Inhibition of Biofilm Formation by Modified Oxylipins from the Shipworm Symbiont Teredinibacter turnerae
Abstract
1. Introduction
2. Results and Discussion
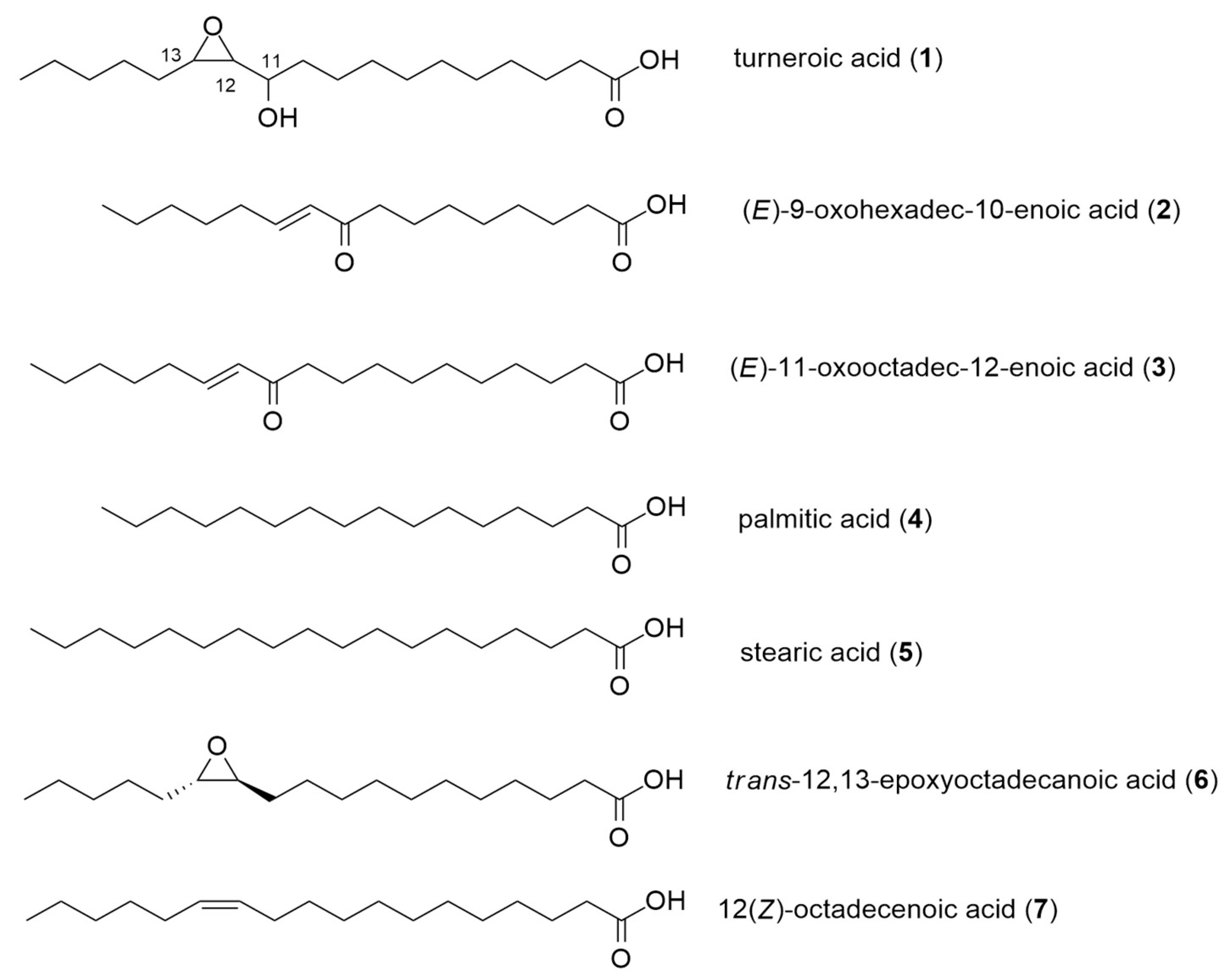
2.1. Structure Elucidation
2.2. Biological Activities of Oxylipins (1–7)
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Culturing and Extraction
3.4. Antimicrobial Microdilution Assay
3.5. Biofilm Inhibition Assay
3.6. Mammalian Antiproliferative Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Donlan, R.M. Role of Biofilms in Antimicrobial Resistance. ASAIO J. 2000, 46, S47–S52. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.L.S.D.; Galdino, A.C.M.; De Mello, T.P.; Ramos, L.D.S.; Branquinha, M.H.; Bolognese, A.M.; Neto, J.C.; Roudbary, M. What are the advantages of living in a community? A microbial biofilm perspective! Memórias do Instituto Oswaldo Cruz 2018, 113, e180212. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal Biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar]
- Otto, M. Staphylococcus epidermidis-The “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef]
- Rossiter, S.E.; Fletcher, M.H.; Wuest, W.M. Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 2017, 117, 12415–12474. [Google Scholar] [CrossRef]
- Schmidt, E.W. Trading molecules and tracking targets in symbiotic interactions. Nat. Chem. Biol. 2008, 4, 466–473. [Google Scholar] [CrossRef]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef]
- Han, A.W.; Sandy, M.; Fishman, B.; Trindade-Silva, A.E.; Soares, C.A.G.; Distel, D.L.; Butler, A.; Haygood, M.G. Turnerbactin, a Novel Triscatecholate Siderophore from the Shipworm Endosymbiont Teredinibacter turnerae T7901. PLoS ONE 2013, 8, e76151. [Google Scholar] [CrossRef]
- Distel, D.L.; Altamia, M.A.; Lin, Z.; Shipway, J.R.; Han, A.; Forteza, I.; Antemano, R.; Limbaco, M.G.J.P.; Tebo, A.G.; Dechavez, R.; et al. Discovery of chemoautotrophic symbiosis in the giant shipwormKuphus polythalamia(Bivalvia: Teredinidae) extends wooden-steps theory. Proc. Natl. Acad. Sci. USA 2017, 114, E3652–E3658. [Google Scholar] [CrossRef]
- Waterbury, J.B.; Calloway, C.B.; Turner, R.D. A Cellulolytic Nitrogen-Fixing Bacterium Cultured from the Gland of Deshayes in Shipworms (Bivalvia: Teredinidae). Science 1983, 221, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Madupu, R.; Durkin, A.S.; Ekborg, N.A.; Pedamallu, C.S.; Hostetler, J.B.; Radune, D.; Toms, B.S.; Henrissat, B.; Coutinho, P.M.; et al. The Complete Genome of Teredinibacter turnerae T7901: An Intracellular Endosymbiont of Marine Wood-Boring Bivalves (Shipworms). PLoS ONE 2009, 4, e6085. [Google Scholar] [CrossRef] [PubMed]
- Elshahawi, S.I.; Trindade-Silva, A.E.; Hanora, A.; Han, A.W.; Flores, M.S.; Vizzoni, V.; Schrago, C.G.; Soares, C.A.G.; Concepcion, G.P.; Distel, D.L.; et al. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc. Natl. Acad. Sci. USA 2013, 110, E295–E304. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.-X.; Majchrzak-Hong, S.; Keyes, G.S.; Iadarola, M.J.; Mannes, A.J.; Ramsden, C.E. Lipidomic profiling of targeted oxylipins with ultra-performance liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2018, 410, 6009–6029. [Google Scholar] [CrossRef]
- Hung, T.D.; Hye, J.L.; Eun, S.Y.; Shinde, P.B.; Yoon, M.L.; Hong, J.; Kim, D.K.; Jung, J.H. Anti-inflammatory constituents of the Red Alga Gracilaria verrucosa and their synthetic analogues. J. Nat. Prod. 2008, 71, 232–240. [Google Scholar]
- Abou-ElWafa, G.S.E.; Shaaban, M.; Shaaban, K.A.; El-Naggar, M.E.E.; Laatsch, H. Three New Unsaturated Fatty Acids from the Marine Green Alga Ulva fasciata Delile. Zeitschrift für Naturforschung B 2009, 64, 1199–1207. [Google Scholar] [CrossRef]
- Davies, D.; Marques, C. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef]
- Marques, C.; Morozoz, A.; Planzos, P.; Zelaya, H. The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl. Environ. Microbiol. 2014, 80, 6976–6991. [Google Scholar] [CrossRef]
- Topa, S.H.; Subramoni, S.; Palombo, E.A.; Kingshott, P.; Rice, S.A.; Blackall, L.L. Cinnamaldehyde disrupts biofilm formation and swarming motility of Pseudomonas aeruginosa. Microbiology 2018, 164, 1087–1097. [Google Scholar] [CrossRef]
- Casillo, A.; Papa, R.; Ricciardelli, A.; Sannino, F.; Ziaco, M.; Tilotta, M.; Selan, L.; Marino, G.; Corsaro, M.M.; Tutino, M.L.; et al. Anti-Biofilm Activity of a Long-Chain Fatty Aldehyde from Antarctic Pseudoalteromonas haloplanktis TAC125 against Staphylococcus epidermidis Biofilm. Front. Cell. Infect. Microbiol. 2017, 7, 46. [Google Scholar] [CrossRef]
- Valliammai, A.; Sethupathy, S.; Priya, A.; Selvaraj, A.; Bhaskar, J.P.; Krishnan, V.; Pandian, S.K. 5-Dodecanolide interferes with biofilm formation and reduces the virulence of Methicillin-resistant Staphylococcus aureus (MRSA) through up regulation of agr system. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Sanabria-Valentín, E.; Vlamakis, H.; Kolter, R. Biofilm Inhibitors that Target Amyloid Proteins. Chem. Biol. 2013, 20, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Camacho, E.T.; Castellanos, L.; Arévalo-Ferro, C.; Duque, C. Disruption in Quorum-Sensing Systems and Bacterial Biofilm Inhibition by Cembranoid Diterpenes Isolated from the Octocoral Eunicea knighti. J. Nat. Prod. 2012, 75, 1637–1642. [Google Scholar] [CrossRef]
- Mueller, M.J.; Berger, S. Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochem. 2009, 70, 1511–1521. [Google Scholar] [CrossRef]
- Wang, B.-Y.; Huang, H.-Q.; Li, S.; Tang, P.; Dai, H.; Xian, J.-A.; Sun, D.-M.; Hu, Y. Thioredoxin H (TrxH) contributes to adversity adaptation and pathogenicity of Edwardsiella piscicida. Veter- Res. 2019, 50, 26. [Google Scholar] [CrossRef] [PubMed]
- Lacerna, I.N.M.; Miller, B.W.; Lim, A.L.; Tun, J.O.; Robes, J.M.D.; Cleofas, M.J.B.; Lin, Z.; Salvador-Reyes, L.A.; Haygood, M.G.; Schmidt, E.W.; et al. Mindapyrroles A–C, Pyoluteorin Analogues from a Shipworm-Associated Bacterium. J. Nat. Prod. 2019, 82, 1024–1028. [Google Scholar] [CrossRef]
- Sarker, S.D.; Nahar, L.; Kumarasamy, Y. Microtitre plate based antibacterial assay incorporatin resazurin as an indicator of cell growth, and its applications in the in vitro antibacterial screening of phytochemicals. Methods 2007, 42, 321–324. [Google Scholar] [CrossRef]
- Skogman, M.E.; Vuorela, P.M.; Fallarero, A. Combining biofilm matrix measurements with biomass and viability assays in susceptibility assessments of antimicrobials against Staphylococcus aureus biofilms. J. Antibiot. 2012, 65, 453–459. [Google Scholar] [CrossRef]
- Yan, Z.; Huang, M.; Melander, C.; Kjellerup, B.V. Dispersal and inhibition of biofilms associated with infections. J. App. Microbiol. 2020, 128(5), 1279–1288. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
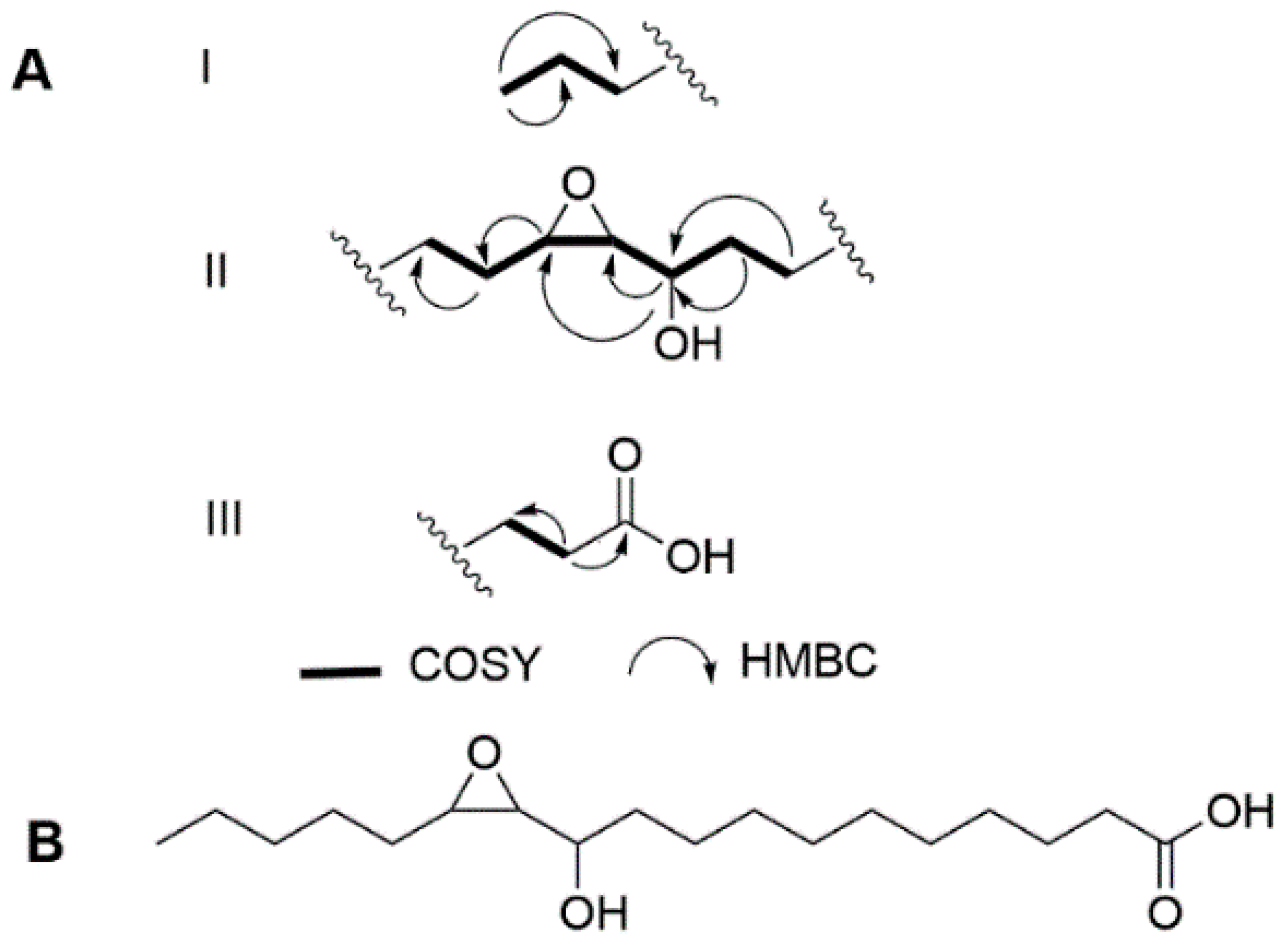
| C/H | δ C a | δ H (J in Hz) b | COSY b | TOCSY | HMBC b |
|---|---|---|---|---|---|
| 1 | 178.1, C | ||||
| 2 | 36.3, CH2 | 2.24, t (7.1) | 3 | 3,4 | 1, 3, 4 |
| 3 | 26.4, CH2 | 1.60, m | 2,4 | 2,4 | 4/5 |
| 4 | 30.4, CH2 | 1.33, m | c | c | 3 |
| 5 | 30.4, CH2 | 1.33, m | c | c | |
| 6 | 30.4, CH2 | 1.33, m | c | c | |
| 7 | 30.4, CH2 | 1.33, m | c | c | |
| 8 | 30.4, CH2 | 1.33, m | c | c | |
| 9 | 26.5, CH2 | 1.46, m | 8, 10 | 7/8, 11 | |
| 10 | 35.2, CH2 | 1.53, 1.34, m | 9, 11 | 11 | 8, 11 |
| 11 | 72.8, CH | 3.30, m | 10, 12 | 10,12 | 9, 10, 12, 13(w) |
| 12 | 63.4, CH | 2.67, dd (6.6, 2.1) | 11, 13 | 11 | 11, 13(w) |
| 13 | 57.7, CH | 2.86, td (5.8, 2.1) | 12, 14 | 14,16,18 | 14 |
| 14 | 32.6, CH2 | 1.54, 1.35, m | 13,15 | 11,12,13,15 | 12, 13 |
| 15 | 26.4, CH2 | 1.35, m | c | c | |
| 16 | 32.8, CH2 | 1.45, m | 17 | 18 | |
| 17 | 23.7, CH2 | 1.35, m | 16, 18 | 18 | 18 |
| 18 | 14.3, CH3 | 0.92, t (7. 1) | 17 | 13,14,15,16,17 | 16, 17 |
| Compounds | MBIC (µg/mL) S. epidermidis RP62A (ATCC® 35984™) | MIC (µg/mL) S. epidermidis RP62A (ATCC® 35984™) | Selectivity Index (MICPlanktonic/MBICBiofilm) | MIC (µg/mL) S. aureus (ATCC® 6538™) | MIC (µg/mL) Methicillin- Resistant S. aureus (ATCC® 43,300™) | IC50 (µg/mL) MDCK NBL-2 (ATCC® CCL-34™) |
|---|---|---|---|---|---|---|
| 1 | 32 | 128 | ~4 | >128 | >128 | 61.7 |
| 2 | >128 | >128 | ~1 | >128 | >128 | 33.0 |
| 3 | 16 | 16 | ~1 | >128 | >128 | 13.6 |
| 4 | >128 | >128 | ~1 | >128 | >128 | >128 |
| 5 | >128 | >128 | ~1 | >128 | >128 | >128 |
| 6 | 16 | >128 | >8 | >128 | >128 | >128 |
| 7 | >128 | >128 | ~1 | 64.0 | >128 | >128 |
| dispersin B | 64 | >>128 | >>2 | n/a | n/a | n/a |
| chloramphenicol | n/a | 8.0 | n/a | n/a | n/a | n/a |
| oxacillin | n/a | n/a | n/a | 0.5 | 8 | n/a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lacerna, N.M., II; Ramones, C.M.V.; Robes, J.M.D.; Picart, M.R.D.; Tun, J.O.; Miller, B.W.; Haygood, M.G.; Schmidt, E.W.; Salvador-Reyes, L.A.; Concepcion, G.P. Inhibition of Biofilm Formation by Modified Oxylipins from the Shipworm Symbiont Teredinibacter turnerae. Mar. Drugs 2020, 18, 656. https://doi.org/10.3390/md18120656
Lacerna NM II, Ramones CMV, Robes JMD, Picart MRD, Tun JO, Miller BW, Haygood MG, Schmidt EW, Salvador-Reyes LA, Concepcion GP. Inhibition of Biofilm Formation by Modified Oxylipins from the Shipworm Symbiont Teredinibacter turnerae. Marine Drugs. 2020; 18(12):656. https://doi.org/10.3390/md18120656
Chicago/Turabian StyleLacerna, Noel M., II, Cydee Marie V. Ramones, Jose Miguel D. Robes, Myra Ruth D. Picart, Jortan O. Tun, Bailey W. Miller, Margo G. Haygood, Eric W. Schmidt, Lilibeth A. Salvador-Reyes, and Gisela P. Concepcion. 2020. "Inhibition of Biofilm Formation by Modified Oxylipins from the Shipworm Symbiont Teredinibacter turnerae" Marine Drugs 18, no. 12: 656. https://doi.org/10.3390/md18120656
APA StyleLacerna, N. M., II, Ramones, C. M. V., Robes, J. M. D., Picart, M. R. D., Tun, J. O., Miller, B. W., Haygood, M. G., Schmidt, E. W., Salvador-Reyes, L. A., & Concepcion, G. P. (2020). Inhibition of Biofilm Formation by Modified Oxylipins from the Shipworm Symbiont Teredinibacter turnerae. Marine Drugs, 18(12), 656. https://doi.org/10.3390/md18120656





