Abstract
Puniceusines A–N (1–14), 14 new isoquinoline alkaloids, were isolated from the extracts of a deep-sea-derived fungus, Aspergillus puniceus SCSIO z021. Their structures were elucidated by spectroscopic analyses. The absolute configuration of 9 was determined by ECD calculations, and the structures of 6 and 12 were further confirmed by a single-crystal X-ray diffraction analysis. Compounds 3–5 and 8–13 unprecedentedly contained an isoquinolinyl, a polysubstituted benzyl or a pyronyl at position C-7 of isoquinoline nucleus. Compounds 3 and 4 showed selective inhibitory activity against protein tyrosine phosphatase CD45 with IC50 values of 8.4 and 5.6 µM, respectively, 4 also had a moderate cytotoxicity towards human lung adenocarcinoma cell line H1975 with an IC50 value of 11.0 µM, and 14, which contained an active center, -C=N+, exhibited antibacterial activity. An analysis of the relationship between the structures, enzyme inhibitory activity and cytotoxicity of 1–14 revealed that the substituents at C-7 of the isoquinoline nucleus could greatly affect their bioactivity.
1. Introduction
Isoquinoline alkaloids are a large group of alkaloids in the plant kingdom, showing diverse pharmacological and biological activities such as anticancer, anti-inflammatory, cholesterol-lowering, antihyperglycemic, antiplasmodial, antifungal, and antimicrobial activity, etc. [1]. According to their structural components, isoquinoline and tetrahydroisoquinoline alkaloids can be classified into over 20 sub-classes, mainly including simple isoquinoline, benzylisoquinoline, bisbenzylisoquinoline, proto-berberine alkaloid, and aporphine alkaloid, etc. [1]. The most famous representative of this group is antidiabetic berberine. However, only a few isoquinoline alkaloids have been isolated from fungi, such as TMC-120A, B and C from Aspergillus ustus [2] and A. insuetus [3]; chaetoindicins A–C from Chaetomium indicum [4]; fusarimine from Fusarium sp. [5]; 8-methoxy-3,5-dimethylisoquinolin-6-ol from Penicillium citrinum [6]; azaphilone from P. sclerotiorum [7]; and spathullins A–B from P. spathulatum [8].
Malfunctions in protein tyrosine phosphatase (PTP) activity are linked to various diseases, ranging from cancer to neurological disorders and diabetes, such as CD45, SHP1, TCPTP, PTP1B and LAR linked to cancer, CD45, SHP1 and LAR also linked to neurological diseases, and PTP1B and LAR also linked to diabetes, etc. [9]. Some PTPs emerged as promising targets for therapeutic intervention in recent years [9,10]. However, achieving the selectivity of PTP inhibitors is a big challenge. In recent years, we obtained a series of natural compounds including anthraquinones [11], meroterpenoids [12], xanthone-type and anthraquinone-type mycotoxins [13], and oxaphenalenones [14] as PTP inhibitors from marine-derived fungi. In order to further explore diversified bioactive compounds from the deep-sea-derived fungus, Aspergillus puniceus SCSIO z021, we changed culture media and further conducted a chemical investigation of this strain cultured with a complex medium, which led to the characterization of 14 undescribed isoquinoline alkaloids, puniceusines A–N (1–14) (Figure 1). Compounds 1–14 were evaluated for their enzyme inhibitory activity against five kinds of PTPs, cytotoxicity towards human lung adenocarcinoma cell line H1975, and antibacterial activity. Herein, we report the isolation, structure elucidation and bioactivities of 1–14.
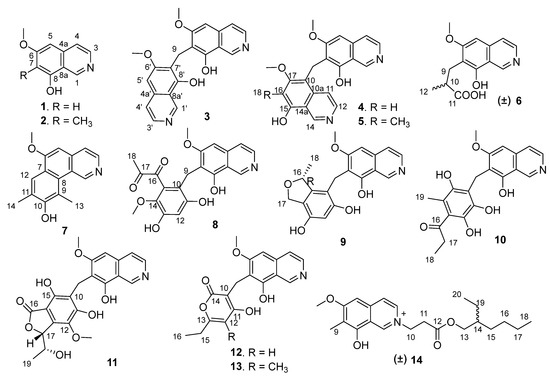
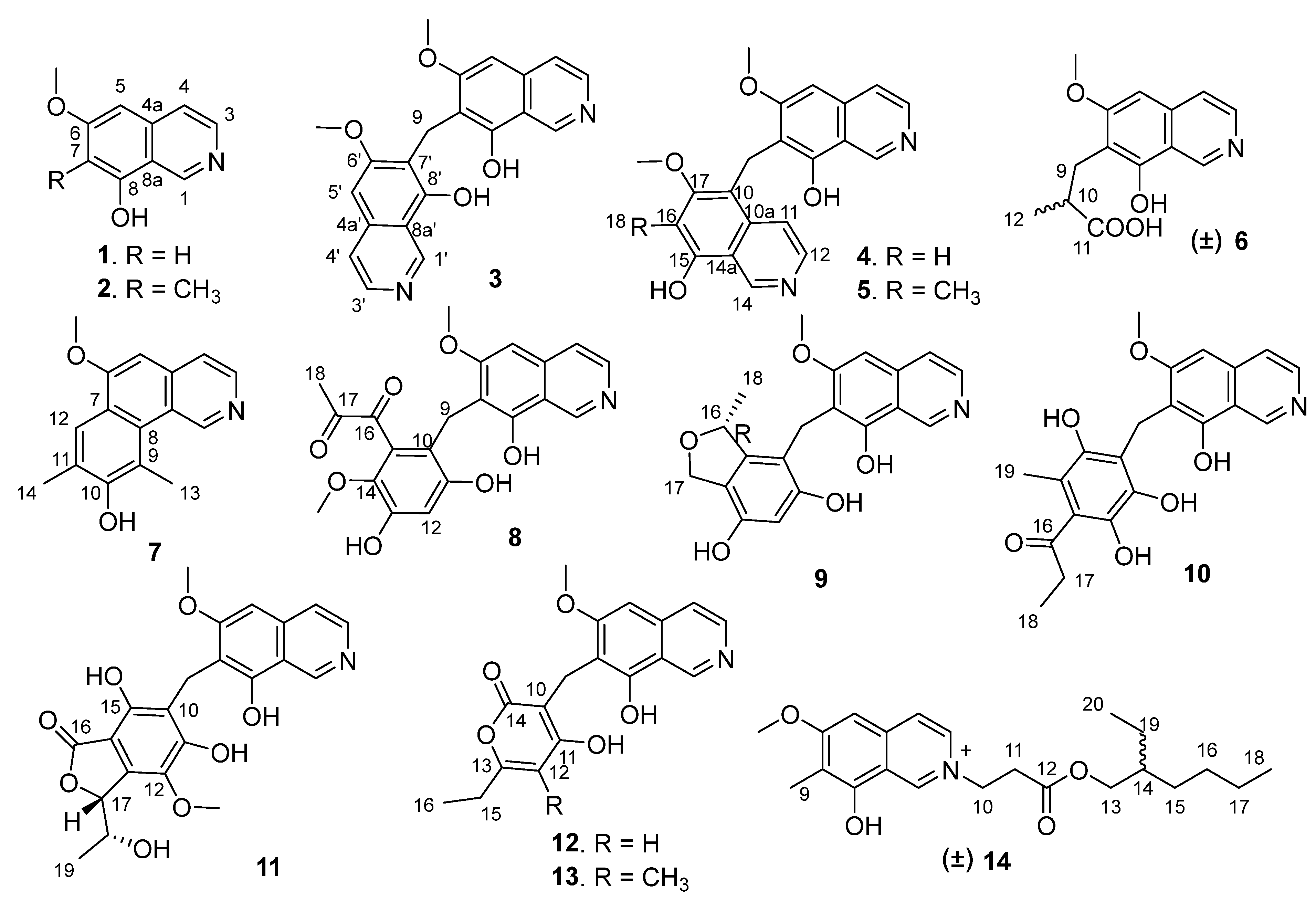
Figure 1.
Structures of the isolated compounds 1–14.
2. Results and Discussion
Puniceusine A (1) has the molecular formula C10H9NO2, as determined by HRESIMS. The 1H NMR spectrum (Table 1) showed the presence of one methoxy group at δH 3.96 (3H, s) and five aromatic hydrogens at δH 9.45 (1H, s), 8.40 (1H, d, J = 5.7 Hz), 8.06 (1H, d, J = 6.4 Hz), 7.13 (1H, s), 6.81 (1H, s). The 13C NMR spectrum (Table 2) showed 10 carbon signals including one methoxy, five aromatic methines, and four aromatic non-protonated carbons. These NMR data were similar to those of 6,8-dimethoxyisoquinolin [15] and papraline [16], and the only clear difference between 1 and 6,8-dimethoxyisoquinolin was the disappearance of one oxygenated methyl, which indicated that 1 was also an isoquinoline alkaloid. This was supported by the HMBC spectrum showing correlations from H-1 to C-3/C-4a/C-8a, H-3 to C-4/C-4a, H-4 to C-3/C-5/C-8a, H-5 to C-4/C-6/C-7/C-8a, and H-7 to C-5/C-6/C-8/C-8a. In addition, the HMBC correlation from δH 3.96 (3H, s) to C-6 suggested that a methoxy group was attached at C-6. Thus, the structure of 1 was determined to be 6-methoxy-8-hydroxy-isoquinolin.

Table 1.
1H NMR Data for Compounds 1–7 (δ in ppm, J in Hz) in DMSO-d6 or Methanol-d4.

Table 2.
13C NMR data for Compounds 1–8 (δ in ppm) in DMSO-d6 or Methanol-d4.
Puniceusine B (2) was assigned the molecular formula C11H11NO2 by HRESIMS. The 1H NMR and 13C NMR data (Table 1 and Table 2) showed great similarity to those of 1, and the main difference between them was the additional presence of one methyl (δH 2.25, 3H, s; δC 9.2) and the disappearance of one aromatic hydrogen in 2. The HMBC correlations from δH 2.25 to C-6/C-7/C-8 suggested the additional methyl attached at C-7. Hence, the structure of 2 was determined to be 6-methoxy-7-methyl-8-hydroxy-isoquinolin.
Puniceusine C (3) was found to have the molecular formula C21H18N2O4 by HRESIMS that was nearly twice that of 2. The 1H and 13C NMR data (Table 1 and Table 2) showed great similarity to those of 2, and the clearest difference between them was the disappearance of a methyl signal and the additional presence of a methylene signal (δH 4.32, 2H, s; δC 19.4) in 3. The 1H NMR spectrum of 3 showed a double increase in the integral areas for H-1, H-3, H-4, H-5, and a methoxy group. The HMBC correlations (Figure 2) from δH 4.32 (H2-9) to C-6/C-7/C-8 suggested a methylene instead of a methyl attached at C-7. These data indicated that 3 was a symmetrical dimer of 1, connected at positions C-7′ and C-7 by a methylene C-9. Thus, the structure of 3 was determined as shown.
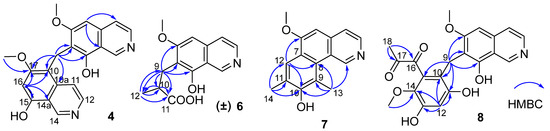
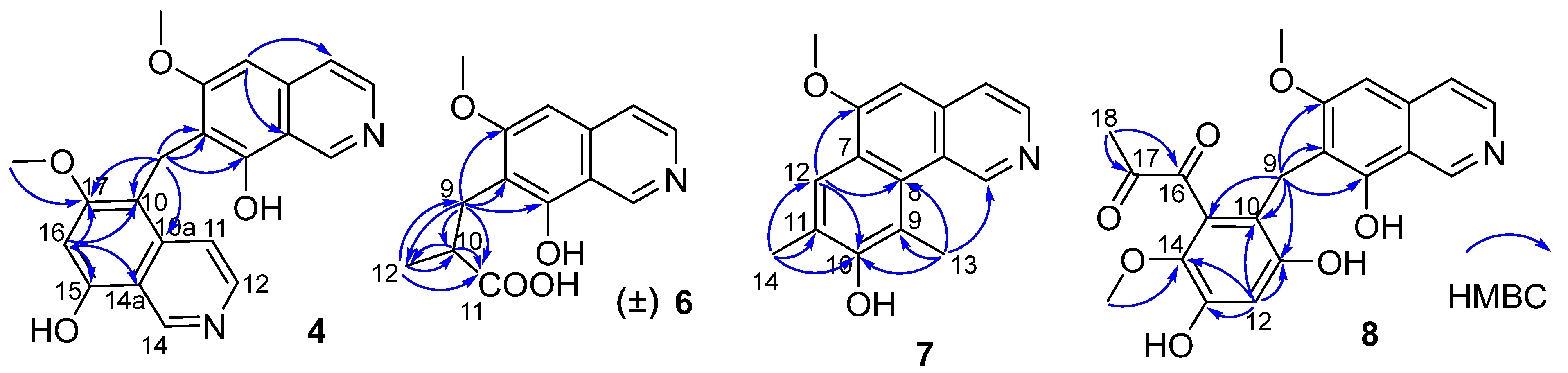
Figure 2.
Key HMBC correlations of compounds 4, 6–8.
Puniceusine D (4) showed the same molecular formula of C21H18N2O4 as that of 3 by analysis of its HRESIMS and NMR data (Table 1 and Table 2). The 1H NMR spectrum showed the presence of two downfield hydrogens at δH 9.59 (1H, s) and 9.52 (1H, s); six aromatic hydrogens at δH 8.58 (1H, d, J = 7.0 Hz), 8.31 (1H, d, J = 7.8 Hz), 8.29 (1H, d, J = 7.0 Hz), 8.05 (1H, d, J = 6.5 Hz), 7.11 (1H, s), and 7.00 (1H, s); two methoxys groups at δH 3.79 (3H, s) and 3.98 (3H, s); and one methylene at δH 4.47 (2H, s). The 13C NMR spectrum showed 21 carbon signals including one methylene, two methoxyls, eight aromatic methines, and ten aromatic non-protonated carbons. These data showed similarity to those of 1–3, which indicated that 4 was also a dimer of 1. The HMBC correlations from H-5 to C-4/C-8a, from H-16 to C-10/C-14a/C-15/C-17, and from H2-9 to C-6/C-7/C-8/C-10/C-10a/C-17 (Figure 2) suggested that 4 was an asymmetric dimer of 1 connected at positions C-7 and C-10 by a methylene C-9. The two methoxy groups were attached at C-6 and C-17 based on the HMBC correlations of δH 3.98 (3H, s) with C-17 and δH 3.79 (3H, s) with C-6, respectively. Therefore, the structure of 4 was established as shown.
The molecular formula of puniceusine E (5) was determined as C22H21N2O4 by HRESIMS. The 1H and 13C NMR data (Table 1 and Table 2) of 5 were greatly similar to those of 4, and the only obvious difference between them was the absence of one aromatic hydrogen and the additional presence of one methyl signal (δH 2.38, 3H, s; δC 10.8) in 5. The HMBC correlations from H3-18 (δH 2.38) to C-15/C-16/C-17 suggested a methyl located at C-16 instead of a hydrogen. Thus, the structure of 5 was established as shown.
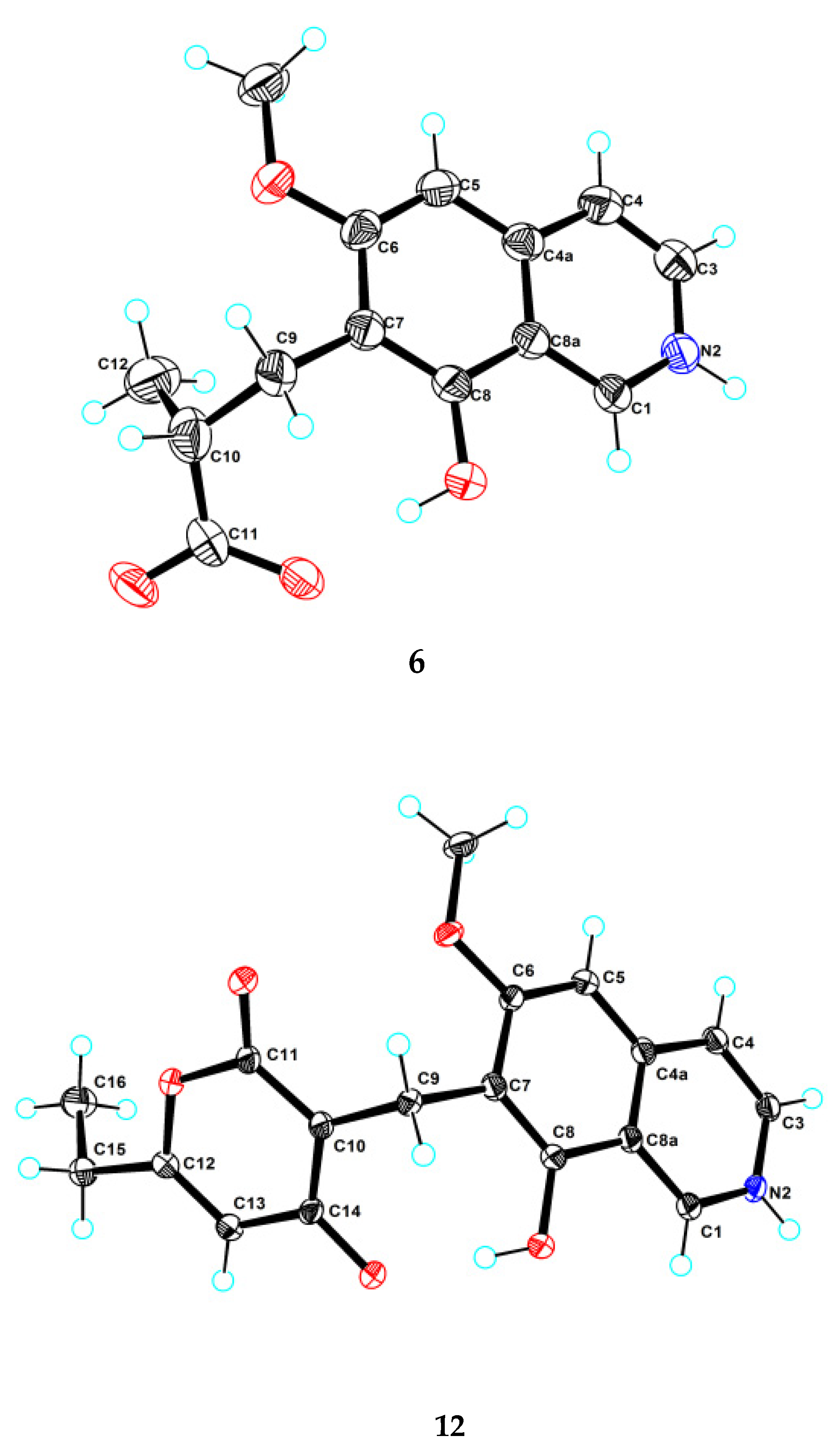
Puniceusine F (6) had the molecular formula C14H15NO4, as determined by HRESIMS. The 1H and 13C NMR data (Table 1 and Table 2) of 6 were similar to those of 1, and the clearest difference between them was the disappearance of one aromatic hydrogen and the additional presence of one methyl (δH 1.27, d, J = 7.5 Hz, 3H), one methylene, one methine, and one carboxyl group in 6. The HMBC correlations (Figure 2) from H2-9 (δH 2.97 (dd, J = 14.0, 5.5 Hz, 1H), 3.17 (dd, J = 14.0, 8.5 Hz, 1H)) to C-6/C-7/C-8/C-10/C-11/C-12, from H-10 (δH 2.85, m, 1H) to C-11/C-12, and from H3-12 (δH 1.27, d, J =7.5 Hz, 3H) to C-9/C-10/C-11 suggested an isobutyric acid group attached at C-7 of the isoquinoline nucleus. The optical rotation and measured CD data of 6 were zero, which indicated that 6 was a racemic mixture. This was supported by the HPLC analysis of 6 with a chiral column (CHIRALPAK IA 4.6 mm × 250 mm column) eluting with n-hexane/ethanol/TFA (68:32:0.2, v/v) (Figure S39). This structure was further confirmed by a single-crystal X-ray diffraction analysis (Figure 3).
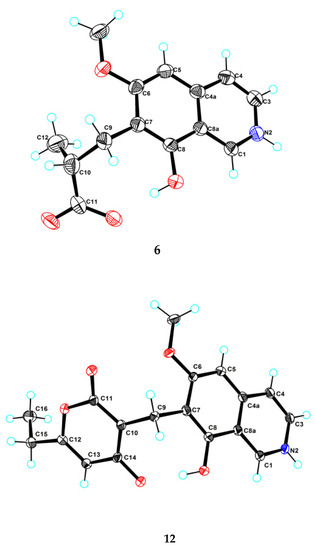
Figure 3.
Ortep plot of the X-ray crystallographic data for 6 and 12.
The molecular formula of puniceusine G (7) was determined to be C16H15NO2, according to its HRESIMS. The 1H and 13C NMR data (Table 1 and Table 2) of 7 were similar to those of 1, and the clearest difference between them was the additional presence of two double bonds (δC 112.0 (C, C-9), 115.0 (CH, C-12), 120.7 (C, C-11), 148.7 (C, C-10)) and two methyls (δH 2.51 (3H, s, H-14), 2.93 (3H, s, H-13); δC 8.0, 8.4) in 7. The HMBC correlations (Figure 2) from H-12 to C-6/C-8/C-10, from H3-13 to C-1/C-8/C-9/C-10, and from H3-14 to C-10/C-11/C-12, suggested a 3,5-dimethyl-4-hydroxyphenyl group attached at the isoquinoline nucleus by sharing C-7 and C-8 to form a benzo[h]isoquinoline unit. Therefore, the structure of 7 was established as shown.
Puniceusine H (8) had the molecular formula of C21H19NO7 as determined by its HRESIMS. The 1H and 13C NMR data (Table 2 and Table 3) of 8 showed similarity to those of 4–6 with the presence of characteristic chemical shifts for a methylene C-9 (δH 4.01 (2H, s), δC 19.3 (CH2)). The HMBC correlations (Figure 2) from H2-9 to C-6/C-7/C-8/C-10/C-11/C-15, from H-12 (δH 6.58, 1H, s) to C-10/C-11/C-13/C-14, and from H3-19 (δH 3.52, 3H, s) to C-14 suggested a 2,4-dihydroxy-5-methoxyphenyl fragment connected with isoquinoline unit by a methylene at position C-7. In addition, the HMBC correlations from H3-18 (δH 2.30, 3H, s) to C-16 (δC 195.1, C)/C-17 (δC 196.9, C) suggested the presence of a 1,2-propanedione group that was exclusively assigned to attach at C-15 of the benzene ring based on the above data. Thus, the structure of 8 was established as shown.

Table 3.
1H NMR data for Compounds 8–14 (δ in ppm, J in Hz) in DMSO-d6 or Methanol-d4.
Puniceusine I (9) had the molecular formula of C20H19NO5 according to its HRESIMS with 12 degrees of unsaturation. The 1H and 13C NMR data (Table 3 and Table 4) of 9 were similar to those of 8, and the clearest difference between them was the disappearance of two keto carbons and one methoxy and the additional presence of one oxygenated methylene (δH 4.84 (1H, dd, J = 11.5, 2.5 Hz), 4.69 (1H, dd, J = 11.5 Hz), δC 68.9, CH2) and one oxygenated methine (δH 5.22 (1H, m), δC 79.3, CH). A detailed analysis of HSQC and HMBC spectra proved that 9 also contained a 1,2,4,5,6-pentasubstituted-benzyl attached at C-7 of isoquinoline nucleus. In addition, the HMBC correlations from H-16 to C-14/C-15/C-17, from H2-17 to C-10/C-11/C-12/C-13/C-14/C-15/C-16/C-18, from H3-18 (δH 1.35, d, J = 6.2 Hz, 3H) to C-15/C-16 (Figure 4), suggested a 2-methyl-2,5-dihydrofuran ring connected with the benzene ring via C-14 and C-15. Therefore, the 2D structure of 9 was established as shown. The absolute configuration of 9 was further determined by electronic circular dichroism (ECD) calculations (Tables S1–S3).

Table 4.
13C NMR data for Compounds 9–14 (δ in ppm) in DMSO-d6 or Methanol-d4.
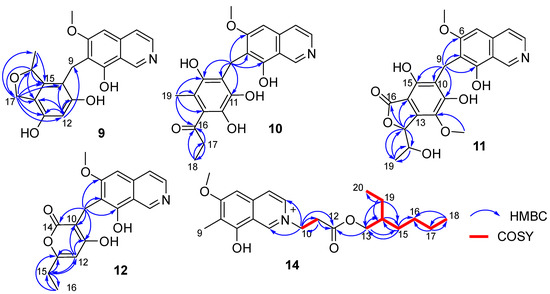
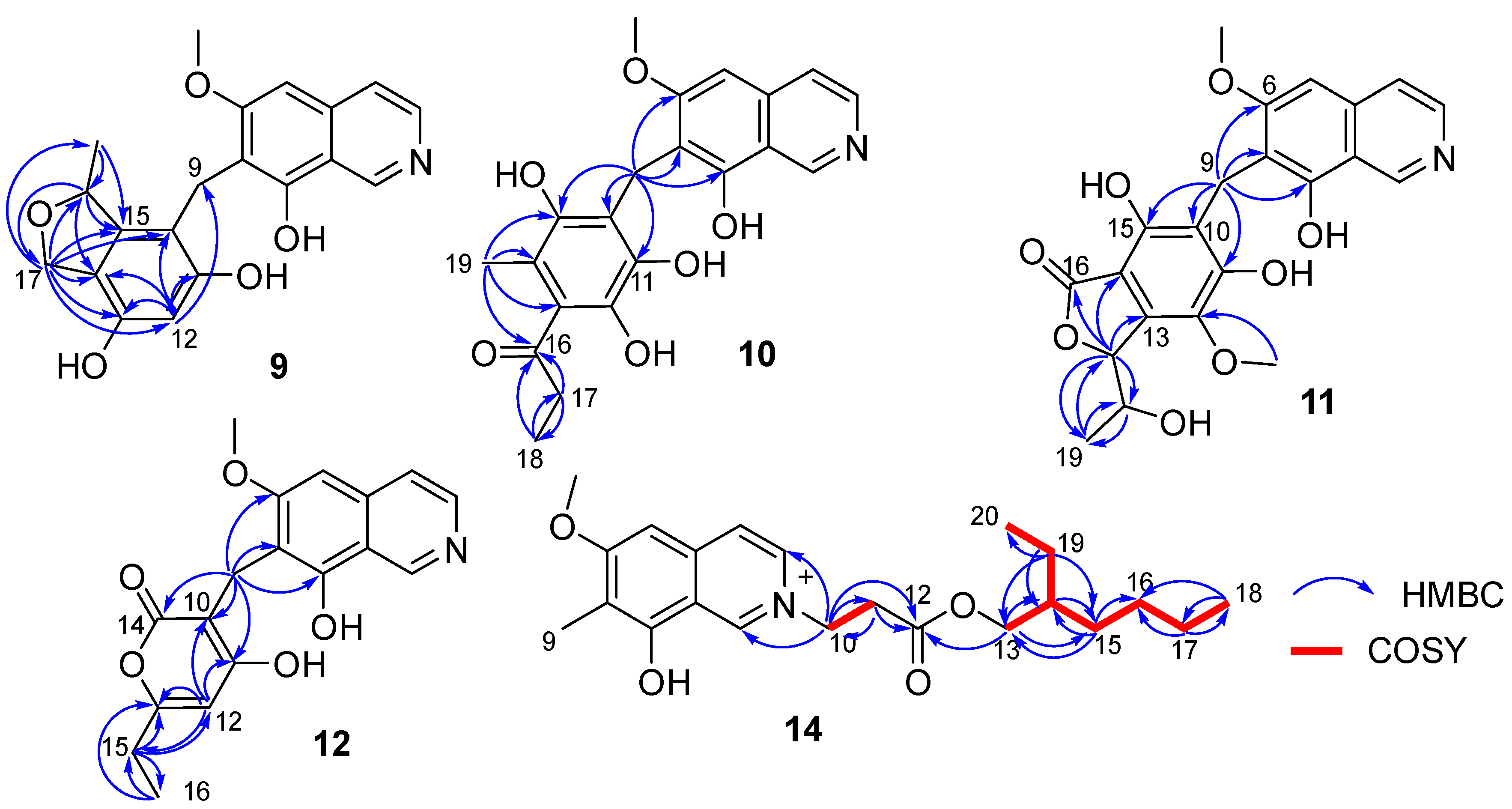
Figure 4.
Key COSY and HMBC correlations of compounds 9–12 and 14.
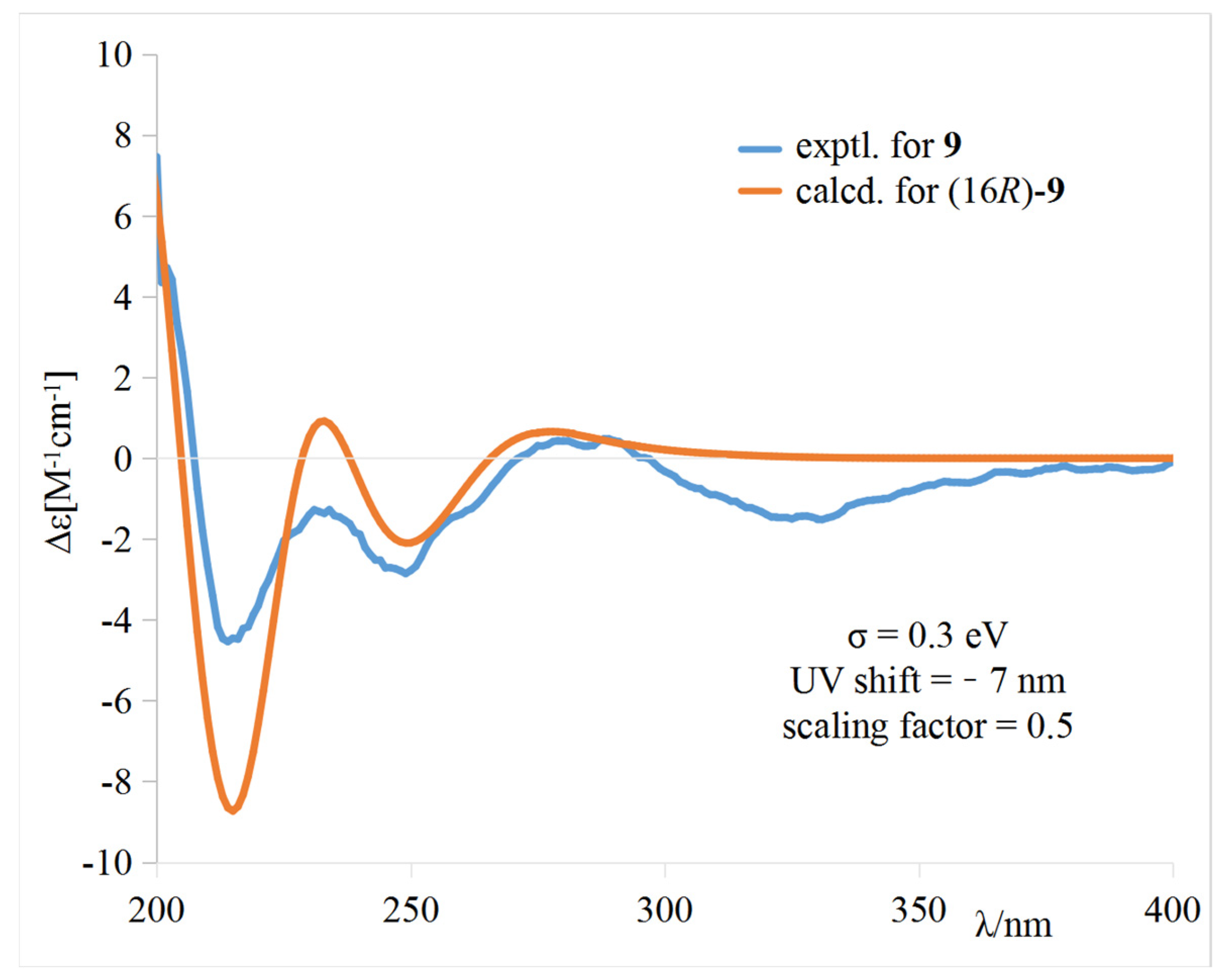
The calculated weighted ECD spectrum of (16R)-9 agreed well with the experimental ECD spectrum of 9 (Figure 5), leading to the assignment of the absolute configuration at C-16.
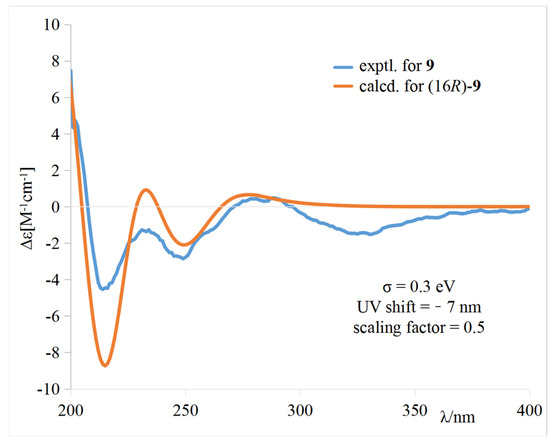
Figure 5.
Comparison of the experimental and calculated ECD spectra of 9 in CH3OH.
Puniceusine J (10) had the molecular formula of C21H21NO6, as determined by its HRESIMS. The 1H and 13C NMR data of 10 (Table 3 and Table 4) were very similar to those of 8 and 9. A detailed analysis of HSQC and HMBC spectra proved that 10 also contained a benzyl attached at position C-7 of isoquinoline unit. The HMBC correlations of H2-9 with C-6/C-7/C-8/C-10/C-11 (δC 170.0, C)/C-15 (δC 171.6, C) (Figure 4) suggested two hydroxyl groups attached at C-11 and C-15, respectively. In addition, the HMBC correlations from H3-19 (δH 1.92, 3H, s) to C-13/C-14/C-15 suggested a methyl attached at C-14. Furthermore, the HMBC correlations (Figure 4) from H2-17 (δH 2.58, 2H, q, J = 7.3 Hz) to C-16/C-18, from H3-18 (δH 1.02, 3H, t, J = 7.3 Hz) to C-16/C-17, and from H3-19 to C-16 suggested an 1-acetonyl group attached at C-13. Lastly, the chemical shift of C-12 (δC 163.1, C) indicated a hydroxy group attached at C-12. Therefore, the structure of 10 was established as shown.
Puniceusine K (11) was found to have the molecular formula C22H21NO8 by HRESIMS. The 1H and 13C NMR data of 11 (Table 3 and Table 4) were very similar to those of 10. A detailed analysis of HSQC and HMBC spectra proved that 11 also contained a hexasubstituted benzyl attached at position C-7 of isoquinoline unit. The HMBC correlations of H2-9 with C-6/C-7/C-8/C-10/C-11 (δC 157.1, C)/C-15 (δC 153.0, C) suggested two hydroxyl groups attached at C-11 and C-15, respectively. Additionally, the HMBC correlations (Figure 4) from H-17 (δH 5.56, d, J = 2.5 Hz) to C-13/C-14/C-16/C-18/C-19, from H-18 (δH 4.36, qd, J = 6.5, 2.5 Hz) to C-19, and from H3-19 (δH 0.94, d, J = 6.5 Hz, 3H) to C-17/C-18, suggested a 5-hydroxy-2-hexene-4-lactone group attached on the benzene ring via C-13 and C-14. In addition, the HMBC correlation of δH 3.76 (s, 3H) with C-12 (δC 136.8, C) suggested a methoxy group attached at C-12 of the benzene ring. The assignment of the substituent groups at positions C-12, C-13 and C-14 of the benzene ring was further supported by comparison of the 1H and 13C NMR data of the same isobenzofuran moiety in 11, embeurekol C [17] and acetophthalidin [18]. In addition, the small 3JHH value (2.5 Hz) between H-17 and H-18 in 11 was closely similar to that of embeurekol C [17]. Furthermore, the specific rotation value of 11 ([α] −9.1 (c 0.1, CH3OH)) was close to that of embeurekol C ([α] −17 (c 0.05, CH3OH)) [17], and the experimental ECD spectrum of 11 (Figure S76) was greatly similar to that of embeurekol C [17]. These data suggested that the absolute configuration of 11 was also 17R, 18S for that of embeurekol C.
Puniceusine L (12) had a molecular formula of C18H17NO5 on the basis of its HRESIMS and NMR data. Its 1H and 13C NMR data (Table 3 and Table 4) showed a similarity to those of 8–11. A detailed analysis of HSQC and HMBC spectra suggested that 12 contained the same isoquinoline unit as 8–11. In addition, considering the molecular formula and unsaturation degrees of 12, the HMBC correlations from H2-9 to C-6/C-7/C-8/C-10/C-11/C-14, from H-12 to C-10/C-11/C-13/C-15, from H2-15 to C-12/C-13/C-16, and from H3-16 to C-13/C-15 (Figure 4), suggested a 6-ethyl-4-hydroxy-2H-pyran-2-one unit attached at the methylene C-9 of isoquinoline unit. The above assignment was further confirmed by a single crystal X-ray diffraction analysis (Figure 3).
Puniceusine M (13) had the molecular formula of C19H19NO5 on the basis of its HRESIMS. The 1H and 13C NMR data (Table 3 and Table 4) were very similar to those of 12. The only difference between them was the disappearance of one aromatic hydrogen and the additional presence of a methyl (δH 2.01 (3H, s), δC 9.9). The HMBC correlations from H3-17 (δH 2.01) to C-11/C-12/C-13 suggested the additional methyl attached at C-12. Therefore, the structure of 13 was established as shown.
Puniceusine N (14) had the molecular formula C22H32NO4+, as determined by HRESIMS. The 1H and 13C NMR (Table 3 and Table 4) data of 14 showed a similarity to those of 2, and the clearest difference between them was the additional presence of two methyls, seven methylenes (one oxygenated), one methine, and one carboxyl in 14. Detailed analysis of the HMBC and COSY spectra proved that 14 contained the same isoquinoline unit as that of 2. In addition, combining with the COSY correlation of H2-10 (δH 4.88, t, J = 6.3 Hz) with H2-11 (δH 3.18, t, J = 6.3 Hz) (Figure 4), the HMBC correlations from H2-10 to C-1/C-3/C-11/C-12 (δC 171.8, C), and from H2-11 to C-9/C-12 (Figure 4), suggested a -CH2-CH2-COO- group attached at the nitrogen atom of isoquinoline unit. Furthermore, the sequential COSY correlations of H2-13/H-14/H2-15/H2-16/H2-17/H2-18, and H-14/H2-19/H3-20 (Figure 4), together with the HMBC correlations from H2-13 to C-12/C-14/C-15, from H2-15 to C-13/C-14/C-16, from H2-17 to C-16/C-18, from H3-18 to C-16/C-17, from H2-19 to C-13/C-14/C-15/C-20, and from H3-20 to C-14/C-19 (Figure 4), suggested that the 2-ethylhexanol group connected with the carboxyl of the -CH2-CH2-COO- group to form an ester. The optical rotation and measured CD data of 14 were zero, which indicated 14 was a racemic mixture. However, an HPLC analysis of 14 with a chiral column (CHIRALPAK IA and IB, respectively, 4.6 mm × 250 mm column), eluting with n-hexane/ethanol/TFA (Figures S99 and S100), showed a big trailing peak. The reason for this could be that the two kinds of chiral columns were not suitable for the chiral separation of 14. Thus, the structure of 14 was determined as shown.
All of the 14 compounds were evaluated for their enzyme inhibitory activity against five PTPs including CD45, SHP1, TCPTP, PTP1B and LAR, cytotoxicity towards human lung adenocarcinoma cell line H1975, and antibacterial activity. The results of protein phosphatase inhibition assays (Table 5) showed that only 3 and 4 selectively exhibited significant inhibitory activity against CD45 with IC50 values of 8.4 and 5.6 µM, respectively, and 1, 8, 9, 10, 12 and 13 showed a mild inhibitory activity against several PTPs. A cytotoxicity assay (Table 5) showed that only 4 had a moderate cytotoxicity towards H1975 cell lines with an IC50 value of 11.0 µM. The analysis of the relationship of their structures, enzyme inhibitory activity and cytotoxicity displayed that the substituents at C-7 of the isoquinoline nucleus could greatly affect their bioactivity. In addition, antibacterial assays exhibited that 14 had medium antibacterial activity towards Staphylococcus aureus, methicillin-resistant S. aureus (MRSA), and Escherichia coli, with MIC values of 100 µg/mL, and 4 could inhibit the growth of E. coli with a MIC value of 100 µg/mL, while other compounds did not show clear antibacterial activity towards the three indicators. The results indicated that -C=N+ unit was an active center for the antibacterial activity of 14.

Table 5.
Inhibition activity against five phosphatases and cytotoxicity of 1–14.
3. Experimental Section
3.1. General Experimental Procedure
The procedures were the same as previously reported [13,14].
3.2. Fungal Material
The strain Aspergillus puniceus SCSIO z021 was isolated from a deep-sea sediment of Okinawa Trough (27°34.01′ N and 126°55.59′ E, ~1589 depth), which was located approximately 4.7 km from an active hydrothermal vent. The strain (GenBank accession number KX258801) was identified as Aspergillus puniceus through DNA extraction, ITS sequence amplification and sequence alignment, which has a 99% similarity to A. puniceus (GenBank accession number GU456970). The strain A. puniceus SCSIO z021 was deposited in the RNAM Center, South China Sea Institute of Oceanology, Chinese Academy of Science.
3.3. Fermentation and Extraction
The fungus strain was cultivated on potato glucose agar (PDA) plate containing 3% sea salt at 28 °C for 7 days. The spores were selected and transferred to a complex culture medium (glucose 1%, D-mannitol 2%, maltose 2%, corn meal 0.05%, monosodium glutamate 1%, KH2PO4 0.05%, MgSO4·7H2O 0.03%, yeast extract 0.3%, sea salt 3%) to obtain a spore suspension that was cultured in a shaker at 28 °C for 3 days at a rotating speed of 180 rmp. The fungus was cultured in 1 L Erlenmeyer flasks each containing 300 mL of 3# medium (glucose 1%, D-mannitol 2%, maltose 2%, corn meal 0.05%, monosodium glutamate 1%, KH2PO4 0.05%, MgSO4·7H2O 0.03%, yeast extract 0.3%, sea salt 3%) at 28 °C for 33 days under static condition. After fermentation, the broth and mycelia were separated with gauze. The broth was extracted with XAD-16 resin and sequentially eluted with H2O and EtOH to obtain crude extract (61.7 g). The mycelia was extracted three times with acetone, and further extracted three times with EtOAc to yield a crude extract (48.2 g).
3.4. Isolation and Purification
The combined extracts (109.9 g) were subjected to a normal-phase silica gel column eluting with a gradient of dichloromethane (DCM)/MeOH (100:0, 98:2, 95:5, 95:5, 90:10, 80:20, 70:30, 50:50, 0:100) to give nine subfractions (Fr.1–Fr.9) based on TLC analysis. Fr.4 (6.6 g) was separated by ODS column using MeOH-H2O-TFA (5:95:0.02 to 100:0:0.02) as eluent to afford 10 subfractions (Fr.4.1–Fr.4.10). Fr.4.1 was separated by Sephadex LH-20 eluting with MeOH followed by semipreparative HPLC (MeOH/H2O/TFA, 28:72:0.03, 3 mL/min) to yield 1 (11.6 mg, tR = 15.8 min). Fr.4.3 was subjected to Sephadex LH-20 using MeOH as mobile phase, which was further purified by semipreparative HPLC (CH3CN/H2O/TFA, 17:83:0.03, 3 mL/min) to yield 2 (30.7 mg, tR = 16.3 min). Fr.4.5 was isolated by Sephadex LH-20 eluting with MeOH, then purified by semipreparative HPLC (CH3CN/H2O/TFA, 25:75:0.03, 3 mL/min) to yield 7 (2.9 mg, tR = 25.9 min). Fr.4.6 was separated by Sephadex LH-20 with a mobile phase of MeOH, and then further purified by semipreparative HPLC (MeOH/H2O/TFA, 45:55:0.03, 3 mL/min) to yield 10 (2.8 mg, tR = 16.1 min) and 12 (15.2 mg, tR = 14.5 min). Fr.4.8 was isolated by silica gel column eluting with a gradient of CH2Cl2/MeOH (100:0, 80:1, 60:1, 40:1, 10:1, 5:1, 1:1, 0:100) to obtain three fractions, then Fr.4.8.1 was further purified by semipreparative HPLC (CH3CN/H2O/TFA, 49:51:0.03, 3 mL/min) to yield 14 (29.4 mg, tR = 20.2 min). Fr.6 (11.0 g) was separated by ODS column using MeOH-H2O-TFA (5:95:0.02 to 100:0:0.02) as mobile phase to yield eight subfractions (Fr.6.1–Fr.6.8). Fr.6.3 was purified by semipreparative HPLC (CH3CN/H2O/TFA, 21:79:0.03, 3 mL/min) to obtain 6 (4.0 mg, tR = 19.5 min), 3 (17.6 mg, tR = 18.4 min) and 4 (12.2 mg, tR = 19.7 min). Fr.6.6 was isolated by semipreparative HPLC (CH3CN/H2O/TFA, 24:76:0.03, 3 mL/min) to yield 11 (7.8 mg, tR = 16.0 min) and 5 (4.3 mg, tR = 14.4 min). Fr.6.7 was isolated by Sephadex LH-20 with MeOH as mobile phase, then further purified by semipreparative HPLC (CH3CN/H2O/TFA, 28:72:0.03, 3 mL/min) to yield 8 (4.1 mg, tR = 39.2 min) and 9 (2.8 mg, tR = 18.4 min). Fr.6.8 was separated by Sephadex LH-20, eluting with MeOH and further purified by semipreparative HPLC (CH3CN/H2O/TFA, 35:65:0.03, 3 mL/min) to get 13 (10.3 mg, tR = 18.0 min).
Puniceusine A (1): white acicular crystal; UV (CH3OH) λmax (log ε) 204 (1.42), 208 (1.42), 241 (1.56), 258 (1.58), 357 (0.74) nm; 1H and 13C NMR, Table 1 and Table 2; HRESIMS m/z 176.0709 [M + H]+ (calcd for C10H10NO2, 176.0706).
Puniceusine B (2): pale yellow powder; UV (CH3OH) λmax (log ε) 206 (1.61), 245 (1.75), 260 (1.83), 305 (0.73), 360 (0.84) nm; 1H and 13C NMR, Table 1 and Table 2; HRESIMS m/z 190.0864 [M + H]+ (calcd for C11H12NO2, 190.0863).
Puniceusine C (3): pale yellow powder; UV (CH3OH) λmax (log ε) 205 (1.87), 244 (1.98), 262 (2.04), 332 (1.07) nm; IR (film) νmax 3415, 1678, 1436, 1382, 1321, 1203, 1184, 1130, 1022, 954, 900, 840, 800, 723 cm−1; 1H and 13C NMR, Table 1 and Table 2; HRESIMS m/z 363.1339 [M + H]+ (calcd for C21H19N2O4, 363.1349).
Puniceusine D (4): pale yellow powder; UV (CH3OH) λmax (log ε) 207 (1.86), 243 (1.88), 260 (1.88) nm; IR (film) νmax 3402, 1678, 1643, 1566, 1384, 1342, 1197, 1132, 1045, 989, 954, 842, 800, 723 cm−1; 1H and 13C NMR, Table 1 and Table 2; HRESIMS m/z 363.1339 [M + H]+ (calcd for C21H19N2O4, 363.1338).
Puniceusine E (5): pale yellow powder; UV (CH3OH) λmax (log ε) 206 (2.07), 248 (1.83), 265 (2.00), 337 (1.06) nm; IR (film) νmax 1703, 1678, 1365, 1178, 0012, 835, 800, 721 cm−1; 1H and 13C NMR, Table 1 and Table 2; HRESIMS m/z 377.1496 [M + H]+ (calcd for C22H21N2O4, 377.1486).
Puniceusine F (6): colorless crystals; [α] 0 (c 0.10, CH3OH); UV (CH3OH) λmax (log ε) 206 (1.63), 244 (1.76), 261 (1.71) nm; IR (film) νmax 3419, 1703, 1681, 1363, 1201, 1180, 1134, 837, 800, 721 cm−1; 1H and 13C NMR, Table 1 and Table 2; HRESIMS m/z 262.1076 [M + H]+ (calcd for C14H16NO4, 262.1074).
Puniceusine G (7): yellow powder; UV (CH3OH) λmax (log ε) 200 (1.26), 246 (1.46) nm; IR (film) νmax 3412, 1680, 1440, 1195, 1136, 1028, 844, 800, 725 cm−1; 1H and 13C NMR, Table 1 and Table 2; HRESIMS m/z 254.1182 [M + H]+ (calcd for C16H16NO2, 254.1176).
Puniceusine H (8): pale yellow powder; UV (CH3OH) λmax (log ε) 191 (1.61), 204 (2.08), 242 (1.06), 272 (1.12) nm; IR (film) νmax 3367, 1680, 1456, 1417, 1394, 1201, 1139, 1020, 839, 802, 721 cm−1; 1H and 13C NMR, Table 2 and Table 3; HRESIMS m/z 398.1244 [M + H]+ (calcd for C21H20NO7, 398.1234).
Puniceusine I (9): yellow powder; [α] + 60.7 (c 0.10, CH3OH); UV (CH3OH) λmax (log ε) 205 (1.30), 237 (0.90), 244 (0.92), 276 (0.63) nm; ECD (CH3OH) λmax (Δε) 201 (+4.35), 202 (+4.71), 214 (−4.54), 232 (−1.32), 246 (−2.71), 279 (+0.44), 322 (−1.45) nm; IR (film) νmax 3402, 1699, 1681, 1361, 1201, 1136, 837, 800, 721 cm−1; 1H and 13C NMR, Table 3 and Table 4; HRESIMS m/z 354.1331 [M + H]+ (calcd for C20H20NO5, 354.1336).
Puniceusine J (10): pale yellow powder; UV (CH3OH) λmax (log ε) 195 (2.08), 208 (1.06), 243 (1.12) nm; IR (film) νmax 3406, 1678, 1392, 1321, 1203, 1132, 840, 800, 723 cm−1; 1H and 13C NMR, Table 3 and Table 4; HRESIMS m/z 384.1455 [M + H]+ (calcd for C21H22NO6, 384.1442).
Puniceusine K (11): pale yellow powder; [α] − 155.6 (c 0.10, CH3OH); UV (CH3OH) λmax (log ε) 205 (1.30), 237 (0.90), 244 (0.92), 276 (0.63) nm; ECD (CH3OH) λmax (Δε) 215 (−2.39), 229 (+0.45), 238 (+0.29), 255 (+0.67), 304 (−0.18) nm; IR (film) νmax 3390, 1674, 1435, 1371, 1319, 1199, 1134, 840, 800, 723 cm−1; 1H and 13C NMR, Table 3 and Table 4; HRESIMS m/z 428.1348 [M + H]+ (calcd for C22H22NO8, 428.1340).
Puniceusine L (12): colorless crystals; UV (CH3OH) λmax (log ε) 206 (2.14), 264 (2.04), 366 (1.12) nm; IR (film) νmax 3080, 1678, 1643, 1396, 1319, 1197, 1130, 839, 798, 721 cm−1; 1H and 13C NMR, Table 3 and Table 4; HRESIMS m/z 328.1195 [M + H]+ (calcd for C18H18NO5, 328.1179).
Puniceusine M (13): pale yellow powder; UV (CH3OH) λmax (log ε) 207 (2.08), 244 (1.74), 267 (1.84) nm; IR (film) νmax 3404, 1678, 1394, 1319, 1201, 1176, 1138, 1026, 837, 800, 721 cm−1; 1H and 13C NMR, Table 3 and Table 4; HRESIMS m/z 342.1332 [M + H]+ (calcd for C19H20NO5, 342.1336).
Puniceusine N (14): pale yellow powder; [α] 0 (c 0.10, CH3OH); UV (CH3OH) λmax (log ε) 206 (1.89), 232 (1.61), 266 (2.08), 309 (1.06), 364 (1.12) nm; IR (film) νmax 3423, 1680, 1363, 1195, 1182, 1128, 839, 800, 719 cm−1; 1H and 13C NMR, Table 3 and Table 4; HRESIMS m/z 374.2329 [M]+ (calcd for C22H32NO4, 374.2326).
3.5. X-ray Crystallographic Analysis of 6 and 12
The crystal data were obtained on a Rigaku MicroMax 007 diffractometer (Rigaku Corporation, Tokyo, Japan)with Cu Kα radiation and a graphite monochromator. The crystal structures of 6 and 12 were solved by direct methods with the SHELXTL and refined by full-matrix, least-squares techniques. Crystallographic data for 6 and 12 were deposited with the Cambridge Crystallographic Data Centre as supplementary publication numbers, CCDC 2112471 and 2112479, respectively.
Crystal data for 6: C14H14.0375NO4, FW = 260.30; colorless crystal from MeOH; crystal size = 0.15 × 0.12 × 0.1 mm3; T = 100.00 (10) K; monoclinic, space group P21/c (no. 14); unit cell parameters: a = 4.82780 (10) Å, b = 13.4963 (3) Å, c = 22.6665 (7) Å, α = 90°, β = 96.025 (3)°, γ = 90°, V = 1468.73 (6) Å3, Z = 4, Dcalc = 1.177 g/cm3, F (000) = 548.0, μ (CuKα) = 0.724 mm−1; 7185 reflections measured (7.634° ≤ 2θ ≤ 147.79°), 2852 unique (Rint = 0.0197, Rsigma = 0.0255), which were used in all calculations. The final R1 was 0.0487 (I > 2σ(I)) and wR2 was 0.1406 (all data).
Crystal data for 12: C18H17NO5, FW = 327.32; colorless crystal from MeOH; crystal size = 0.13 × 0.12 × 0.1 mm3; T = 100.00 (10) K; triclinic, space group P-1 (no. 2); unit cell parameters: a = 7.6037 (4) Å, b = 9.9253 (5) Å, c = 10.5261 (6) Å, α = 70.543 (5)°, β = 85.628 (4)°, γ = 81.605 (4)°, V = 740.68(7) Å3, Z = 2, Dcalc = 1.468 g/cm3, F (000) = 344.0, μ (CuKα) = 0.897 mm−1; 7019 reflections measured (8.914° ≤ 2θ ≤ 148.202°), 2880 unique (Rint = 0.0259, Rsigma = 0.0317) which were used in all calculations. The final R1 was 0.0419 (I > 2σ(I)) and wR2 was 0.1150 (all data).
3.6. ECD Calculations
The ECD calculation for 9 was performed by Gaussian 16 program package. The procedures were the same as described in our previous study [13,14]. Briefly, the conformational search was performed by a MMFF model, then the conformers with lower relative energies (<10 kcal/mol) were subjected to geometry optimization with the DFT method at the B3LYP/6-311G(d) level. Vibrational frequency calculations were carried out at the same level to evaluate their relative thermal (ΔE) and free energies (ΔG) at 298.15 K. The geometry-optimized conformers were further calculated at the M06-2X/def2-TZVP level and the solvent (methanol) effects were taken into consideration by using SMD. The optimized conformers with a Boltzmann distribution of more than 1% population were subjected to ECD calculation, which were performed by TDDFT methodology at the PBE1PBE/TZVP level. The ECD spectrum was generated by the software SpecDis using a Gaussian band shape with 0.3 eV exponential half-width from dipole-length dipolar and ratational strengths. The calculated spectrum of 9 was generated from the low-energy conformers according to the Boltzmann distribution of each conformer in MeOH solution. Details regarding optimized conformation geometries, thermodynamic parameters, and Boltzmann distributions (Tables S1–S3) of all conformations are provided in the Supporting Information.
3.7. Protein Phosphatase Inhibition Assays
The same methods as described in our previous study [13,14] were applied to test the inhibition activity of compounds 1–14 against five human protein tyrosine phosphatases (CD45, SHP1, TCPTP, PTP1B and LAR).
3.8. Cytotoxicity
Cytotoxic activity was evaluated using human lung adenocarcinoma cell line H1975 by CCK-8 method. Briefly, each of the test compounds was dissolved in DMSO and further diluted to give final concentrations of 80, 40, 20, 10, 5, 2.5, and 1.25 µg/mL, respectively. H1975 cells (5 × 103 cells/plate) were seeded in 96-well plates and treated with compounds at the indicated concentration for 24 h, and then 10 μL CCK-8 reagent was added to each well, and the plates were incubated at 37 °C for another 4 h. Next, the optical density was measured at a wavelength of 450 nm with the Bio-Rad (Hercules, CA, USA) microplate reader. Dose–response curves were generated, and the IC50 values were calculated from the linear portion of log dose–response curves.
3.9. Antibacterial Assays
Antibacterial activities of 1–14 against E. coli, S. aureus and MRSA were evaluated using the 2-fold dilution assay in 96-microwell plates. Briefly, all the indicator bacteria were cultured on Luria−Bertani (LB) agar plates at 37 °C for 12 h, and then a single colony was picked into LB liquid medium and cultivated on a rotary shaker at 37 °C for 12 h. Then, the bacterial suspensions with LB medium were diluted until the difference of the OD600 values between the bacterial suspensions and the medium was 0.01~0.02. Each of the tested compounds was dissolved in DMSO to give an initial concentration of 5 mg/mL, and further diluted with the bacterial suspensions by twofold serial dilution to give a final concentration of 100, 50, 25, 12.5, 6.25, and 3.125 μg/mL, respectively. The 96-well plates were incubated at 37 °C for 12 h. MIC value was determined as the lowest concentration with no visible bacterial growth. Ampicillin was used as the positive control and DMSO as the negative control. All experiments were performed three times.
4. Conclusions
Summarily, 14 new isoquinoline alkaloids (1–14) were obtained from the deep-sea-derived fungus, A. puniceus SCSIO z021. Compounds 3–5 and 8–13 unprecedentedly contained an isoquinolinyl, a polysubstituted benzyl or a pyronyl at position C-7 of the isoquinoline nucleus, which was different from 1-benzylisoquinoline analogues and other isoquinoline alkaloids from plants commonly containing substituents at positions C-1, N-2, C-3, C-4 or C-8 of isoquinoline skeleton. In addition, 3 and 4 showed selective inhibitory activity against CD45; 4 also had moderate cytotoxicity towards human lung adenocarcinoma cell line H1975, and 14 contained an active center -C=N+ which had evident antibacterial activity towards three indicator bacteria. An analysis of the relationship of the structures, enzyme inhibitory activity and cytotoxicity of 1–14 displayed that the substituents at C-7 of the isoquinoline nucleus could greatly affect their bioactivity. The results greatly enrich the structural diversity of isoquinoline alkaloids from fungi, and provide a potential lead compound for the development of a selective CD45 inhibitor and anticancer drug.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md20010078/s1, Figures S1–S126: the 1D NMR, 2D NMR, HRESIMS, IR, and UV spectra of compounds 1–14, Tables S1–S3: details of ECD calculation of compound 9, Tables S4 and S5: X-ray crystallographic data for compound 6 and X-ray crystallographic data for compound 12.
Author Contributions
Performing experiments, analyze data, and writing—original draft preparation, C.-M.L. and F.-H.Y.; test the activities, X.-H.L., X.-X.Z. and L.-X.L.; formal analysis, X.L.; resources, analyze data, writing—review and editing, supervision, and funding acquisition, S.-H.Q. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are grateful for the financial support provided by the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0406), National Natural Science Foundation of China (82173709), Key Science and Technology Project of Hainan Province (ZDKJ202018), Key-Area Research and Development Program of Guangdong Province (2020B1111030005), Institution of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences (ISEE2021PY05), and Guangdong Provincial-level Special Funds for Promoting High-quality Economic Development (2020032).
Acknowledgments
The authors also appreciate the analytical facility center (Z.H.X., A.J.S., Y.Z., X.M., X.H.Z.) of the South China Sea Institute of Oceanology, Chinese Academy of Sciences, for acquiring NMR, HRESIMS, experimental ECD data, and X-ray crystallographic data.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Singh, S.; Pathak, N.; Fatima, E.; Negi, A.S. Plant isoquinoline alkaloids: Advances in the chemistry and biology of berberine. Eur. J. Med. Chem. 2021, 226, 113839. [Google Scholar] [CrossRef] [PubMed]
- Kohno, J.; Hiramatsu, H.; Nishio, M.; Sakurai, M.; Okuda, T.; Komatsubara, S. Structures of TMC-120A, B and C, novel isoquinoline alkaloids from Aspergillus ustus TC 1118. Tetrahedron 1999, 55, 11247–11252. [Google Scholar] [CrossRef]
- Copmans, D.; Kildgaard, S.; Rasmussen, S.A.; Ṡlezak, M.; Dirkx, N.; Partoens, M.; Esguerra, C.V.; Crawford, A.D.; Larsen, T.O.; de Witte, P.A.M. Zebrafish-based discovery of antiseizure compounds from the North Sea: Isoquinoline alkaloids TMC-120A and TMC-120B. Mar. Drugs 2019, 17, 607. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Li, B.G.; Yang, T.; Liu, G.Y.; Zhang, G.L. Chaetoindicins A-C, three isoquinoline alkaloids from the fungus Chaetomium indicum. Org. Lett. 2006, 8, 3613–3615. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.X.; Xiao, J.; Laatsch, H.; Holstein, J.J.; Dittrich, B.; Zhang, Q.; Gao, J.M. Fusarimine, a novel polyketide isoquinoline alkaloid, from the endophytic fungus Fusarium sp. ln12, isolated from Melia azedarach. Tetrahedron Lett. 2012, 53, 6372–6375. [Google Scholar] [CrossRef]
- El-Neketi, M.; Ebrahim, W.; Lin, W.; Gedara, S.; Badria, F.; Saad, H.E.; Lai, D.; Proksch, P. Alkaloids and polyketides from Penicillium citrinum, an endophyte isolated from the Moroccan plant Ceratonia siliqua. J. Nat. Prod. 2013, 76, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.C.; Yuan, J.; Tan, Q.; Chen, Y.; Zhu, Y.J.; Jiang, H.M.; Zou, G.; Zang, Z.Z.; Wang, B.; She, Z.G. Peniazaphilones A–I, produced by co-culturing of mangrove endophytic fungi, Penicillium sclerotiorum THSH-4 and Penicillium sclerotiorum ZJHJJ-18. Chin. J. Chem. 2021, 39, 3404–3412. [Google Scholar] [CrossRef]
- Nord, C.; Levenfors, J.J.; Bjerketorp, J.; Sahlberg, C.; Guss, B.; Öberg, B.; Broberg, A. Antibacterial isoquinoline alkaloids from the fungus Penicillium spathulatum Em19. Molecules 2019, 24, 4616. [Google Scholar] [CrossRef] [PubMed]
- Bialy, L.; Waldmann, H. Inhibitors of protein tyrosine phosphatases: Next-generation drugs? Angew. Chem. Int. Ed. Engl. 2005, 44, 3814–3839. [Google Scholar] [CrossRef] [PubMed]
- Barr, A.J. Protein tyrosine phosphatases as drug targets: Strategies and challenges of inhibitor development. Future Med. Chem. 2010, 2, 1563–1576. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.Y.; Zhang, X.X.; Zheng, H.Z.; Zheng, Z.H.; Nong, X.H.; Liang, X.; Ma, X.; Qi, S.H. Novel anthraquinone derivatives as inhibitors of protein tyrosine phosphatases and indoleamine 2,3-dioxygenase 1 from the deep-sea derived fungus Alternaria tenuissima DFFSCS013. Org. Chem. Front. 2019, 6, 3252–3258. [Google Scholar] [CrossRef]
- Cheng, X.; Liang, X.; Zheng, Z.H.; Zhang, X.X.; Lu, X.H.; Yao, F.H.; Qi, S.H. Penicimeroterpenoids A–C, meroterpenoids with rearrangement skeletons from the marine-derived fungus Penicillium sp. SCSIO 41512. Org. Lett. 2020, 16, 6330–6333. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Huang, Z.H.; Ma, X.; Zheng, Z.H.; Zhang, X.X.; Lu, X.H.; Qi, S.H. Mycotoxins as inhibitors of protein tyrosine phosphatases from the deep-sea-derived fungus Aspergillus puniceus SCSIO z021. Bioorg. Chem. 2021, 107, 104571. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Huang, Z.H.; Shen, W.B.; Lu, X.H.; Zhang, X.X.; Ma, X.; Qi, S.H. Talaromyoxaones A and B: Unusual oxaphenalenone spirolactones as phosphatase inhibitors from the marine-derived fungus Talaromyces purpureogenus SCSIO 41517. J. Org. Chem. 2021, 86, 12831–12839. [Google Scholar] [CrossRef] [PubMed]
- White, J.D.; Straus, D.S. Gattermann reaction of 3,5-dimethoxyphenylacetonitrile. Synthesis of 6,8-dioxyisoquinolines. J. Org. Chem. 2002, 32, 2689–2692. [Google Scholar] [CrossRef]
- Atta-ur-Rahman; Ahmad, S.; Bhatti, M.K.; Choudhary, M.I. Alkaloidal constituents of Fumaria indica. Phytochemistry 1995, 40, 593–596. [Google Scholar] [CrossRef]
- Ebrahim, W.; Aly, A.H.; Mándi, A.; Wray, V.; Essassi, E.; Ouchbani, T.; Bouhfid, R.; Lin, W.; Proksch, P.; Kurtán, T.; et al. O-heterocyclic embeurekols from Embellisia eureka, an endophyte of Cladanthus arabicus. Chirality 2013, 25, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.B.; Ubukata, M.; Kakeya, H.; Onose, R.; Okada, G.; Takahashi, I.; Isono, K.; Osada, H. Acetophthalidin, a novel inhibitor of mammalian cell cycle, produced by a fungus isolated from a sea sediment. J. Antibiot. 1996, 49, 216–219. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).