Discovery of New Secondary Metabolites from Marine Bacteria Hahella Based on an Omics Strategy
Abstract
:1. Introduction
2. Results
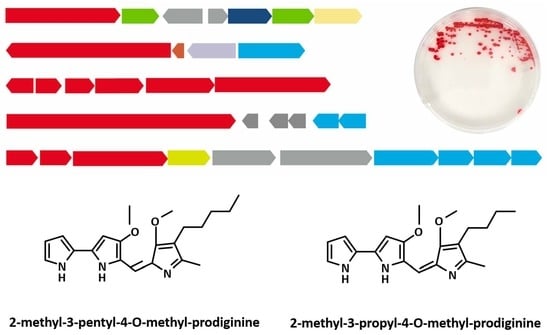
2.1. SMBGC Analysis in Hahella
2.2. New Strain Isolation and Identification
2.3. Genome Sequencing and SMBGCs Prediction in NBU794
2.4. Prodiginines Identification
2.5. Anti-Microbial Assay
2.6. Production of Prodigiosin
2.7. Discovery of Chejuenolide A–C
3. Discussion
4. Materials and Methods
4.1. Anti-SMASH Analysis
4.2. Sampling and Strain Isolation
4.3. Strain Culture Condition
4.4. Strain Identification
4.5. Compound Purification
4.6. LC-MS Analysis
4.7. Molecular Networking Analysis
4.8. NMR
4.9. Antimicrobial Activity
4.10. Fermentation of Prodigiosin
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Casillo, A.; Lanzetta, R.; Parrilli, M.; Corsaro, M.M. Exopolysaccharides from marine and marine extremophilic bacteria: Structures, properties, ecological roles and applications. Mar. Drugs 2018, 16, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef] [PubMed]
- Baik, K.S.; Seong, C.N.; Kim, E.M.; Yi, H.; Bae, K.S.; Chun, J. Hahella ganghwensis sp. nov., isolated from tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 2, 681–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.K.; Chun, J.; Moon, E.Y.; Ko, S.H.; Lee, D.S.; Lee, H.S.; Bae, K.S. Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int. J. Syst. Evol. Microbiol. 2001, 51 Pt 2, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lee, H.K.; Cho, J.C. Hahella antarctica sp. nov., isolated from Antarctic seawater. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 2, 353–356. [Google Scholar] [CrossRef] [Green Version]
- Senapin, S.; Dong, H.T.; Meemetta, W.; Siriphongphaew, A.; Charoensapsri, W.; Santimanawong, W.; Turner, W.A.; Rodkhum, C.; Withyachumnarnkul, B.; Vanichviriyakit, R. Hahella chejuensis is the etiological agent of a novel red egg disease in tilapia (Oreochromis spp.) hatcheries in Thailand. Aquaculture 2016, 454, 1–7. [Google Scholar] [CrossRef]
- Jeong, H.; Yim, J.H.; Lee, C.; Choi, S.H.; Park, Y.K.; Yoon, S.H.; Hur, C.G.; Kang, H.Y.; Kim, D.; Lee, H.H.; et al. Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucl. Acids Res. 2005, 33, 7066–7073. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.S.; Park, Y.K.; Kim, J.F.; Jeong, H.; Oh, T.K.; Kim, B.S.; Lee, C.H. Biosynthesis of antibiotic prodiginines in the marine bacterium Hahella chejuensis KCTC 2396. J. Appl. Microbiol. 2007, 102, 937–944. [Google Scholar] [CrossRef]
- Sakai-Kawada, F.E.; Ip, C.G.; Hagiwara, K.A.; Awaya, J.D. Biosynthesis and Bioactivity of Prodiginine Analogs in Marine Bacteria, Pseudoalteromonas: A Mini Review. Front. Microbiol. 2019, 10, 1715. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.Y.; Yang, P.Z.; Jiang, S.T.; Zheng, G.; Cao, L.L.; Cao, X.M.; Zhu, X.X.; Zhang, D.F.; Liu, G.Q. Experimental study of acute oral toxicity and genetic toxicity of natural red pigment prodigiosin. Food Sci. 2017, 38, 224–228. [Google Scholar]
- Lin, S.R.; Chen, Y.H.; Tseng, F.J.; Weng, C.F. The production and bioactivity of prodigiosin: Quo vadis? Drug Discov. Today 2020, 25, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Sohn, J.H.; Lee, D.; Kim, J.K.; Kong, I.S.; Ahn, S.C.; Oh, H. Chejuenolides A and B, new macrocyclic tetraenes from the marine bacterium Hahella chejuensis. Tetrahedron Lett. 2008, 49, 7128–7131. [Google Scholar] [CrossRef]
- Seo, C.; Oh, H. Chejuenolide C: A new macrocyclic metabolite from the marine bacterium Hahella chejuensis. B Korean Chem. Soc. 2009, 30, 1181–1183. [Google Scholar]
- Ng, B.G.; Han, J.W.; Lee, D.W.; Choi, G.J.; Kim, B.S. The chejuenolide biosynthetic gene cluster harboring an iterative trans-AT PKS system in Hahella chejuensis strain MB-1084. J. Antibiot. 2018, 71, 495–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucl. Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H. The year 2020 in natural product bioinformatics: An overview of the latest tools and databases. Nat. Prod. Rep. 2021, 38, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [Green Version]
- Walsh, C.T. The chemical versatility of natural-product assembly lines. Acc. Chem. Res. 2008, 41, 4–10. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef] [Green Version]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Munoz, J.C.; Terlouw, B.R.; van der Hooft, J.J.J.; van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucl. Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.S.; Kim, Y.S.; Park, S.; Kim, J.; Kang, S.J.; Lee, M.H.; Ryu, S.; Choi, J.M.; Oh, T.K.; Yoon, J.H. Exceptional production of both prodigiosin and cycloprodigiosin as major metabolic constituents by a novel marine bacterium, Zooshikella rubidus S1-1. Appl. Environ. Microbiol. 2011, 77, 4967–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, Y.; Kim, H.J.; Kim, S.; Park, S.Y.; Kim, H.; Jeong, S.; Lee, S.J.; Lee, M.S. Enhanced large-scale production of Hahella chejuensis-derived prodigiosin and evaluation of its bioactivity. J. Microbiol. Biotechnol. 2021, 31, 1624–1631. [Google Scholar] [CrossRef]
- Wang, F.P.; Li, M.; Huang, L.; Zhang, X.H. Cultivation of uncultured marine microorganisms. Mar. Life Sci. Technol. 2021, 3, 117–120. [Google Scholar] [CrossRef]
- Pye, C.R.; Bertin, M.J.; Lokey, R.S.; Gerwick, W.H.; Linington, R.G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 2017, 114, 5601–5606. [Google Scholar] [CrossRef] [Green Version]
- Kautsar, S.A.; van der Hooft, J.J.J.; de Ridder, D.; Medema, M.H. BiG-SLiCE: A highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. GigaScience 2021, 10, giaa154. [Google Scholar] [CrossRef]
- Moran, N.A. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 2873–2878. [Google Scholar] [CrossRef] [Green Version]
- O’Fallon, B. Population structure, levels of selection, and the evolution of intracellular symbionts. Evolution 2008, 62, 361–373. [Google Scholar] [CrossRef]
- Pettersson, M.E.; Berg, O.G. Muller’s ratchet in symbiont populations. Genetica 2007, 130, 199–211. [Google Scholar] [CrossRef]
- Wernegreen, J.J.; Richardson, A.O.; Moran, N.A. Parallel acceleration of evolutionary rates in symbiont genes underlying host nutrition. Mol. Phylogenet. Evol. 2001, 19, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Williamson, N.R.; Fineran, P.C.; Leeper, F.J.; Salmond, G.P. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 2006, 4, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.X.; Withall, D.M.; Challis, G.L.; Thomson, R.J. Structure, chemical synthesis, and biosynthesis of prodiginine natural products. Chem. Rev. 2016, 116, 7818–7853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, G.; Deleu, M.; Jourdan, E.; Thonart, P.; Ongena, M. The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell Microbiol. 2011, 13, 1824–1837. [Google Scholar] [CrossRef]
- Steigenberger, J.; Verleysen, Y.; Geudens, N.; Martins, J.C.; Heerklotz, H. The optimallipid chain length of a membrane-permeabilizing lipopeptide results from the balance of membrane partitioning and local damage. Front. Microbiol. 2021, 12, 669709. [Google Scholar] [CrossRef]
- Tabbene, O.; Kalai, L.; Ben Slimene, I.; Karkouch, I.; Elkahoui, S.; Gharbi, A.; Cosette, P.; Mangoni, M.L.; Jouenne, T.; Limam, F. Anti-candida effect of bacillomycin D-like lipopeptides from Bacillus subtilis B38. FEMS Microbiol. Lett. 2011, 316, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Berdy, B.; Spoering, A.L.; Ling, L.L.; Epstein, S.S. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat. Protoc. 2017, 12, 2232–2242. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.Y.; Xu, H.J.; Shi, X.Q.; Nie, Z.; Yu, Y.; Zhang, X.M.; Bai, Y.L.; Qiao, M.Q.; Gao, C.C. The study of optimal conditions of electroporation in Pseudomonas aeruginosa. Yi Chuan Xue Bao 2004, 31, 311–316. [Google Scholar] [PubMed]





| Compound | Minimum Inhibitory Concentration (µg/mL) | ||||
|---|---|---|---|---|---|
| Methicillin-Resistant Staphylococcus aureus | Bacillus subtilis | Escherichia coli | Salmonella paratyphi B | Candida albicans | |
| 2-methyl-3-propyl-prodiginine | 50 | 25 | - | - | 200 |
| 2-methyl-3-butyl-prodiginine | 50 | 12.5 | - | - | - |
| Prodigiosin | 1.56 | 1.56 | 3.12 | 12.5 | 1.56 |
| 2-methyl-3-heptyl-prodiginine | 1.56 | 6.25 | 12.5 | 25 | 6.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, S.; Li, P.; Wang, J.; Zhang, Y.; Lu, H.; Shi, L.; Huang, T.; Zhang, W.; Ding, L.; He, S.; et al. Discovery of New Secondary Metabolites from Marine Bacteria Hahella Based on an Omics Strategy. Mar. Drugs 2022, 20, 269. https://doi.org/10.3390/md20040269
He S, Li P, Wang J, Zhang Y, Lu H, Shi L, Huang T, Zhang W, Ding L, He S, et al. Discovery of New Secondary Metabolites from Marine Bacteria Hahella Based on an Omics Strategy. Marine Drugs. 2022; 20(4):269. https://doi.org/10.3390/md20040269
Chicago/Turabian StyleHe, Shufen, Peishan Li, Jingxuan Wang, Yanzhu Zhang, Hongmei Lu, Liufei Shi, Tao Huang, Weiyan Zhang, Lijian Ding, Shan He, and et al. 2022. "Discovery of New Secondary Metabolites from Marine Bacteria Hahella Based on an Omics Strategy" Marine Drugs 20, no. 4: 269. https://doi.org/10.3390/md20040269
APA StyleHe, S., Li, P., Wang, J., Zhang, Y., Lu, H., Shi, L., Huang, T., Zhang, W., Ding, L., He, S., & Liu, L. (2022). Discovery of New Secondary Metabolites from Marine Bacteria Hahella Based on an Omics Strategy. Marine Drugs, 20(4), 269. https://doi.org/10.3390/md20040269