Bias ASsessment Tool | 2019 | Studies developing, validating, or updating a diagnostic or prognostic prediction model | Evaluates study risk of bias and applicability | No; PROBAST-AI 3 is in development |
| CLAIM 2 | Checklist for AI 1 in Medical Imaging | 2020 | AI 1 studies in medical imaging | Guides authors in presenting (and aids reviewers in evaluating) their research | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merkaj, S.; Bahar, R.C.; Zeevi, T.; Lin, M.; Ikuta, I.; Bousabarah, K.; Cassinelli Petersen, G.I.; Staib, L.; Payabvash, S.; Mongan, J.T.; et al. Machine Learning Tools for Image-Based Glioma Grading and the Quality of Their Reporting: Challenges and Opportunities. Cancers 2022, 14, 2623. https://doi.org/10.3390/cancers14112623
Merkaj S, Bahar RC, Zeevi T, Lin M, Ikuta I, Bousabarah K, Cassinelli Petersen GI, Staib L, Payabvash S, Mongan JT, et al. Machine Learning Tools for Image-Based Glioma Grading and the Quality of Their Reporting: Challenges and Opportunities. Cancers. 2022; 14(11):2623. https://doi.org/10.3390/cancers14112623
Chicago/Turabian StyleMerkaj, Sara, Ryan C. Bahar, Tal Zeevi, MingDe Lin, Ichiro Ikuta, Khaled Bousabarah, Gabriel I. Cassinelli Petersen, Lawrence Staib, Seyedmehdi Payabvash, John T. Mongan, and et al. 2022. "Machine Learning Tools for Image-Based Glioma Grading and the Quality of Their Reporting: Challenges and Opportunities" Cancers 14, no. 11: 2623. https://doi.org/10.3390/cancers14112623
APA StyleMerkaj, S., Bahar, R. C., Zeevi, T., Lin, M., Ikuta, I., Bousabarah, K., Cassinelli Petersen, G. I., Staib, L., Payabvash, S., Mongan, J. T., Cha, S., & Aboian, M. S. (2022). Machine Learning Tools for Image-Based Glioma Grading and the Quality of Their Reporting: Challenges and Opportunities. Cancers, 14(11), 2623. https://doi.org/10.3390/cancers14112623








