Coral Holobionts Possess Distinct Lipid Profiles That May Be Shaped by Symbiodiniaceae Taxonomy
Abstract
:1. Introduction
2. Results
2.1. Genetic Identification of Coral Endosymbionts
2.2. Thylakoid Membrane Lipidome of Coral Endosymbionts
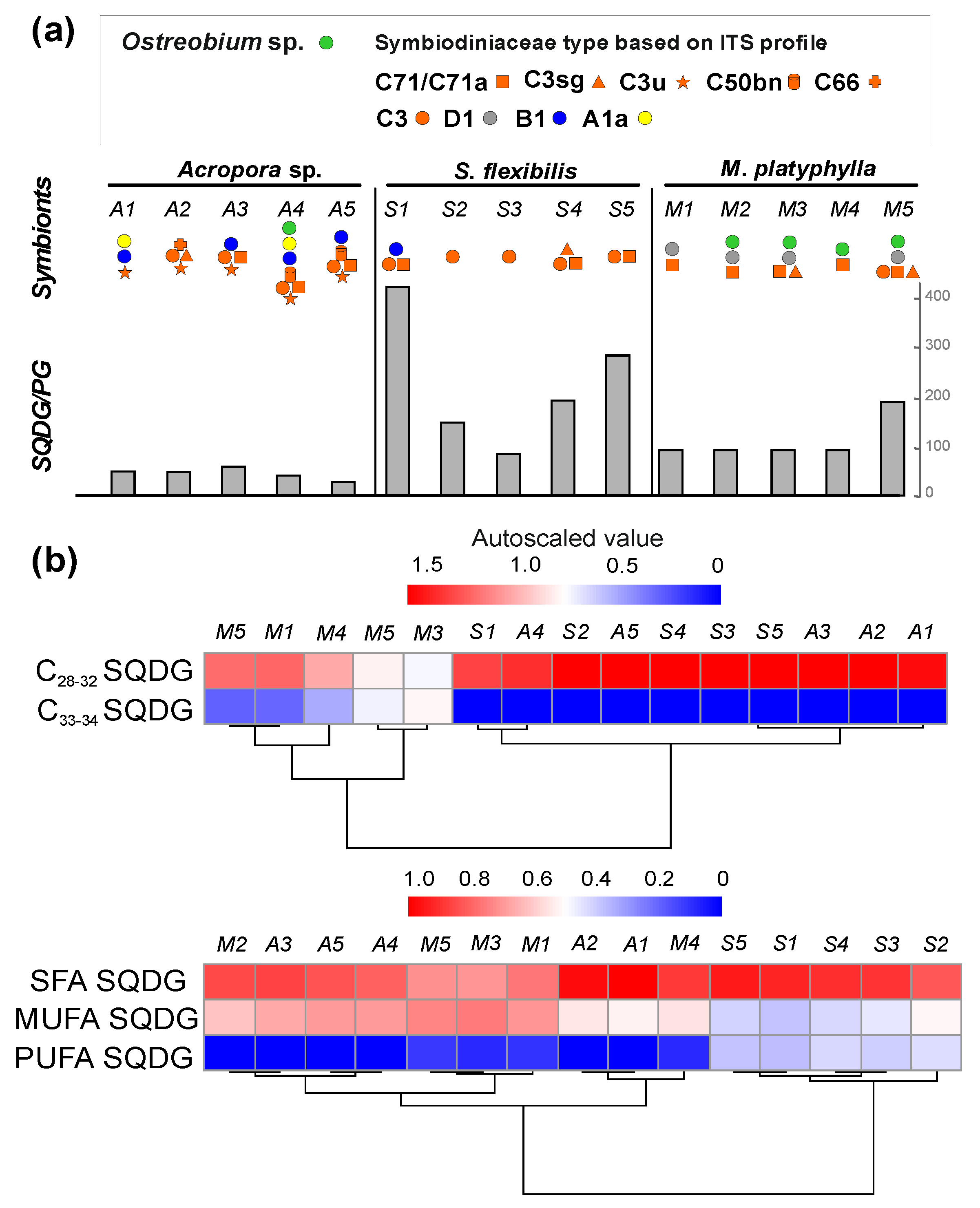
2.2.1. Phospholipid PG and Glycolipid SQDG
2.2.2. Galactolipids MGDG and DGDG
3. Discussion
3.1. Thylakoid Lipidome Features of Coral Endosymbionts
3.1.1. Lipid Head Groups
3.1.2. Lipid Acyl Chains: Degree of Unsaturation
3.1.3. Lipid Acyl Chains: Chain Length
3.2. Symbionts with Thermotolerant Features of Lipidome May Help Corals Adapt to Climate Change in the SCS
4. Conclusions
5. Materials and Methods
5.1. Specimen Collection
5.2. Molecular Genetics Analysis
5.3. Lipid Analysis
5.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 2016, 70, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Baird, A.H.; Bhagooli, R.; Ralph, P.J.; Takahashi, S. Coral bleaching: The role of the host. Trends Ecol. Evol. 2008, 24, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Spalding, M.D.; Fox, H.E.; Halpern, B.S.; McManus, M.A.; Molnar, J.; Allen, G.R.; Davidson, N.; Jorge, Z.A.; Lombana, A.L.; Lourie, S.A.; et al. Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. Bioscience 2007, 57, 573–583. [Google Scholar] [CrossRef] [Green Version]
- Clifton, J. Science, funding and participation: Key issues for marine protected area networks and the Coral Triangle Initiative. Environ. Conserv. 2009, 36, 91–96. [Google Scholar] [CrossRef]
- Huang, D.W.; Licuanan, W.Y.; Hoeksema, B.W.; Chen, C.A.; Ang, P.O.; Huang, H.; Lane, D.J.W.; Vo, S.T.; Waheed, Z.; Affendi, Y.A.; et al. Extraordinary diversity of reef corals in the South China Sea. Mar. Biodivers. 2015, 45, 157–168. [Google Scholar] [CrossRef]
- Muir, P.R.; Pichon, M. Biodiversity of Reef-Building, Scleractinian Corals. Mesophotic Coral Ecosyst. 2019, 12, 589–620. [Google Scholar]
- Veron, J.E.N.; Stafford-Smith, M.G.; Turak, E.; De Vantier, L.M. Corals of the World. Available online: http://www.coralsoftheworld.org/coral_geographic/interactive_map/ (accessed on 27 July 2022).
- Lewis, J.B. Biology and Ecology of the Hydrocoral Millepora on Coral Reefs. In Advances in Marine Biology; Academic Press: Cambridge, MA, USA, 2006; Volume 50, pp. 1–55. [Google Scholar]
- Benayahu, Y.; Bridge, T.C.L.; Colin, P.L.; Liberman, R.; McFadden, C.S.; Pizarro, O.; Schleyer, M.H.; Shoham, E.; Reijnen, B.T.; Weis, M.; et al. Octocorals of the Indo-Pacific. Mesophotic Coral Ecosyst. 2019, 12, 709–728. [Google Scholar]
- Bayer, F.M. Key to the genera of Octocorallia exclusive of Pennatulacea (Coelenterata: Anthozoa), with diagnoses of new taxa. Proc. Biol. Soc. Wash. 1981, 94, 902–947. [Google Scholar]
- Coffroth, M.A.; Santos, S.R. Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 2005, 156, 19–34. [Google Scholar] [CrossRef]
- Ebenezer, V.; Medlin, L.K.; Ki, J.S. Molecular detection, quantification, and diversity evaluation of microalgae. Mar. Biotechnol. 2012, 14, 129–142. [Google Scholar]
- LaJeunesse, T.C.; Parkinson, J.E.; Gabrielson, P.W.; Jeong, H.J.; Reimer, J.D.; Voolstra, C.R.; Santos, S.R. Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018, 28, 2570–2580.e6. [Google Scholar] [CrossRef] [Green Version]
- LaJeunesse, T.C.; Wiedenmann, J.; Casado-Amezúa, P.; D’Ambra, I.; Turnham, K.E.; Nitschke, M.R.; Oakley, C.A.; Goffredo, S.; Spano, C.A.; Cubillos, V.M.; et al. Revival of Philozoon Geddes for host-specialized dinoflagellates, ‘zooxanthellae’, in animals from coastal temperate zones of northern and southern hemispheres. Eur. J. Phycol. 2022, 57, 166–180. [Google Scholar] [CrossRef]
- Pochon, X.; Gates, R.D. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol. Phylogenet. Evol. 2010, 56, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Pochon, X.; LaJeunesse, T.C. Miliolidium n. gen, a new symbiodiniacean genus whose members associate with Soritid Foraminifera or are free-living. J. Eukaryot. Microbiol. 2021, 68, e12856. [Google Scholar] [CrossRef]
- Ziegler, M.; Arif, C.; Voolstra, C.R. Symbiodiniaceae Diversity in Red Sea Coral Reefs & Coral Bleaching. Coral Reefs World 2019, 11, 69–89. [Google Scholar]
- Berkelmans, R.; van Oppen, M.J.H. The role of zooxanthellae in the thermal tolerance of corals: A ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. B-Biol. Sci. 2006, 273, 2305–2312. [Google Scholar]
- Silverstein, R.N.; Cunning, R.; Baker, A.C. Tenacious D: Symbiodinium in clade D remain in reef corals at both high and low temperature extremes despite impairment. J. Exp. Biol. 2017, 220, 1192–1196. [Google Scholar] [PubMed] [Green Version]
- Hume, B.C.C.; D’Angelo, C.; Smith, E.G.; Stevens, J.R.; Burt, J.A.; Wiedenmann, J. Validation of the binary designation Symbiodinium thermophilum (dinophyceae). J. Phycol. 2018, 54, 762–764. [Google Scholar] [CrossRef] [PubMed]
- LaJeunesse, T.C.; Wham, D.C.; Pettay, D.T.; Parkinson, J.E.; Keshavmurthy, S.; Chen, C.A. Ecologically differentiated stress-tolerant endosymbionts in the dinoflagellate genus Symbiodinium (Dinophyceae) Clade D are different species. Phycologia 2014, 53, 305–319. [Google Scholar]
- Chakravarti, L.J.; Beltran, V.H.; van Oppen, M.J.H. Rapid thermal adaptation in photosymbionts of reef-building corals. Glob. Chang. Biol. 2017, 23, 4675–4688. [Google Scholar]
- Russnak, V.; Rodriguez-Lanetty, M.; Karsten, U. Photophysiological tolerance and thermal plasticity of genetically different Symbiodiniaceae endosymbiont species of Cnidaria. Front. Mar. Sci. 2021, 8, 657348. [Google Scholar] [CrossRef]
- Abrego, D.; Ulstrup, K.E.; Willis, B.L.; van Oppen, M.J.H. Species-specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proc. R. Soc. B-Biol. Sci. 2008, 275, 2273–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howells, E.J.; Beltran, V.H.; Larsen, N.W.; Bay, L.K.; Willis, B.L.; van Oppen, M.J.H. Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat. Clim. Chang. 2012, 2, 116–120. [Google Scholar] [CrossRef]
- del Campo, J.; Pombert, J.F.; Slapeta, J.; Larkum, A.; Keeling, P.J. The ‘other’ coral symbiont: Ostreobium diversity and distribution. ISME J. 2017, 11, 296–299. [Google Scholar] [CrossRef] [Green Version]
- Kuhl, M.; Holst, G.; Larkum, A.W.D.; Ralph, P.J. Imaging of oxygen dynamics within the endolithic algal community of the massive coral Porites lobata. J. Phycol. 2008, 44, 541–550. [Google Scholar] [CrossRef]
- Fine, M.; Loya, Y. Endolithic algae: An alternative source of photoassimilates during coral bleaching. Proc. R. Soc. B-Biol. Sci. 2002, 269, 1205–1210. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.S.; Bachok, Z.; Hii, Y.S. Effects of supplementary polyunsaturated fatty acids on the health of the scleractinian coral Galaxea fascicularis (Linnaeus, 1767). J. Exp. Mar. Biol. Ecol. 2017, 491, 1–8. [Google Scholar] [CrossRef]
- Treignier, C.; Grover, R.; Ferrier-Pages, C.; Tolosa, I. Effect of light and feeding on the fatty acid and sterol composition of zooxanthellae and host tissue isolated from the scleractinian coral Turbinaria reniformis. Limnol. Oceanogr. 2008, 53, 2702–2710. [Google Scholar] [CrossRef] [Green Version]
- Dalsgaard, J.; John, M.S.; Kattner, G.; Muller-Navarra, D.; Hagen, W. Fatty acid trophic markers in the pelagic marine environment. Adv. Mar. Biol. 2003, 46, 225–340. [Google Scholar]
- Imbs, A.B.; Latyshev, N.A.; Dautova, T.N.; Latypov, Y.Y. Distribution of lipids and fatty acids in corals by their taxonomic position and presence of zooxanthellae. Mar. Ecol.-Prog. Ser. 2010, 409, 65–75. [Google Scholar] [CrossRef]
- Sikorskaya, T.V.; Ermolenko, E.V.; Efimova, K.V. Lipids of Indo-Pacific gorgonian corals are modified under the influence of microbial associations. Coral Reefs 2022, 41, 277–291. [Google Scholar] [CrossRef]
- Awai, K.; Matsuoka, R.; Shioi, Y. Lipid and fatty acid compositions of Symbiodinium strains. In Proceedings of the 12th International Coral Reef Symposium, Cairns, Australia, 9–13 July 2012. [Google Scholar]
- Leblond, J.D.; Khadka, M.; Duong, L.; Dahmen, J.L. Squishy lipids: Temperature effects on the betaine and galactolipid profiles of a C-18/C-18 peridinin-containing dinoflagellate, Symbiodinium microadriaticum (Dinophyceae), isolated from the mangrove jellyfish, Cassiopea xamachana. Phycol. Res. 2015, 63, 219–230. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Thelen, J.J.; Fedosejevs, E.; Harwood, J.L. The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 2019, 74, 31–68. [Google Scholar] [CrossRef] [PubMed]
- Boudiere, L.; Michaud, M.; Petroutsos, D.; Rebeille, F.; Falconet, D.; Bastien, O.; Roy, S.; Finazzi, G.; Rolland, N.; Jouhet, J.; et al. Glycerolipids in photosynthesis: Composition, synthesis and trafficking. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 470–480. [Google Scholar] [CrossRef]
- Bishop, D.G.; Kenrick, J.R. Fatty acid composition of symbiotic zooxanthellae in relation to their hosts. Lipids 1980, 15, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Leblond, J.D.; Chapman, P.J. Lipid class distribution of highly unsaturated long chain fatty acids in marine dinoflagellates. J. Phycol. 2000, 36, 1103–1108. [Google Scholar] [CrossRef]
- Tchernov, D.; Gorbunov, M.Y.; de Vargas, C.; Yadav, S.N.; Milligan, A.J.; Haggblom, M.; Falkowski, P.G. Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proc. Natl. Acad. Sci. USA 2004, 101, 13531–13535. [Google Scholar] [PubMed] [Green Version]
- Rosset, S.; Koster, G.; Brandsma, J.; Hunt, A.N.; Postle, A.D.; D’Angelo, C. Lipidome analysis of Symbiodiniaceae reveals possible mechanisms of heat stress tolerance in reef coral symbionts. Coral Reefs 2019, 38, 1241–1253. [Google Scholar] [CrossRef] [Green Version]
- Sikorskaya, T.V.; Efimova, K.V.; Imbs, A.B. Lipidomes of phylogenetically different symbiotic dinoflagellates of corals. Phytochemistry 2021, 181, 112579. [Google Scholar] [PubMed]
- Hume, B.C.C.; Smith, E.G.; Ziegler, M.; Warrington, H.J.M.; Burt, J.A.; LaJeunesse, T.C.; Wiedenmann, J.; Voolstra, C.R. SymPortal: A novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 2019, 19, 1063–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verbruggen, H.; Marcelino, V.R.; Guiry, M.D.; Cremen, M.C.M.; Jackson, C.J. Phylogenetic position of the coral symbiont Ostreobium (Ulvophyceae) inferred from chloroplast genome data. J. Phycol. 2017, 53, 790–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tortorelli, G.; Rautengarten, C.; Bacic, A.; Segal, G.; Ebert, B.; Davy, S.K.; van Oppen, M.J.H.; McFadden, G.I. Cell surface carbohydrates of symbiotic dinoflagellates and their role in the establishment of cnidarian–dinoflagellate symbiosis. ISME J. 2022, 16, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, D.J.; Howells, E.J.; Wham, D.C.; Steury, T.D.; Santos, S.R. Population genetics of reef coral endosymbionts (Symbiodinium, Dinophyceae). Mol. Ecol. 2017, 26, 2640–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.J.; Yu, K.F.; Chen, B.; Wang, Y.H.; Liang, J.Y.; Luo, W.W.; Xu, L.J.; Huang, X.Y. Diversity of Symbiodiniaceae in 15 coral species from the southern South China Sea: Potential relationship with coral thermal adaptability. Front. Microbiol. 2019, 10, 2343. [Google Scholar] [CrossRef]
- Chauka, L.J.; Steinert, G.; Mtolera, M.S.P. Influence of local environmental conditions and bleaching histories on the diversity and distribution of Symbiodinium in reef-building corals in Tanzania. Afr. J. Mar. Sci. 2016, 38, 57–64. [Google Scholar] [CrossRef]
- Qin, Z.J.; Yu, K.F.; Chen, S.C.; Chen, B.; Liang, J.Y.; Yao, Q.C.; Yu, X.P.; Liao, Z.H.; Deng, C.Q.; Liang, Y.T. Microbiome of juvenile corals in the outer reef slope and lagoon of the South China Sea: Insight into coral acclimatization to extreme thermal environments. Environ. Microbiol. 2021, 23, 4389–4404. [Google Scholar] [CrossRef]
- Wicks, L.C.; Sampayo, E.; Gardner, J.P.A.; Davy, S.K. Local endemicity and high diversity characterise high-latitude coral-Symbiodinium partnerships. Coral Reefs 2010, 29, 989–1003. [Google Scholar] [CrossRef]
- Chwastek, G.; Surma, M.A.; Rizk, S.; Grosser, D.; Lavrynenko, O.; Rucinska, M.; Jambor, H.; Saenz, J. Principles of membrane adaptation revealed through environmentally induced bacterial lipidome remodeling. Cell Rep. 2020, 32, 108165. [Google Scholar]
- Jeucken, A.; Molenaar, M.R.; van de Lest, C.; Jansen, J.W.A.; Helms, J.B.; Brouwers, J.F. A Comprehensive functional characterization of escherichia coli lipid genes. Cell Rep. 2019, 27, 1597–1606.e2. [Google Scholar] [CrossRef] [Green Version]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous Adaptation and the regulation of membrane lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef] [Green Version]
- Funakoshi, Y.; Iwao, Y.; Noguchi, S.; Itai, S. Effect of alkyl chain length and unsaturation of the phospholipid on the physicochemical properties of lipid nanoparticles. Chem. Pharm. Bull. 2015, 63, 731–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimojima, M. Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 2011, 50, 234–239. [Google Scholar] [CrossRef]
- Yu, B.; Xu, C.C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737. [Google Scholar] [CrossRef] [Green Version]
- Dormann, P.; Benning, C. Galactolipids rule in seed plants. Trends Plant Sci. 2002, 7, 112–118. [Google Scholar] [CrossRef]
- Bruce, B.D. The role of lipids in plastid protein transport. Plant Mol. Biol. 1998, 38, 223–246. [Google Scholar] [CrossRef]
- Garrett, T.A.; Schmeitzel, J.L.; Klein, J.A.; Hwang, J.J.; Schwarz, J.A. Comparative lipid profiling of the cnidarian Aiptasia pallida and its dinoflagellate symbiont. PLoS ONE 2013, 8, e57975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imbs, A.B.; Grigorchuk, V.P. Lipidomic study of the influence of dietary fatty acids on structural lipids of cold-water nudibranch molluscs. Sci. Rep. 2019, 9, 20013. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Jensen, M.O.; Mouritsen, O.G. Lipids do influence protein function—The hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta (BBA)—Biomembr. 2004, 1666, 205–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, R.; Ursell, T.; Wiggins, P.; Sens, P. Emerging roles for lipids in shaping membrane-protein function. Nature 2009, 459, 379–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, W.F.D.; Sapay, N.; Tieleman, D.P. Atomistic simulations of pore formation and closure in lipid bilayers. Biophys. J. 2014, 106, 210–219. [Google Scholar] [CrossRef] [Green Version]
- Orsi, M.; Essex, J.W. Permeability of drugs and hormones through a lipid bilayer: Insights from dual-resolution molecular dynamics. Soft Matter 2010, 6, 3797–3808. [Google Scholar] [CrossRef]
- Lepetit, B.; Goss, R.; Jakob, T.; Wilhelm, C. Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth. Res. 2012, 111, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Büchel, C. Light-Harvesting Complexes of diatoms: Fucoxanthin-chlorophyll proteins. In Photosynthesis in Algae: Biochemical and Physiological Mechanisms; Larkum, A.W.D., Grossman, A.R., Raven, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 441–457. [Google Scholar]
- Mansour, J.S.; Pollock, F.J.; Diaz-Almeyda, E.; Iglesias-Prieto, R.; Medina, M. Intra- and interspecific variation and phenotypic plasticity in thylakoid membrane properties across two Symbiodinium clades. Coral Reefs 2018, 37, 841–850. [Google Scholar] [CrossRef]
- Loh, W.K.W.; Loi, T.; Carter, D.; Hoegh-Guldberg, O. Genetic variability of the symbiotic dinoflagellates from the wide-ranging coral species Seriatopora hystrix and Acropora longicyathus in the Indo-West Pacific. Mar. Ecol.-Prog. Ser. 2001, 222, 97–107. [Google Scholar] [CrossRef]
- Crabbe, M.J.C.; Carlin, J.P. Multiple Symbiodinium clades in Acropora species scleractinian corals from the Ningaloo reef, Australia. Int. J. Integr. Biol. 2009, 5, 72–74. [Google Scholar]
- Rouzé, H.; Lecellier, G.J.; Saulnier, D.; Planes, S.; Gueguen, Y.; Wirshing, H.H.; Berteaux-Lecellier, V. An updated assessment of Symbiodinium spp. that associate with common scleractinian corals from Moorea (French Polynesia) reveals high diversity among background symbionts and a novel finding of clade B. PeerJ 2017, 5, e2856. [Google Scholar] [CrossRef] [Green Version]
- Abrego, D.; Van Oppen, M.J.H.; Willis, B.L. Highly infectious symbiont dominates initial uptake in coral juveniles. Mol. Ecol. 2009, 18, 3518–3531. [Google Scholar] [CrossRef]
- Yamashita, H.; Koike, K.; Shinzato, C.; Jimbo, M.; Suzuki, G. Can Acropora tenuis larvae attract native Symbiodiniaceae cells by green fluorescence at the initial establishment of symbiosis? PLoS ONE 2021, 16, e0252514. [Google Scholar] [CrossRef] [PubMed]
- Mies, M.; Francini-Filho, R.B.; Zilberberg, C.; Garrido, A.G.; Longo, G.O.; Laurentino, E.; Güth, A.Z.; Sumida, P.Y.G.; Banha, T.N.S. South atlantic coral reefs are major global warming refugia and less susceptible to bleaching. Front. Mar. Sci. 2020, 7, 514. [Google Scholar] [CrossRef]
- Thomas, L.; Kendrick, G.A.; Kennington, W.J.; Richards, Z.T.; Stat, M. Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western Australia with 454 amplicon pyrosequencing. Mol. Ecol. 2014, 23, 3113–3126. [Google Scholar] [CrossRef]
- Stimson, J.; Sakai, K.; Sembali, H. Interspecific comparison of the symbiotic relationship in corals with high and low rates of bleaching-induced mortality. Coral Reefs 2002, 21, 409–421. [Google Scholar] [CrossRef]
- Davy, S.K.; Turner, J.R. Early development and acquisition of zooxanthellae in the temperate symbiotic sea anemone Anthopleura ballii (Cocks). Biol. Bull. 2003, 205, 66–72. [Google Scholar] [CrossRef]
- Chauka, L.J.; Macdonald, A.H.H. Status of coral-Symbiodiniaceae research in Western Indian Ocean. Symbiosis 2019, 77, 207–215. [Google Scholar] [CrossRef]
- Sebastian, C.R.; Sink, K.J.; McClanahan, T.R.; Cowan, D.A. Bleaching response of corals and their Symbiodinium communities in southern Africa. Mar. Biol. 2009, 156, 2049–2062. [Google Scholar] [CrossRef]
- Shoguchi, E.; Shinzato, C.; Kawashima, T.; Gyoja, F.; Mungpakdee, S.; Koyanagi, R.; Takeuchi, T.; Hisata, K.; Tanaka, M.; Fujiwara, M.; et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013, 23, 1399–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Oppen, M.J.H.; Bongaerts, P.; Underwood, J.N.; Peplow, L.M.; Cooper, T.F. The role of deep reefs in shallow reef recovery: An assessment of vertical connectivity in a brooding coral from west and east Australia. Mol. Ecol. 2011, 20, 1647–1660. [Google Scholar] [CrossRef]
- Chakravarti, L.J.; van Oppen, M.J.H. Experimental evolution in coral photosymbionts as a tool to increase thermal tolerance. Front. Mar. Sci. 2018, 5, 227. [Google Scholar] [CrossRef] [Green Version]
- Correa, A.M.S.; Brandt, M.E.; Smith, T.B.; Thornhill, D.J.; Baker, A.C. Symbiodinium associations with diseased and healthy scleractinian corals. Coral Reefs 2009, 28, 437–448. [Google Scholar] [CrossRef]
- Saad, O.S.; Lin, X.; Ng, T.Y.; Li, L.; Ang, P.; Lin, S. Species richness and generalists–specialists mosaicism of symbiodiniacean symbionts in corals from Hong Kong revealed by high-throughput ITS sequencing. Coral Reefs 2022, 41, 1–12. [Google Scholar] [CrossRef]
- Yamashita, H.; Suzuki, G.; Hayashibara, T.; Koike, K. Do corals select zooxanthellae by alternative discharge? Mar. Biol. 2010, 158, 87–100. [Google Scholar] [CrossRef]
- Hunter, C.L.; Morden, C.W.; Smith, C.M. The utility of ITS sequences in assessing relationships among zooxanthellae and corals. Proc. 8th Int. Coral Reef Symp. 1997, 2, 1599–1602. [Google Scholar]
- Palumbi, S.R. Nucleic Acids II: The polymerase chain reaction. In Molecular Systematics, 2nd ed.; Hillis, D.M., Moritz, C., Mable, B.K., Eds.; Sinauer: Sunderland, MA, USA, 1996. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Snisky, J.J., White, T.J., Eds.; Academic Press Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- LaJeunesse, T.; Trench, R. Biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal sea anemone Anthopleura elegantissima (Brandt). Biol. Bull. 2000, 199, 126–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, A.W.; Suarez, A.; Goff, L.J. Molecular delineation of species and syngens in volvocacean green algae (chlorophyta). J. Phycol. 1994, 30, 80–90. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folch, J.; Lees, M.; Sloane-Stanley, G.A. A simple methods for the isolation and pyrification of total lipid extraction from animal tissue. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Sikorskaya, T.V.; Ermolenko, E.V.; Imbs, A.B. Effect of experimental thermal stress on lipidomes of the soft coral Sinularia sp. and its symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 2020, 524, 151295. [Google Scholar] [CrossRef]
- Zar, J. Biostatistical Analysis; Prentice Hall: Upper Saddle River, NJ, USA, 1999. [Google Scholar]

| Colony | Symbiodiniaceae Type Based on ITS Profile | Ostreobium sp. (23S rRNA) | Nucleotide Sequences per Coral Colony | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1a | B1 | C3 | C3sg | C3u | C50bn | C66 | C71/C71a | D1 | |||
| A1 | + | + | + | 3 | |||||||
| A2 | + | + | + | + | 4 | ||||||
| A3 | + | + | + | + | 4 | ||||||
| A4 | + | + | + | + | + | + | + | 7 | |||
| A5 | + | + | + | + | + | 5 | |||||
| M1 | + | + | 2 | ||||||||
| M2 | + | + | + | 3 | |||||||
| M3 | + | + | + | + | 4 | ||||||
| M4 | + | + | 2 | ||||||||
| M5 | + | + | + | + | + | 5 | |||||
| S1 | + | + | + | 3 | |||||||
| S2 | + | 1 | |||||||||
| S3 | + | 1 | |||||||||
| S4 | + | + | + | 3 | |||||||
| S5 | + | + | 2 | ||||||||
| MGDG | DGDG | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coral Colonies | C30–34 | C36–38 | C40–42 | With 0–5 d.b. | With 6–8 d.b. | With 9–11 d.b. | C30–34 | C36–38 | C40–42 | With 0–5 d.b. | With 6–8 d.b. | With 9–11 d.b. | |
| A1 | Content, % of sum in lipid class | 3.26 | 91.57 | 4.72 | 3.26 | 50.52 | 45.77 | 0.00 | 91.69 | 5.66 | 0.00 | 40.83 | 56.52 |
| A2 | 5.37 | 84.75 | 4.99 | 5.37 | 45.38 | 44.36 | 0.00 | 87.32 | 8.29 | 0.00 | 35.75 | 59.85 | |
| A3 | 7.37 | 87.56 | 5.40 | 7.37 | 49.07 | 43.88 | 0.00 | 91.92 | 8.08 | 0.00 | 46.05 | 53.95 | |
| A4 | 4.67 | 90.68 | 2.93 | 4.67 | 49.24 | 44.37 | 0.00 | 88.91 | 10.74 | 0.00 | 51.60 | 48.05 | |
| A5 | 5.04 | 90.87 | 4.09 | 5.04 | 50.83 | 44.12 | 0.00 | 89.01 | 11.36 | 0.00 | 51.50 | 48.87 | |
| S1 | 63.04 | 36.96 | 0.00 | 42.39 | 41.01 | 16.60 | 43.51 | 55.43 | 2.41 | 29.97 | 45.45 | 25.93 | |
| S2 | 57.98 | 42.02 | 0.00 | 37.95 | 43.95 | 18.09 | 32.42 | 66.59 | 1.28 | 14.21 | 52.85 | 33.22 | |
| S3 | 55.34 | 44.66 | 0.00 | 31.13 | 52.78 | 16.09 | 36.04 | 63.01 | 0.95 | 19.58 | 53.69 | 26.73 | |
| S4 | 50.97 | 48.36 | 0.00 | 29.36 | 51.72 | 18.26 | 46.85 | 52.61 | 0.17 | 16.94 | 44.28 | 38.42 | |
| S5 | 53.59 | 46.02 | 0.00 | 33.33 | 48.93 | 17.35 | 43.74 | 55.95 | 0.12 | 16.98 | 50.39 | 32.43 | |
| M1 | 17.05 | 80.87 | 0.00 | 16.29 | 17.69 | 63.94 | 10.29 | 50.62 | 34.99 | 11.26 | 3.77 | 82.34 | |
| M2 | 1.53 | 98.29 | 0.00 | 0.32 | 9.32 | 90.18 | 2.61 | 54.73 | 36.47 | 2.61 | 9.34 | 82.82 | |
| M3 | 51.57 | 48.40 | 0.00 | 23.76 | 42.45 | 33.76 | 52.24 | 17.78 | 27.45 | 58.74 | 1.39 | 37.34 | |
| M4 | 40.16 | 60.70 | 0.00 | 32.40 | 24.56 | 43.90 | 27.50 | 49.36 | 23.31 | 31.69 | 5.27 | 63.21 | |
| M5 | 45.45 | 53.71 | 0.00 | 23.19 | 40.11 | 35.86 | 51.59 | 20.73 | 27.08 | 56.90 | 0.73 | 42.18 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikorskaya, T.V.; Ermolenko, E.V.; Efimova, K.V.; Dang, L.T.P. Coral Holobionts Possess Distinct Lipid Profiles That May Be Shaped by Symbiodiniaceae Taxonomy. Mar. Drugs 2022, 20, 485. https://doi.org/10.3390/md20080485
Sikorskaya TV, Ermolenko EV, Efimova KV, Dang LTP. Coral Holobionts Possess Distinct Lipid Profiles That May Be Shaped by Symbiodiniaceae Taxonomy. Marine Drugs. 2022; 20(8):485. https://doi.org/10.3390/md20080485
Chicago/Turabian StyleSikorskaya, Tatyana V., Ekaterina V. Ermolenko, Kseniya V. Efimova, and Ly T. P. Dang. 2022. "Coral Holobionts Possess Distinct Lipid Profiles That May Be Shaped by Symbiodiniaceae Taxonomy" Marine Drugs 20, no. 8: 485. https://doi.org/10.3390/md20080485
APA StyleSikorskaya, T. V., Ermolenko, E. V., Efimova, K. V., & Dang, L. T. P. (2022). Coral Holobionts Possess Distinct Lipid Profiles That May Be Shaped by Symbiodiniaceae Taxonomy. Marine Drugs, 20(8), 485. https://doi.org/10.3390/md20080485






