Characterization of a New Mixture of Mono-Rhamnolipids Produced by Pseudomonas gessardii Isolated from Edmonson Point (Antarctica)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Bacterial Isolation and Biosurfactant Activity Screening
2.2. Bacterial Identification
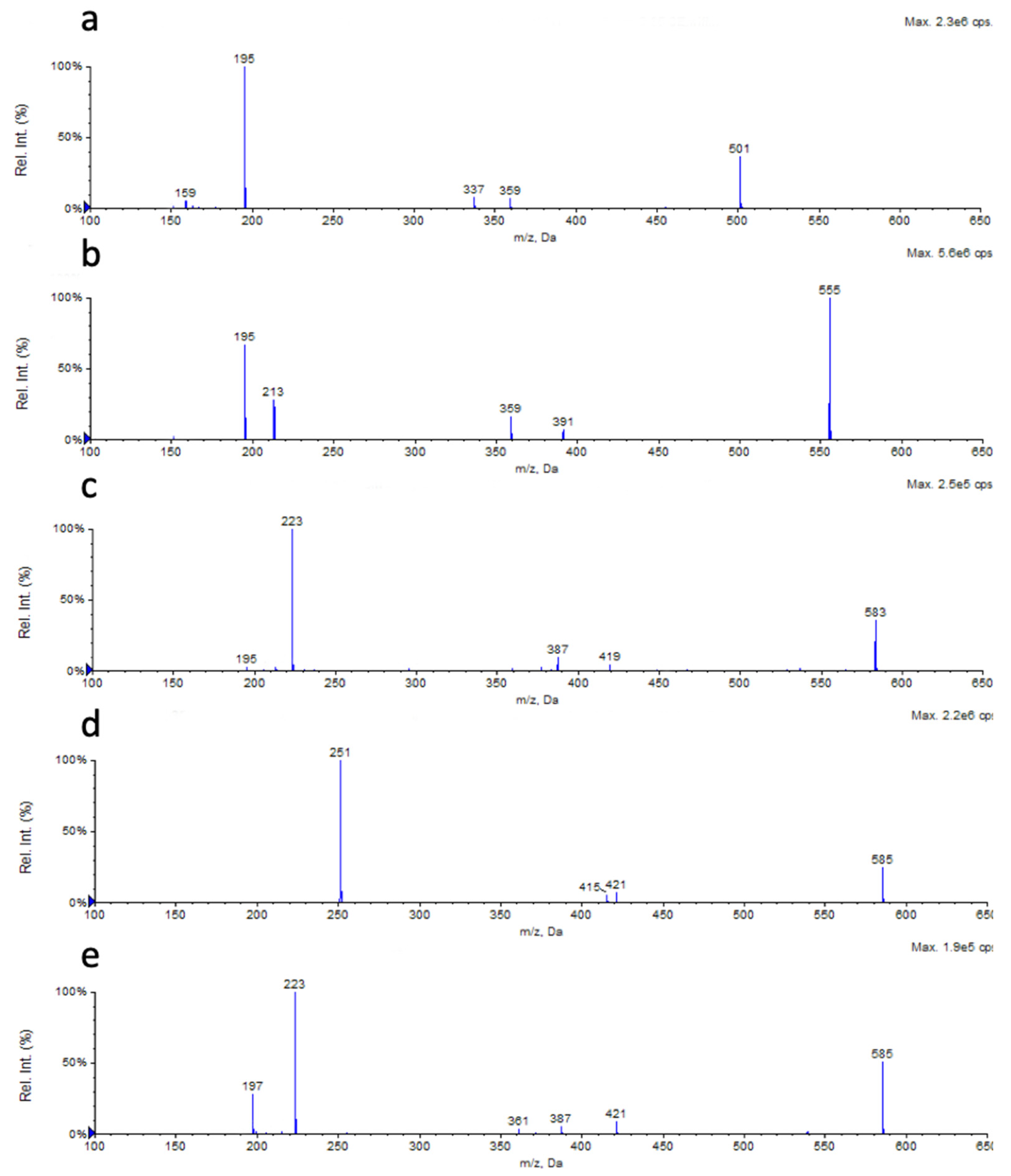
2.3. Chemical Characterization
2.4. Analysis of the Influence of the Carbon Source on the RL Chemical Composition
2.5. Antimicrobial Activity
3. Materials and Methods
3.1. Isolation of Microorganisms
3.2. Media and Buffers
3.3. Extract Preparation
3.4. Biosurfactant Screening
3.4.1. CTAB Agar Method
3.4.2. Emulsification Capacity Assay
3.4.3. Oil-Spreading Test
3.5. Bacterial Identification
3.6. Biosurfactant Fraction Enrichment
3.7. Chemical Profiling and Structural Analysis of Biosurfactants
3.8. Antimicrobial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Soberón-Chávez, G.; Maier, R.M. Biosurfactants: A general overview. In Biosurfactants: From Genes to Applications; Soberón-Chávez, G., Ed.; Springer: Berlin, Germany, 2011; Volume 20, pp. 1–11. [Google Scholar]
- Tiso, T.; Germer, A.; Küpper, B.; Wichmann, R.; Blank, L. Methods for Recombinant Rhamnolipid Production; Springer: Berlin, Germany, 2016. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wu, Q.; Hua, Y.; Chen, J.; Zhang, H.; Wang, H. Potential applications of biosurfactant rhamnolipids in agriculture and biomedicine. App. Microbiol. Biotechnol. 2017, 101, 8309–8319. [Google Scholar] [CrossRef] [PubMed]
- Caiazza, N.C.; Shanks, R.M.; O’Toole, G.A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, A.K.; Kappeli, O.; Fiechter, A.; Reiser, J. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J. Bacteriol. 1991, 173, 4212–4219. [Google Scholar] [CrossRef] [Green Version]
- Davey, M.E.; Caiazza, N.C.; O’Toole, G.A. Rhamnolipid surfactant production affects biofilm architecture in pseudomonas aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef] [Green Version]
- Abalos, A.; Pinazo, A.; Infante, M.R.; Casals, M.; García, F.; Manresa, A. Physicochemical and antimicrobial properties of new rhamnolipids produced by pseudomonas aeruginosa AT10 from Soybean Oil Refinery Wastes. Langmuir 2001, 17, 1367–1371. [Google Scholar] [CrossRef]
- Remichkova, M.; Galabova, D.; Roeva, I.; Karpenko, E.; Shulga, A.; Galabov, A.S. Anti-herpesvirus activities of Pseudomonas sp. S-17 rhamnolipid and its complex with alginate. Z. Nat. C J. Biosci. 2008, 63, 75–81. [Google Scholar] [CrossRef]
- Wang, X.; Gong, L.; Liang, S.; Han, X.; Zhu, C.; Li, Y. Algicidal activity of rhamnolipid biosurfactants produced by Pseudomonas aeruginosa. Harmful Algae 2005, 4, 433–443. [Google Scholar] [CrossRef]
- Goswami, D.; Borah, S.N.; Lahkar, J.; Handique, P.J.; Deka, S. Antifungal properties of rhamnolipid produced by Pseudomonas aeruginosa DS9 against Colletotrichum falcatum. J. Basic Microbiol. 2015, 55, 1265–1274. [Google Scholar] [CrossRef]
- Mishra, A.; Dubey, R.; Yabaji, S.; Jaiswal, S. Evaluation of antimycobacterial rhamnolipid production from non-cytotoxic strains of Pseudomonas aeruginosa isolated from rhizospheric soil of medicinal plants. Int. J. Biol. Res. 2016, 4, 112. [Google Scholar] [CrossRef] [Green Version]
- de Freitas Ferreira, J.; Vieira, E.A.; Nitschke, M. The antibacterial activity of rhamnolipid biosurfactant is pH dependent. Food Res. Int. 2019, 116, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Borah, S.N.; Goswami, D.; Sarma, H.K.; Cameotra, S.S.; Deka, S. Rhamnolipid biosurfactant against fusarium verticillioides to control stalk and ear rot disease of maize. Front. Microbiol. 2016, 7, 1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampelotto, H.P. Polar microbiology: Recent advances and future perspectives. Biology 2014, 3, 81–84. [Google Scholar] [CrossRef] [Green Version]
- Gudiña, E.J.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactants produced by marine microorganisms with therapeutic applications. Mar. Drugs 2016, 14, 38. [Google Scholar] [CrossRef] [Green Version]
- Perfumo, A.; Banat, I.M.; Marchant, R. Going green and cold: Biosurfactants from low-temperature environments to biotechnology applications. Trends Biotechnol. 2018, 36, 277–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tytgat, B.; Verleyen, E.; Obbels, D.; Peeters, K.; De Wever, A.; D’hondt, S.; De Meyer, T.; Van Criekinge, W.; Vyverman, W.; Willems, A. Bacterial diversity assessment in antarctic terrestrial and aquatic microbial mats: A comparison between bidirectional pyrosequencing and cultivation. PLoS ONE 2014, 9, e97564. [Google Scholar] [CrossRef] [Green Version]
- Wynn-Williams, D.D. Antarctic microbial diversity: The basis of polar ecosystem processes. Biodivers. Conserv. 1996, 5, 1271–1293. [Google Scholar] [CrossRef]
- Tedesco, P.; Maida, I.; Palma Esposito, F.; Tortorella, E.; Subko, K.; Ezeofor, C.; Zhang, Y.; Tabudravu, J.; Jaspars, M.; Fani, R.; et al. Antimicrobial activity of monoramnholipids produced by bacterial strains isolated from the Ross Sea (Antarctica). Mar. Drugs 2016, 14, 83. [Google Scholar] [CrossRef]
- Rahman, K.S.M.; Rahman, T.J.; McClean, S.; Marchant, R.; Banat, I.M. Rhamnolipid biosurfactant production by strains of pseudomonas aeruginosa using low-cost raw materials. Biotechnol. Prog. 2008, 18, 1277–1281. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure–function relationship of lipopeptide biosurfactants. BBA Mol. Cell Biol. Lipids 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Siegmund, I.; Wagner, F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 1991, 5, 265–268. [Google Scholar] [CrossRef]
- Walter, V.; Syldatk, C.; Hausmann, R. Screening Concepts for the Isolation of Biosurfactant Producing Microorganisms; Springer: New York, NY, USA, 2010; Volume 672, pp. 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Verhille, S.; Baida, N.; Dabboussi, F.; Hamze, M.; Izard, D.; Leclerc, H. Pseudomonas gessardii sp. nov. and Pseudomonas migulae sp. nov., two new species isolated from natural mineral waters. Int. J. Syst. Bacteriol. 1999, 49, 1559–1572. [Google Scholar] [CrossRef] [Green Version]
- Irorere, V.U.; Tripathi, L.; Marchant, R.; McClean, S.; Banat, I.M. Microbial rhamnolipid production: A critical re-evaluation of published data and suggested future publication criteria. App. Microbiol. Biotechnol. 2017, 101, 3941–3951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristoffersen, V.; Rämä, T.; Isaksson, J.; Andersen, H.J.; Gerwick, H.W.; Hansen, E. Characterization of rhamnolipids produced by an arctic marine bacterium from the pseudomonas fluorescence group. Mar. Drugs 2018, 16, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hošková, M.; Schreiberová, O.; Ježdík, R.; Chudoba, J.; Masák, J.; Sigler, K.; Řezanka, T. Characterization of rhamnolipids produced by non-pathogenic Acinetobacter and Enterobacter bacteria. Bioresour. Technol. 2013, 130, 510–516. [Google Scholar] [CrossRef]
- Řezanka, T.; Siristova, L.; Sigler, K. Rhamnolipid-producing thermophilic bacteria of species Thermus and Meiothermus. Extremophiles 2011, 15, 697. [Google Scholar] [CrossRef]
- Sharma, A.; Jansen, R.; Nimtz, M.; Johri, B.N.; Wray, V. Rhamnolipids from the rhizosphere bacterium pseudomonas sp. GRP3 that reduces damping-off disease in chilli and tomato nurseries. J. Nat. Prod. 2007, 70, 941–947. [Google Scholar] [CrossRef]
- Behrens, B.; Engelen, J.; Tiso, T.; Blank, L.M.; Hayen, H. Characterization of rhamnolipids by liquid chromatography/mass spectrometry after solid-phase extraction. Anal. Bioanal. Chem. 2016, 408, 2505–2514. [Google Scholar] [CrossRef]
- Wittgens, A.; Santiago-Schuebel, B.; Henkel, M.; Tiso, T.; Blank, L.M.; Hausmann, R.; Hofmann, D.; Wilhelm, S.; Jaeger, K.-E.; Rosenau, F. Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida—A step forward to tailor-made rhamnolipids. Appl. Microbiol. Biotechnol. 2018, 102, 1229–1239. [Google Scholar] [CrossRef]
- Ramani, K.; Jain, S.C.; Mandal, A.B.; Sekaran, G. Microbial induced lipoprotein biosurfactant from slaughterhouse lipid waste and its application to the removal of metal ions from aqueous solution. Colloids Surf. B 2012, 97, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Díaz De Rienzo, M.A.; Kamalanathan, I.D.; Martin, P.J. Comparative study of the production of rhamnolipid biosurfactants by B. thailandensis E264 and P. aeruginosa ATCC 9027 using foam fractionation. Process. Biochem. 2016, 51, 820–827. [Google Scholar] [CrossRef]
- Kumari, B.; Singh, S.N.; Singh, D.P. Characterization of two biosurfactant producing strains in crude oil degradation. Process. Biochem. 2012, 47, 2463–2471. [Google Scholar] [CrossRef]
- Nicolò, M.S.; Cambria, M.G.; Impallomeni, G.; Rizzo, M.G.; Pellicorio, C.; Ballistreri, A.; Guglielmino, S.P.P. Carbon source effects on the mono/dirhamnolipid ratio produced by Pseudomonas aeruginosa L05, a new human respiratory isolate. New Biotechnol. 2017, 39, 36–41. [Google Scholar] [CrossRef]
- Patowary, K.; Das, M.; Patowary, R.; Kalita, M.; Deka, S. Recycling of bakery waste as an alternative carbon source for rhamnolipid biosurfactant production. J. Surfacatnts Deterg. 2018, 373–384. [Google Scholar] [CrossRef]
- Tan, Y.N.; Li, Q. Microbial production of rhamnolipids using sugars as carbon sources. Microb. Cell Fact. 2018, 17, 89. [Google Scholar] [CrossRef]
- Eraqi, W.A.; Yassin, A.S.; Ali, A.E.; Amin, M.A. Utilization of crude glycerol as a substrate for the production of rhamnolipid by pseudomonas aeruginosa. Biotechnol. Res. Int. 2016, 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Chhetri, B.A.; Watts, C.K.; Islam, R.M. Waste cooking oil as an alternate feedstock for biodiesel production. Energies 2008, 1, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Tiwary, B.N. Isolation and characterization of glycolipid biosurfactant produced by a Pseudomonas otitidis strain isolated from Chirimiri coal mines, India. Bioresour. Bioprocess. 2016, 3, 42. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Mawgoud, A.M.; Hausmann, R.; Lépine, F.; Müller, M.M.; Déziel, E. Rhamnolipids: Detection, analysis, biosynthesis, genetic regulation, and bioengineering of production. In Biosurfactants: From Genes to Applications; Soberón-Chávez, G., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 13–55. [Google Scholar]
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. A stereospecific pathway diverts β-Oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chem. Biol. 2014, 21, 156–164. [Google Scholar] [CrossRef]
- Peng, X.; Yuan, X.-Z.; Liu, H.; Zeng, G.-M.; Chen, X.-H. Degradation of polycyclic aromatic hydrocarbons (PAHs) by laccase in rhamnolipid reversed micellar system. App. Biochem. Biotechnol. 2015, 176, 45–55. [Google Scholar] [CrossRef]
- Zhu, S.; Liang, S.; You, X. Effect of rhamnolipid biosurfactant on solubilization and biodegradation of polycyclic aromatic hydrocarbons. In Proceedings of the 2013 Third International Conference on Intelligent System Design and Engineering Applications, Hong Kong, China, 16–18 January 2013; pp. 651–654. [Google Scholar] [CrossRef]
- Magalhães, L.; Nitschke, M. Antimicrobial activity of rhamnolipids against Listeria monocytogenes and their synergistic interaction with nisin. Food Control 2013, 29, 138–142. [Google Scholar] [CrossRef] [Green Version]
- Onbasli, D.; Aslim, B. Determination of antimicrobial activity and production of some metabolites by Pseudomonas aeruginosa B1 and B2 in sugar beet molasses. Afr. J. Biotechnol. 2009, 7, 4614–4619. [Google Scholar]
- Sotirova, A.V.; Spasova, D.I.; Galabova, D.N.; Karpenko, E.; Shulga, A. Rhamnolipid–biosurfactant permeabilizing effects on gram-positive and gram-negative bacterial strains. Curr. Microbiol. 2008, 56, 639–644. [Google Scholar] [CrossRef]
- Das, P.; Yang, X.-P.; Ma, L.Z. Analysis of biosurfactants from industrially viable Pseudomonas strain isolated from crude oil suggests how rhamnolipids congeners affect emulsification property and antimicrobial activity. Front. Microbiol. 2014, 5, 696. [Google Scholar] [CrossRef]
- Zhang, L.; Jiao, L.; Zhong, J.; Guan, W.; Lu, C. Light up the interactions between bacteria and surfactants with aggregation-induced emission characteristics. Mater. Chem. Front. 2017, 1, 1829–1835. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Li, X.-Z.; Li, J. Antimicrobial Resistance in Stenotrophomonas Maltophilia: Mechanisms and Clinical Implications; Springer International: Cham, Switzerland, 2017; Volume 2, pp. 937–958. [Google Scholar] [CrossRef]
- Looney, W.J.; Narita, M.; Mühlemann, K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Corvec, S.; Poirel, L.; Naas, T.; Drugeon, H.; Nordmann, P. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 1530–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, N.; Sorokin, A.; Anderson, I.; Galleron, N.; Candelon, B.; Kapatral, V.; Bhattacharyya, A.; Reznik, G.; Mikhailova, N.; Lapidus, A.; et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 2003, 423, 87–91. [Google Scholar] [CrossRef]
- Soriano, F.; Huelves, L.; Naves, P.; Rodríguez-Cerrato, V.; del Prado, G.; Ruiz, V.; Ponte, C. In vitro activity of ciprofloxacin, moxifloxacin, vancomycin and erythromycin against planktonic and biofilm forms of Corynebacterium urealyticum. J. Antimicrob. Chemother. 2008, 63, 353–356. [Google Scholar] [CrossRef] [Green Version]
- Shahi, S.K.; Kumar, A. Isolation and genetic analysis of multidrug resistant bacteria from diabetic foot ulcers. Front. Microbiol. 2016, 6, 1464. [Google Scholar] [CrossRef] [Green Version]
- Alanazi, A.S.; Qureshi, K.A.; Elhassan, G.O.; Elsayed El-Agamy, E.I. Isolation, purification and characterization of antimicrobial agent antagonistic to escherichia coli ATCC 10536 produced by bacillus pumilus safr-032 isolated from the soil of unaizah, al qassim province of Saudi Arabia. Pak. J. Biol. Sci. 2016, 19, 191–201. [Google Scholar] [CrossRef]
- Werbrouck, H.; Vermeulen, A.; Van Coillie, E.; Messens, W.; Herman, L.; Devlieghere, F.; Uyttendaele, M. Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 2009, 134, 140–146. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, characterization, and application of bacteriophage LPSE1 against salmonella enterica in ready to eat (RTE) Foods. Front. Microbiol. 2018, 9, 1046. [Google Scholar] [CrossRef]
- Azizkhani, M.; Misaghi, A.; Basti, A.A.; Gandomi, H.; Hosseini, H. Corrigendum to “effects of zataria multiflora boiss. essential oil on growth and gene expression of enterotoxins A, C and E in Staphylococcus aureus ATCC 29213.” [Int. J. Food Microbiol. 163 (2013) 159–165]. Int. J. Food Microbiol. 2013, 166, 125. [Google Scholar] [CrossRef]
- Barry, A.L.; Jones, R.N. Criteria for disk susceptibility tests and quality control guidelines for the cefoperazone-sulbactam combination. J. Clin. Microbiol. 1988, 26, 13–17. [Google Scholar] [CrossRef] [Green Version]
- Lima, D.B.; Torres, A.F.C.; Mello, C.P.; de Menezes, R.R.P.P.B.; Sampaio, T.L.; Canuto, J.A.; da Silva, J.J.A.; Freire, V.N.; Quinet, Y.P.; Havt, A.; et al. Antimicrobial effect of Dinoponera quadriceps (Hymenoptera: Formicidae) venom against Staphylococcus aureus strains. J. Appl. Microbiol. 2014, 117, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; Deboy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piessens, V.; De Vliegher, S.; Verbist, B.; Braem, G.; Van Nuffel, A.; De Vuyst, L.; Heyndrickx, M.; Van Coillie, E. Characterization of coagulase-negative staphylococcus species from cows’ milk and environment based on bap, icaA, and mecA genes and phenotypic susceptibility to antimicrobials and teat dips. J. Dairy Sci. 2012, 95, 7027–7038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Li, X.-Z.; Poole, K. Fluoroquinolone susceptibilities of efflux-mediated multidrug-resistant Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia. J. Antimicrob. Chemother. 2001, 48, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Huertas Méndez, N.D.J.; Vargas Casanova, Y.; Gómez Chimbi, A.K.; Hernández, E.; Leal Castro, A.L.; Melo Diaz, J.M.; Rivera Monroy, Z.J.; García Castañeda, J.E. Synthetic peptides derived from bovine lactoferricin exhibit antimicrobial activity against E. coli ATCC 11775, S. maltophilia ATCC 13636 and S. enteritidis ATCC 13076. Molecules 2017, 22, 452. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, S.; Biehler, K.; Jonas, D. A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J. Bacteriol. 2009, 191, 2934–2943. [Google Scholar] [CrossRef] [Green Version]



| No. | Retention Time (min) | Measured [M − H]− (m)/z | Δ ppm vs. Theoretical Value | Molecular Formula | Key Fragment (Rha-FA1) | RL | n1 | n2 | Structure |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 14.22 | 475.2779 | −26.9 | C24H44O9 | 305.1290 | Rha-C8-C10 | 1 | 3 |  |
| 2 | 16.76 | 489.3087 | +4.7 | C25H46O9 | 319.1520 | Rha-C9-C10 | 2 | 3 | |
| 3 | 17.21 | 501.2946 | −23.5 | C26H46O9 | 359.2195 | Rha-C12:1-C8 | 5(-2H) | 1 | |
| 4 | 19.38 | 503.3036 | −36.6 | C26H48O9 | 333.1660 | Rha-C10-C10 | 3 | 3 | |
| 5 | 21.12 | 517.3278 | −12.1 | C27H50O9 | 381.2199 | Rha-C11-C10 | 4 | 3 | |
| 6 | 23.00 | 529.3102 | −52.0 | C28H50O9 | 359.1872 | Rha-C12:1-C10 | 5(-2H) | 3 | |
| 7 | 24.75 | 531.3319 | −40.3 | C28H52O9 | 361.2039 | Rha-C12-C10 | 5 | 3 | |
| 8 | 25.26 | 555.3454 | −14.2 | C30H52O9 | 359.1942 | Rha-C12:1-C12:1 | 5(-2H) | 5(-2H) | |
| 9 | 26.63 | 557.3553 | −24.6 | C30H54O9 | 387.2223 | Rha-C14:1-C10 | 7(-2H) | 3 | |
| 10 | 27.17 | 557.3553 | −24.6 | C30H54O9 | 361.2039 | Rha-C12-C12:1 | 5 | 5(-2H) | |
| 11 | 27.94 | 557.3641 | −8.8 | C30H54O9 | 359.1872 | Rha-C12:1-C12 | 5(-2H) | 5 | |
| 12 | 29.52 | 583.3678 | −28.8 | C32H5609 | 387.2296 | Rha-C14:1-C12:1 | 7(-2H) | 5(-2H) | |
| 13 | 29.84 | 559.3688 | −28.2 | C30H56O9 | 389.2431 | Rha-C14-C10 | 7 | 3 | |
| 14 | 31.46 | 585.3919 | −14.3 | C32H5809 | 415.2556 | Rha-C16:1-C10 | 9(-2H) | 3 | |
| 15 | 32.20 | 585.3919 | −14.3 | C32H5809 | 387.2296 | Rha-C14:1-C12 | 7(-2H) | 5 | |
| Rha-C12-C14:1 | 5 | 7(-2H) | |||||||
| 16 | 35.85 | 587.4017 | −24.2 | C32H6009 | 417.2728 | Rha-C16-C10 | 9 | 3 |
| RL Relative Abundance (%) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon Source | 1 | 2 | 3 * | 4 | 5 | 6 | 7 | 8 * | 9 | 10 | 11 | 12 * | 13 | 14 * | 15 * | 16 |
| Monosaccharides | ||||||||||||||||
| Glucose | 10.4 | 1.6 | 15.1 | 23.4 | 25.1 | 3.4 | 8.5 | 1.4 | 2.8 | 0.2 | 6.2 | 0.6 | 0.2 | 1.1 | ||
| Mannose | 27.9 | 0.1 | 0.4 | 5.9 | 29.3 | 25.1 | 2.4 | 5.1 | 0.6 | 0.8 | 0.1 | 1.5 | 0.5 | 0.3 | ||
| Rhamnose | 13.2 | 0.6 | 0.2 | 6.1 | 42.9 | 32.4 | 1.2 | 2.4 | 0.6 | 0.4 | ||||||
| Polysaccharides | ||||||||||||||||
| Starch | 29.6 | 9.2 | 36.4 | 19.8 | 0.6 | 2.6 | 0.2 | 0.3 | 0.9 | 0.4 | ||||||
| Xylan | 18.9 | 9.7 | 50.1 | 18.8 | 0.9 | 1.6 | ||||||||||
| Fatty acids and derivatives | ||||||||||||||||
| Glycerol | 5.2 | 0.2 | 5.3 | 9.5 | 0.5 | 19.1 | 14.0 | 8.0 | 12.5 | 1.9 | 5.0 | 0.3 | 11.8 | 4.7 | 1.0 | 1.0 |
| UCO | 5.9 | 3.2 | 13.4 | 17.4 | 17.0 | 10.7 | 5.4 | 2.6 | 6.4 | 1.3 | 12.2 | 3.3 | 1.0 | 0.2 | ||
| Petroleum derivatives | ||||||||||||||||
| Benzene | 24.1 | 0.3 | 0.2 | 6.4 | 37.3 | 25.5 | 2.0 | 2.0 | 0.3 | 0.6 | 1.3 | |||||
| Diesel | 22.7 | 0.6 | 2.2 | 10.8 | 0.5 | 30.4 | 22.8 | 1.9 | 2.7 | 0.8 | 1.1 | 0.3 | 2.7 | 0.5 | ||
| PAHs | ||||||||||||||||
| Anthracene | 4.6 | 9.0 | 1.2 | 28.4 | 50.2 | 1.0 | 2.1 | 0.3 | 0.6 | 2.4 | 0.2 | |||||
| Phenanthrene | 100.0 | |||||||||||||||
| Pyrene | ||||||||||||||||
| Miscellaneous | ||||||||||||||||
| TYP CE | 8.5 | 0.1 | 0.5 | 11.1 | 0.8 | 21.7 | 24.1 | 4.0 | 7.4 | 4.9 | 4.0 | 0.3 | 9.4 | 1.7 | 0.5 | 1.0 |
| TYP 80% | 10.4 | 2.5 | 2.1 | 12.5 | 1.3 | 16.9 | 22.2 | 6.2 | 7.8 | 2.0 | 4.2 | 0.1 | 10.7 | 0.9 | 0.1 | 0.1 |
| TYP 100% | 6.1 | 2.6 | 16.9 | 21.9 | 7.9 | 8.5 | 10.4 | 6.1 | 1.4 | 11.2 | 4.4 | 0.6 | 2.0 | |||
| Control | ||||||||||||||||
| Minimum Inhibitory Concentration (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|
| Strains | Fractions | Strains | Fractions | ||||
| Gram-Positive | 60% | 80% | 100% | Gram-Negative | 60% | 80% | 100% |
| B. cereus | - | 6.25 | 3.13 | S. maltophilia 700475 | - | 12.5 | 25.0 |
| L. monocytogenes | - | 25.0 | 12.5 | S. maltophilia 13637 | - | 12.5 | 12.5 |
| S. aureus 29213 | - | 12.5 | 25.0 | S. maltophilia 13636 | - | 12.5 | 25.0 |
| S. aureus 23235 | - | 12.5 | 12.5 | A. baumannii | - | - | - |
| S. aureus 6538P | - | 25.0 | 25.0 | B. metallica | - | - | - |
| S. epidermidis | - | 50.0 | 100 | E. coli | - | - | - |
| S. xylosus | - | - | - | K. pneumoniae | - | - | - |
| P. aeruginosa | - | - | - | ||||
| S. Enteritidis | - | - | - | ||||
| S. Typhimurium | - | - | - | ||||
| Minimum Inhibitory Concentration (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|
| Strains | Glucose | Mannose | Rhamnose | Glycerol | TYP | Xylan | Starch |
| Gram-positive | |||||||
| B. cereus | 6.25 | 100 | - | 3.13 | 7.81 | - | - |
| L. monocytogenes | 25.0 | - | - | 3.13 | 62.5 | - | - |
| S. aureus 6538P | 37.5 | 100 | - | 6.25 | 98.3 | - | - |
| S. epidermidis | - | - | - | 3.13 | 62.5 | - | - |
| Gram-negative | |||||||
| S. maltophilia 13637 | 50.0 | - | - | 3.13 | 62.5 | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buonocore, C.; Tedesco, P.; Vitale, G.A.; Palma Esposito, F.; Giugliano, R.; Monti, M.C.; D’Auria, M.V.; de Pascale, D. Characterization of a New Mixture of Mono-Rhamnolipids Produced by Pseudomonas gessardii Isolated from Edmonson Point (Antarctica). Mar. Drugs 2020, 18, 269. https://doi.org/10.3390/md18050269
Buonocore C, Tedesco P, Vitale GA, Palma Esposito F, Giugliano R, Monti MC, D’Auria MV, de Pascale D. Characterization of a New Mixture of Mono-Rhamnolipids Produced by Pseudomonas gessardii Isolated from Edmonson Point (Antarctica). Marine Drugs. 2020; 18(5):269. https://doi.org/10.3390/md18050269
Chicago/Turabian StyleBuonocore, Carmine, Pietro Tedesco, Giovanni Andrea Vitale, Fortunato Palma Esposito, Rosa Giugliano, Maria Chiara Monti, Maria Valeria D’Auria, and Donatella de Pascale. 2020. "Characterization of a New Mixture of Mono-Rhamnolipids Produced by Pseudomonas gessardii Isolated from Edmonson Point (Antarctica)" Marine Drugs 18, no. 5: 269. https://doi.org/10.3390/md18050269
APA StyleBuonocore, C., Tedesco, P., Vitale, G. A., Palma Esposito, F., Giugliano, R., Monti, M. C., D’Auria, M. V., & de Pascale, D. (2020). Characterization of a New Mixture of Mono-Rhamnolipids Produced by Pseudomonas gessardii Isolated from Edmonson Point (Antarctica). Marine Drugs, 18(5), 269. https://doi.org/10.3390/md18050269









